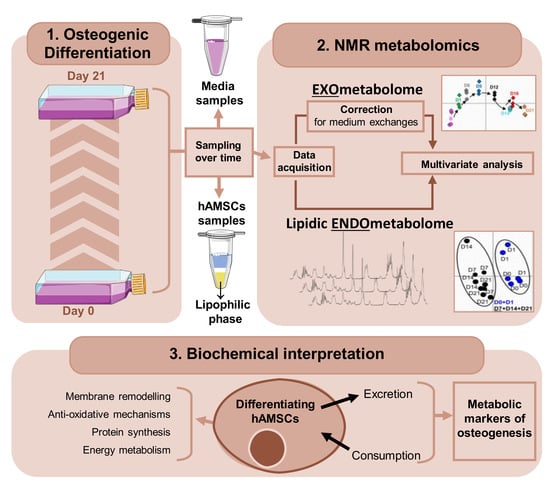
Endo- and Exometabolome Crosstalk in Mesenchymal Stem Cells Undergoing Osteogenic Differentiation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expansion of Human Adipose-Derived Mesenchymal Stem Cells (hAMSCs)
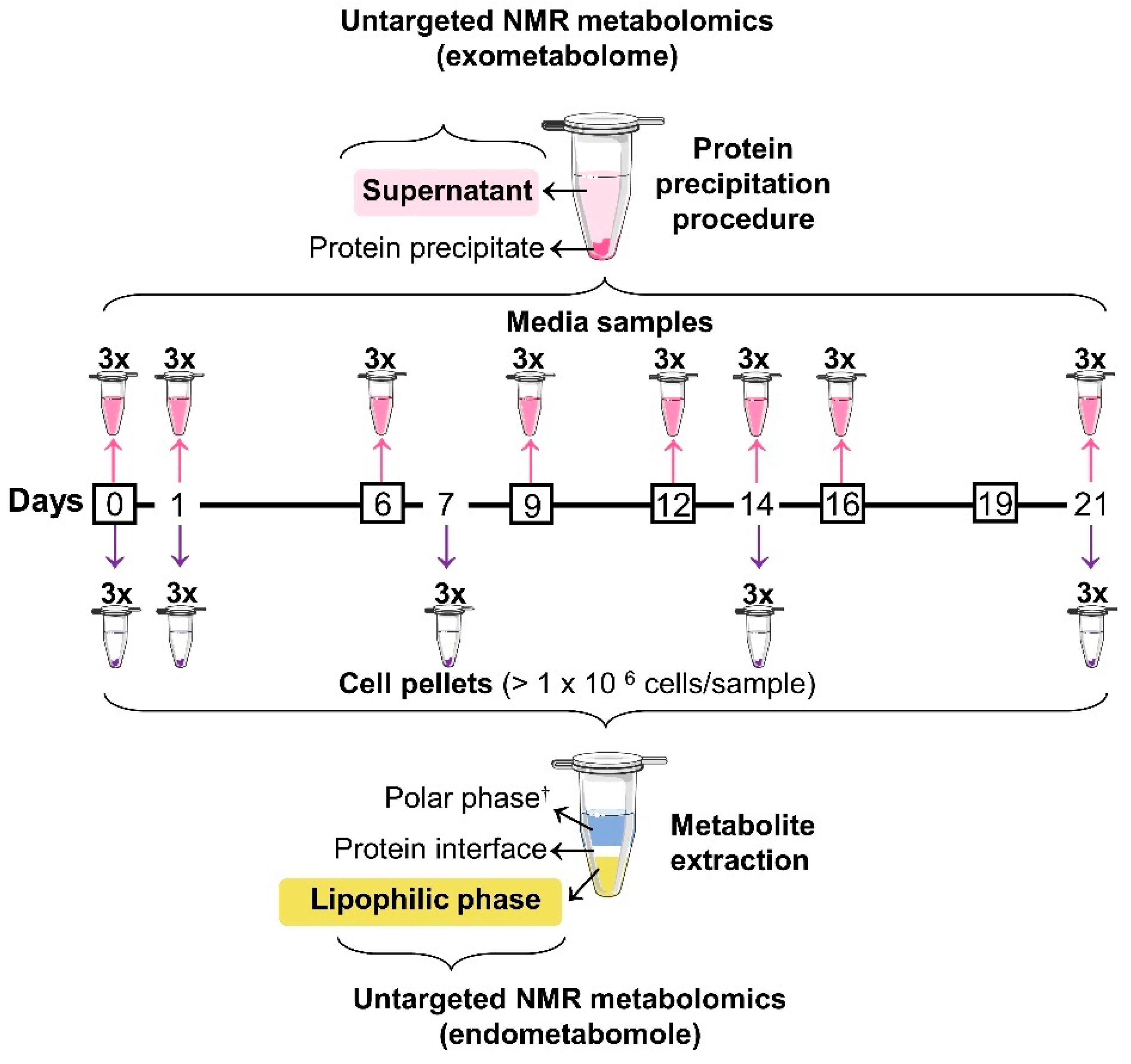
2.2. Osteogenic Differentiation and Sampling
2.3. Lipidic Extraction
2.4. Protein Precipitation in Media Samples
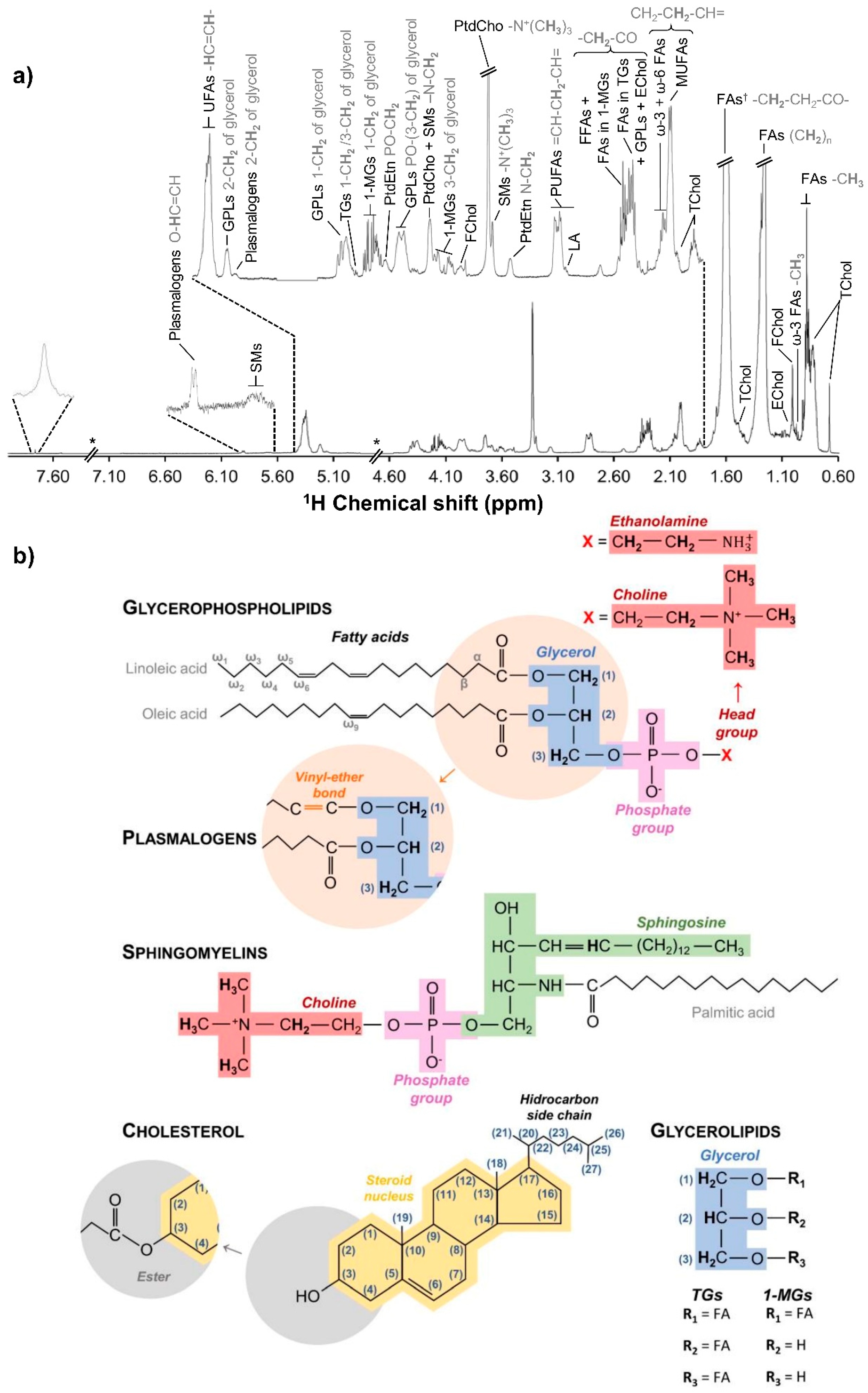
2.5. NMR Spectroscopy
2.6. Data Analysis and Statistics
3. Results
3.1. hAMSC Lipophilic Endometabolome Changes
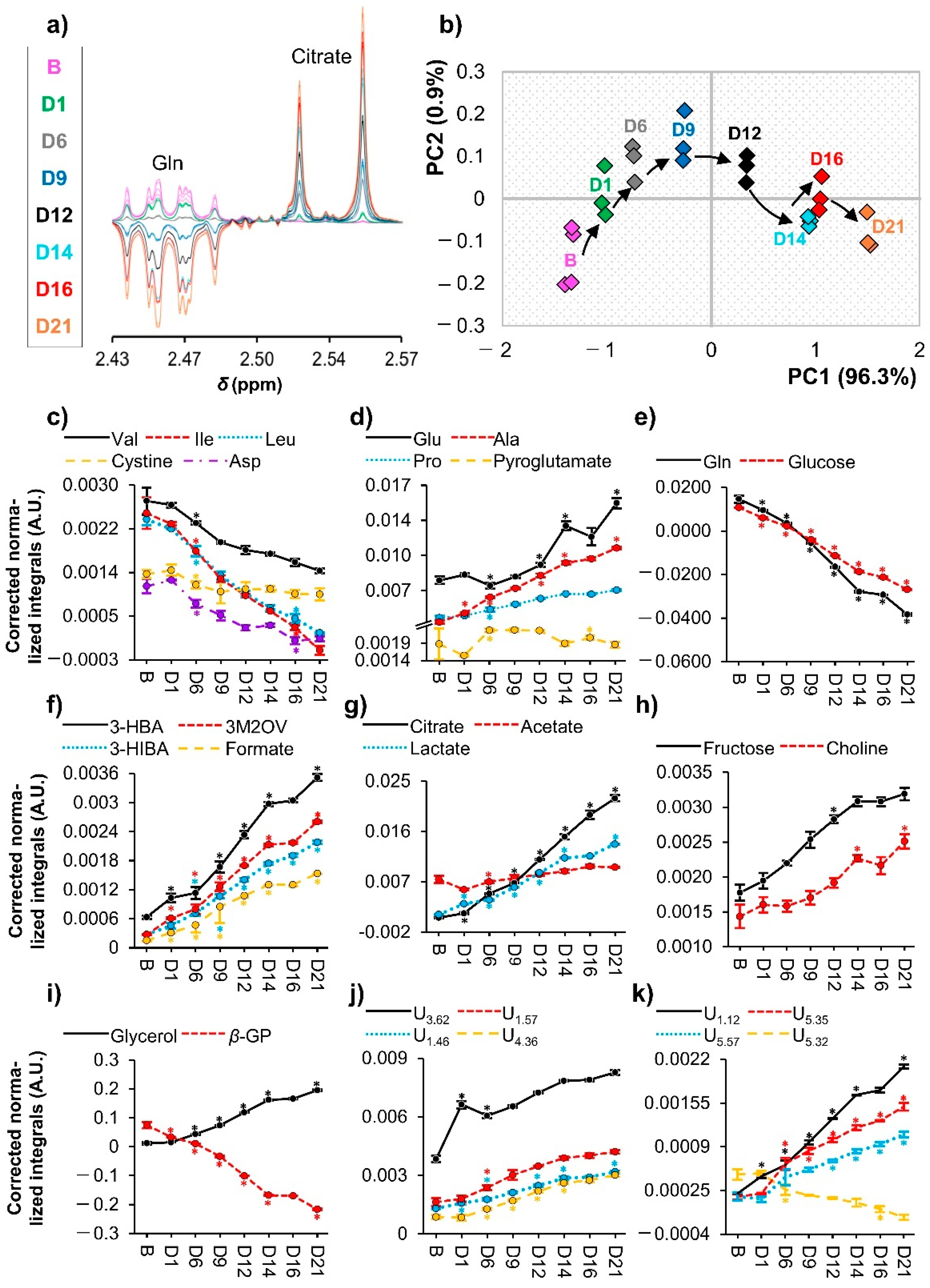
3.2. hAMSC Exometabolome Changes
4. Discussion
4.1. The Dynamics of hAMSC Endometabolome during Osteogenic Differentiation
4.2. Articulation between the Endo- and Exometabolomes of hAMSCs during Osteogenic Differentiation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Iaquinta, M.R.; Mazzoni, E.; Bononi, I.; Rotondo, J.C.; Mazziotta, C.; Montesi, M.; Sprio, S.; Tampieri, A.; Tognon, M.; Martini, F. Adult stem cells for bone regeneration and repair. Front. Cell Dev. Biol. 2019, 7, 268. [Google Scholar] [CrossRef] [Green Version]
- Bispo, D.S.C.; Jesus, C.S.H.; Marques, I.M.C.; Romek, K.M.; Oliveira, M.B.; Mano, J.F.; Gil, A.M. Metabolomic applications in stem cell research: A review. Stem Cell Rev. Rep. 2021, 17, 2003–2024. [Google Scholar] [CrossRef] [PubMed]
- Coman, C.; Solari, F.A.; Hentschel, A.; Sickmann, A.; Zahedi, R.P.; Ahrends, R. Simultaneous Metabolite, Protein, Lipid Extraction (SIMPLEX): A combinatorial multimolecular omics approach for systems biology. Mol. Cell. Proteom. 2016, 15, 1453–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villaret-Cazadamont, J.; Poupin, N.; Tournadre, A.; Batut, A.; Gales, L.; Zalko, D.; Cabaton, N.J.; Bellvert, F.; Bertrand-Michel, J. An optimized dual extraction method for the simultaneous and accurate analysis of polar metabolites and lipids carried out on single biological samples. Metabolites 2020, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Surrati, A.; Evseev, S.; Jourdan, F.; Kim, D.-H.; Sottile, V. Osteogenic response of human mesenchymal stem cells analysed using combined intracellular and extracellular metabolomic monitoring. Cell. Physiol. Biochem. 2021, 55, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Casati, S.; Giannasi, C.; Niada, S.; Bergamaschi, R.F.; Orioli, M.; Brini, A.T. Bioactive lipids in mscs biology: State of the art and role in inflammation. Int. J. Mol. Sci. 2021, 22, 1481. [Google Scholar] [CrossRef] [PubMed]
- Clémot, M.; Demarco, R.S.; Jones, D.L. Lipid mediated regulation of adult stem cell behavior. Front. Cell Dev. Biol. 2020, 8, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurowski, K.; Kochan, K.; Walczak, J.; Barańska, M.; Piekoszewski, W.; Buszewski, B. Analytical techniques in lipidomics: State of the art. Crit. Rev. Anal. Chem. 2017, 47, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Vosegaard, T.; Guo, Z. Applications of nuclear magnetic resonance in lipid analyses: An emerging powerful tool for lipidomics studies. Prog. Lipid Res. 2017, 68, 37–56. [Google Scholar] [CrossRef]
- Dai, H.; Hong, B.; Xu, Z.; Ma, L.; Chen, Y.; Xiao, Y.; Wu, R. Nuclear magnetic resonance spectroscopy is highly sensitive for lipid-soluble metabolites. Neural Regen. Res. 2013, 8, 2103–2110. [Google Scholar] [CrossRef]
- Bhinderwala, F.; Wase, N.; DiRusso, C.; Powers, R. Combining mass spectrometry and NMR improves metabolite detection and annotation. J. Proteome Res. 2018, 17, 4017–4022. [Google Scholar] [CrossRef] [PubMed]
- Kiamehr, M.; Viiri, L.E.; Vihervaara, T.; Koistinen, K.M.; Hilvo, M.; Ekroos, K.; Käkelä, R.; Aalto-Setälä, K. Lipidomic profiling of patient-specific iPSC-derived hepatocyte-like cells. Dis. Model. Mech. 2017, 10, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Tanosaki, S.; Tohyama, S.; Fujita, J.; Someya, S.; Hishiki, T.; Matsuura, T.; Nakanishi, H.; Ohto-Nakanishi, T.; Akiyama, T.; Morita, Y.; et al. Fatty acid synthesis is indispensable for survival of human pluripotent stem cells. iScience 2020, 23, 101535. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, K.; Xing, G.; Li, L.; Ma, B.; Hu, Z.; Duan, L.; Liu, X. Phospholipid remodeling is critical for stem cell pluripotency by facilitating mesenchymal-to-epithelial transition. Sci. Adv. 2019, 5, eaax7525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Lee, H.-R.; Kim, H.-Y.; Lee, H.; Kim, H.-J.; Choi, H.-K. Characterization and classification of rat neural stem cells and differentiated cells by comparative metabolic and lipidomic profiling. Anal. Bioanal. Chem. 2019, 411, 5423–5436. [Google Scholar] [CrossRef] [PubMed]
- Rudan, M.V.; Mishra, A.; Klose, C.; Eggert, U.S.; Watt, F.M. Human epidermal stem cell differentiation is modulated by specific lipid subspecies. Proc. Natl. Acad. Sci. USA 2020, 117, 22173–22182. [Google Scholar] [CrossRef]
- Li, J.; Cui, Z.; Zhao, S.; Sidman, R.L. Unique glycerophospholipid signature in retinal stem cells correlates with enzymatic functions of diverse long-chain Acyl-CoA synthetases. Stem Cells 2007, 25, 2864–2873. [Google Scholar] [CrossRef]
- da Silva, C.G.; de Sá Barretto, L.S.; Lo Turco, E.G.; de Lima Santos, A.; Lessio, C.; Martins, H.A., Jr.; de Almeida, F.G. Lipidomics of mesenchymal stem cell differentiation. Chem. Phys. Lipids 2020, 232, 104964. [Google Scholar] [CrossRef]
- Rampler, E.; Egger, D.; Schoeny, H.; Rusz, M.; Pacheco, M.P.; Marino, G.; Kasper, C.; Naegele, T.; Koellensperger, G. The power of LC-MS based multiomics: Exploring adipogenic differentiation of human mesenchymal stem/stromal cells. Molecules 2019, 24, 3615. [Google Scholar] [CrossRef] [Green Version]
- Liaw, L.; Prudovsky, I.; Koza, R.A.; Anunciado-Koza, R.V.; Siviski, M.E.; Lindner, V.; Friesel, R.E.; Rosen, C.J.; Baker, P.R.S.; Simons, B.; et al. Lipid profiling of in vitro cell models of adipogenic differentiation: Relationships with mouse adipose tissues. J. Cell. Biochem. 2016, 117, 2182–2193. [Google Scholar] [CrossRef] [Green Version]
- Bojin, F.M.; Gruia, A.T.; Cristea, M.I.; Ordodi, V.L.; Paunescu, V.; Mic, F.A. Adipocytes differentiated in vitro from rat mesenchymal stem cells lack essential free fatty acids compared to adult adipocytes. Stem Cells Dev. 2012, 21, 507–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruia, A.T.; Suciu, M.; Barbu-Tudoran, L.; Azghadi, S.M.R.; Cristea, M.I.; Nica, D.V.; Vaduva, A.; Muntean, D.; Mic, A.A.; Mic, F.A. Mesenchymal stromal cells differentiating to adipocytes accumulate autophagic vesicles instead of functional lipid droplets. J. Cell. Physiol. 2016, 231, 863–875. [Google Scholar] [CrossRef]
- Alakpa, E.V.; Jayawarna, V.; Lampel, A.; Burgess, K.V.; West, C.C.; Bakker, S.C.J.; Roy, S.; Javid, N.; Fleming, S.; Lamprou, D.A.; et al. Tunable supramolecular hydrogels for selection of lineage-guiding metabolites in stem cell cultures. Chem 2016, 1, 298–319. [Google Scholar] [CrossRef] [Green Version]
- Levental, K.R.; Surma, M.A.; Skinkle, A.D.; Lorent, J.H.; Zhou, Y.; Klose, C.; Chang, J.T.; Hancock, J.F.; Levental, I. W-3 polyunsaturated fatty acids direct differentiation of the membrane phenotype in mesenchymal stem cells to potentiate osteogenesis. Sci. Adv. 2017, 3, eaao1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orapiriyakul, W.; Tsimbouri, M.P.; Childs, P.; Campsie, P.; Wells, J.; Fernandez-Yague, M.A.; Burgess, K.; Tanner, K.E.; Tassieri, M.; Meek, D.; et al. Nanovibrational stimulation of mesenchymal stem cells induces therapeutic reactive oxygen species and inflammation for three-dimensional bone tissue engineering. ACS Nano 2020, 14, 10027–10044. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, T.; Monica Tsimbouri, P.; Llopis-Hernandez, V.; Campsie, P.; Scurr, D.; Childs, P.G.; Phillips, D.; Donnelly, S.; Wells, J.A.; O’Brien, F.J.; et al. The use of nanovibration to discover specific and potent bioactive metabolites that stimulate osteogenic differentiation in mesenchymal stem cells. Sci. Adv. 2021, 7, eabb7921. [Google Scholar] [CrossRef] [PubMed]
- Georgi, N.; Cillero-Pastor, B.; Eijkel, G.B.; Periyasamy, P.C.; Kiss, A.; van Blitterswijk, C.; Post, J.N.; Heeren, R.M.A.; Karperien, M. Differentiation of mesenchymal stem cells under hypoxia and normoxia: Lipid profiles revealed by time-of-flight secondary ion mass spectrometry and multivariate analysis. Anal. Chem. 2015, 87, 3981–3988. [Google Scholar] [CrossRef]
- Rocha, B.; Cillero-Pastor, B.; Eijkel, G.; Bruinen, A.L.; Ruiz-Romero, C.; Heeren, R.M.A.; Blanco, F.J. Characterization of lipidic markers of chondrogenic differentiation using mass spectrometry imaging. Proteomics 2015, 15, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Kilpinen, L.; Tigistu-Sahle, F.; Oja, S.; Greco, D.; Parmar, A.; Saavalainen, P.; Nikkilä, J.; Korhonen, M.; Lehenkari, P.; Käkelä, R.; et al. Aging bone marrow mesenchymal stromal cells have altered membrane glycerophospholipid composition and functionality. J. Lipid Res. 2013, 54, 622–635. [Google Scholar] [CrossRef] [Green Version]
- Chatgilialoglu, A.; Rossi, M.; Alviano, F.; Poggi, P.; Zannini, C.; Marchionni, C.; Ricci, F.; Tazzari, P.L.; Taglioli, V.; Calder, P.C.; et al. Restored in vivo-like membrane lipidomics positively influence in vitro features of cultured mesenchymal stromal/stem cells derived from human placenta. Stem Cell Res. Ther. 2017, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Chen, Y.; Wang, H.; Bai, Y.; Zhao, J.; Zhang, X.; Liang, L.; Chen, Y.; Ye, C.; Li, Y.; et al. Integrated lipidomics and transcriptomics characterization upon aging-related changes of lipid species and pathways in human bone marrow mesenchymal stem cells. J. Proteome Res. 2019, 18, 2065–2077. [Google Scholar] [CrossRef]
- Mastrangelo, A.; Panadero, M.I.; Perez, L.M.; Galvez, B.G.; Garcia, A.; Barbas, C.; Ruperez, F.J. New insight on obesity and adipose-derived stem cells using comprehensive metabolomics. Biochem. J. 2016, 473, 2187–2203. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Yi, T.G.; Ahn, S.H.; Lim, D.K.; Kim, S.-n.; Lee, H.J.; Cho, Y.K.; Lim, J.Y.; Sung, J.H.; Yun, J.H.; et al. Comparative study on metabolite level in tissue-specific human mesenchymal stem cells by an ultra-performance liquid chromatography quadrupole time of flight mass spectrometry. Anal. Chim. Acta 2018, 1024, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Burk, J.; Melzer, M.; Hagen, A.; Lips, K.S.; Trinkaus, K.; Nimptsch, A.; Leopold, J. Phospholipid profiles for phenotypic characterization of adipose-derived multipotent mesenchymal stromal cells. Front. Cell Dev. Biol. 2021, 9, 784405. [Google Scholar] [CrossRef] [PubMed]
- DeVeaux, S.A.; Ogle, M.E.; Vyshnya, S.; Chiappa, N.F.; Leitmann, B.; Rudy, R.; Day, A.; Mortensen, L.J.; Kurtzberg, J.; Roy, K.; et al. Characterizing human mesenchymal stromal cells’ immune-modulatory potency using targeted lipidomic profiling of sphingolipids. Cytotherapy 2022, in press. [CrossRef] [PubMed]
- Lv, M.; Zhang, S.; Jiang, B.; Cao, S.; Dong, Y.; Cao, L.; Guo, S. Adipose-derived stem cells regulate metabolic homeostasis and delay aging by promoting mitophagy. FASEB J. 2021, 35, e21709. [Google Scholar] [CrossRef] [PubMed]
- Surrati, A.; Linforth, R.; Fisk, I.D.; Sottile, V.; Kim, D.H. Non-destructive characterisation of mesenchymal stem cell differentiation using LC-MS-based metabolite footprinting. Analyst 2016, 141, 3776–3787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amer, M.H.; Alvarez-Paino, M.; McLaren, J.; Pappalardo, F.; Trujillo, S.; Wong, J.Q.; Shrestha, S.; Abdelrazig, S.; Stevens, L.A.; Lee, J.B.; et al. Designing topographically textured microparticles for induction and modulation of osteogenesis in mesenchymal stem cell engineering. Biomaterials 2021, 266, 120450. [Google Scholar] [CrossRef] [PubMed]
- Sigmarsdottir, T.B.; McGarrity, S.; Yurkovich, J.T.; Rolfsson, Ó.; Sigurjónsson, Ó.E. Analyzing metabolic states of adipogenic and osteogenic differentiation in human mesenchymal stem cells via genome scale metabolic model reconstruction. Front. Cell Dev. Biol. 2021, 9, 642681. [Google Scholar] [CrossRef]
- Bispo, D.S.C.; Jesus, C.S.H.; Correia, M.; Ferreira, F.; Bonifazio, G.; Goodfellow, B.J.; Oliveira, M.B.; Mano, J.F.; Gil, A.M. NMR metabolomics assessment of osteogenic differentiation of adipose-tissue-derived mesenchymal stem cells. J. Proteome Res. 2022, articles in press. [CrossRef]
- Wu, H.; Southam, A.D.; Hines, A.; Viant, M.R. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef]
- Tukiainen, T.; Tynkkynen, T.; Mäkinen, V.P.; Jylänki, P.; Kangas, A.; Hokkanen, J.; Vehtari, A.; Gröhn, O.; Hallikainen, M.; Soininen, H.; et al. A multi-metabolite analysis of serum by 1H NMR spectroscopy: Early systemic signs of Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2008, 375, 356–361. [Google Scholar] [CrossRef]
- Dais, P.; Misiak, M.; Hatzakis, E. Analysis of marine dietary supplements using NMR spectroscopy. Anal. Methods 2015, 7, 5226–5238. [Google Scholar] [CrossRef]
- Subramanian, A.; Joshi, B.S.; Roy, A.D.; Roy, R.; Gupta, V.; Dang, R.S. NMR spectroscopic identification of cholesterol esters, plasmalogen and phenolic glycolipids as fingerprint markers of human intracranial tuberculomas. NMR Biomed. 2008, 21, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Nieva-Echevarría, B.; Goicoechea, E.; Manzanos, M.J.; Guillén, M.D. Usefulness of 1H NMR in assessing the extent of lipid digestion. Food Chem. 2015, 179, 182–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefevre, C.; Panthu, B.; Naville, D.; Guibert, S.; Pinteur, C.; Elena-Herrmann, B.; Vidal, H.; Rautureau, G.J.P.; Mey, A. Metabolic phenotyping of adipose-derived stem cells reveals a unique signature and intrinsic differences between fat pads. Stem Cells Int. 2019, 2019, 9323864. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Veselkov, K.A.; Lindon, J.C.; Ebbels, T.M.D.; Crockford, D.; Volynkin, V.V.; Holmes, E.; Davies, D.B.; Nicholson, J.K. Recursive segment-wise peak alignment of biological 1 H NMR spectra for improved metabolic biomarker recovery. Anal. Chem. 2009, 81, 56–66. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef]
- Bridge, P.D.; Sawilowsky, S.S. Increasing physicians’ awareness of the impact of statistics on research outcomes: Comparative power of the t-test and Wilcoxon rank-sum test in small samples applied research. J. Clin. Epidemiol. 1999, 52, 229–235. [Google Scholar] [CrossRef]
- Berben, L.; Sereika, S.M.; Engberg, S. Effect size estimation: Methods and examples. Int. J. Nurs. Stud. 2012, 49, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Mosconi, E.; Fontanella, M.; Sima, D.M.; Van Huffel, S.; Fiorini, S.; Sbarbati, A.; Marzola, P. Investigation of adipose tissues in Zucker rats using in vivo and ex vivo magnetic resonance spectroscopy. J. Lipid Res. 2011, 52, 330–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rendina-Ruedy, E.; Guntur, A.R.; Rosen, C.J. Intracellular lipid droplets support osteoblast function. Adipocyte 2017, 6, 250–258. [Google Scholar] [CrossRef] [Green Version]
- Gillet, C.; Spruyt, D.; Rigutto, S.; Dalla Valle, A.; Berlier, J.; Louis, C.; Debier, C.; Gaspard, N.; Malaisse, W.J.; Gangji, V.; et al. Oleate abrogates palmitate-induced lipotoxicity and proinflammatory response in human bone marrow-derived mesenchymal stem cells and osteoblastic cells. Endocrinology 2015, 156, 4081–4093. [Google Scholar] [CrossRef] [Green Version]
- Else, P.L. The highly unnatural fatty acid profile of cells in culture. Prog. Lipid Res. 2020, 77, 101017. [Google Scholar] [CrossRef]
- Casado-Díaz, A.; Santiago-Mora, R.; Dorado, G.; Quesada-Gómez, J.M. The omega-6 arachidonic fatty acid, but not the omega-3 fatty acids, inhibits osteoblastogenesis and induces adipogenesis of human mesenchymal stem cells: Potential implication in osteoporosis. Osteoporos. Int. 2013, 24, 1647–1661. [Google Scholar] [CrossRef]
- Abshirini, M.; Ilesanmi-Oyelere, B.L.; Kruger, M.C. Potential modulatory mechanisms of action by long-chain polyunsaturated fatty acids on bone cell and chondrocyte metabolism. Prog. Lipid Res. 2021, 83, 101113. [Google Scholar] [CrossRef]
- Kashirina, A.S.; López-Duarte, I.; Kubánková, M.; Gulin, A.A.; Dudenkova, V.V.; Rodimova, S.A.; Torgomyan, H.G.; Zagaynova, E.V.; Meleshina, A.V.; Kuimova, M.K. Monitoring membrane viscosity in differentiating stem cells using BODIPY-based molecular rotors and FLIM. Sci. Rep. 2020, 10, 14063. [Google Scholar] [CrossRef]
- Klontzas, M.E.; Vernardis, S.I.; Heliotis, M.; Tsiridis, E.; Mantalaris, A. Metabolomics analysis of the osteogenic differentiation of umbilical cord blood mesenchymal stem cells reveals differential sensitivity to osteogenic agents. Stem Cells Dev. 2017, 26, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Klontzas, M.E.; Reakasame, S.; Silva, R.; Morais, J.C.F.; Vernardis, S.; MacFarlane, R.J.; Heliotis, M.; Tsiridis, E.; Panoskaltsis, N.; Boccaccini, A.R.; et al. Oxidized alginate hydrogels with the GHK peptide enhance cord blood mesenchymal stem cell osteogenesis: A paradigm for metabolomics-based evaluation of biomaterial design. Acta Biomater. 2019, 88, 224–240. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guo, H.; Li, H. Cholesterol loading affects osteoblastic differentiation in mouse mesenchymal stem cells. Steroids 2013, 78, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xiu, C.; Zhou, Q.; Ni, L.; Du, J.; Gong, T.; Li, M.; Saijilafu, H.Y.; Chen, J. A dual role of cholesterol in osteogenic differentiation of bone marrow stromal cells. J. Cell. Physiol. 2019, 234, 2058–2066. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.J.; Han, X.; Chung, K.-N.; Gross, R.W. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: A quantitative electrospray ionization/mass spectrometric analysis. Biochemistry 2002, 41, 2075–2088. [Google Scholar] [CrossRef]
- Baker, N.; Sohn, J.; Tuan, R.S. Promotion of human mesenchymal stem cell osteogenesis by PI3-kinase/Akt signaling, and the influence of caveolin-1/cholesterol homeostasis. Stem Cell Res. Ther. 2015, 6, 238. [Google Scholar] [CrossRef] [Green Version]
- van de Peppel, J.; van Leeuwen, J.P.T.M. Vitamin D and gene networks in human osteoblasts. Front. Physiol. 2014, 5, 137. [Google Scholar] [CrossRef] [Green Version]
- Zoeller, R.A.; Lake, A.C.; Nagan, N.; Gaposchkin, D.P.; Legner, M.A.; Lieberthal, W. Plasmalogens as endogenous antioxidants: Somatic cell mutants reveal the importance of the vinyl ether. Biochem. J. 1999, 338, 769–776. [Google Scholar] [CrossRef]
- Luthringer, B.J.C.; Katha, U.M.R.; Willumeit, R. Phosphatidylethanolamine biomimetic coating increases mesenchymal stem cell osteoblastogenesis. J. Mater. Sci. Mater. Med. 2014, 25, 2561–2571. [Google Scholar] [CrossRef]
- Butler, W.T. The nature and significance of osteopontin. Connect. Tissue Res. 1989, 23, 123–136. [Google Scholar] [CrossRef]
- Suzuki, A.; Iwata, J. Amino acid metabolism and autophagy in skeletal development and homeostasis. Bone 2021, 146, 115881. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.Y.; Rawal, A.; Schmidt-Rohr, K. Strongly bound citrate stabilizes the apatite nanocrystals in bone. Proc. Natl. Acad. Sci. USA 2010, 107, 22425–22429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uno, K.; Takarada, T.; Takarada-Iemata, M.; Nakamura, Y.; Fujita, H.; Hinoi, E.; Yoneda, Y. Negative regulation of osteoblastogenesis through downregulation of runt-related transcription factor-2 in osteoblastic MC3T3-E1 cells with stable overexpression of the cystine/glutamate antiporter xCT subunit. J. Cell. Physiol. 2011, 226, 2953–2964. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.H.; Yang, R.S.; Tang, C.H.; Wu, M.Y.; Fu, W.M. Regulation of the maturation of osteoblasts and osteoclastogenesis by glutamate. Eur. J. Pharmacol. 2008, 589, 37–44. [Google Scholar] [CrossRef]
- Takarada-Iemata, M.; Takarada, T.; Nakamura, Y.; Nakatani, E.; Hori, O.; Yoneda, Y. Glutamate preferentially suppresses osteoblastogenesis than adipogenesis through the cystine/glutamate antiporter in mesenchymal stem cells. J. Cell. Physiol. 2011, 226, 652–665. [Google Scholar] [CrossRef]
- Pattappa, G.; Heywood, H.K.; de Bruijn, J.D.; Lee, D.A. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J. Cell. Physiol. 2011, 226, 2562–2570. [Google Scholar] [CrossRef]
- Chen, C.-T.; Shih, Y.-R.V.; Kuo, T.K.; Lee, O.K.; Wei, Y.-H. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 2008, 26, 960–968. [Google Scholar] [CrossRef]
- Shum, L.C.; White, N.S.; Mills, B.N.; De Mesy Bentley, K.L.; Eliseev, R.A. Energy metabolism in mesenchymal stem cells during osteogenic differentiation. Stem Cells Dev. 2016, 25, 114–122. [Google Scholar] [CrossRef] [Green Version]
- Zubiría, M.G.; Alzamendi, A.; Moreno, G.; Rey, M.A.; Spinedi, E.; Giovambattista, A. Long-term fructose intake increases adipogenic potential: Evidence of direct effects of fructose on adipocyte precursor cells. Nutrients 2016, 8, 198. [Google Scholar] [CrossRef] [Green Version]
- Legeza, B.; Balázs, Z.; Odermatt, A. Fructose promotes the differentiation of 3T3-L1 adipocytes and accelerates lipid metabolism. FEBS Lett. 2014, 588, 490–496. [Google Scholar] [CrossRef] [Green Version]
- Felice, J.I.; Gangoiti, M.V.; Molinuevo, M.S.; McCarthy, A.D.; Cortizo, A.M. Effects of a metabolic syndrome induced by a fructose-rich diet on bone metabolism in rats. Metabolism 2014, 63, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.P.; Li, Z.; Zoch, M.L.; Frey, J.L.; Bowman, C.E.; Kushwaha, P.; Ryan, K.A.; Goh, B.C.; Scafidi, S.; Pickett, J.E.; et al. Fatty acid oxidation by the osteoblast is required for normal bone acquisition in a sex- and diet-dependent manner. JCI Insight 2017, 2, e92704. [Google Scholar] [CrossRef] [PubMed]
- Bermeo, S.; Al Saedi, A.; Vidal, C.; Khalil, M.; Pang, M.; Troen, B.R.; Myers, D.; Duque, G. Treatment with an inhibitor of fatty acid synthase attenuates bone loss in ovariectomized mice. Bone 2019, 122, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Nováková, S.; Danchenko, M.; Okajčeková, T.; Baranovičová, E.; Kováč, A.; Grendár, M.; Beke, G.; Pálešová, J.; Strnádel, J.; Janíčková, M.; et al. Comparative proteomic and metabolomic analysis of human osteoblasts, differentiated from dental pulp stem cells, hinted crucial signaling pathways promoting osteogenesis. Int. J. Mol. Sci. 2021, 22, 7908. [Google Scholar] [CrossRef]
- Newman, J.C.; Verdin, E. Ketone bodies as signaling metabolites. Trends Endocrinol. Metab. 2014, 25, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zou, B.; Shi, Z.; Wu, Q.; Chen, G.Q. The effect of 3-hydroxybutyrate on the in vitro differentiation of murine osteoblast MC3T3-E1 and in vivo bone formation in ovariectomized rats. Biomaterials 2007, 28, 3063–3073. [Google Scholar] [CrossRef]
- Crown, S.B.; Marze, N.; Antoniewicz, M.R. Catabolism of branched chain amino acids contributes significantly to synthesis of odd-chain and even-chain fatty acids in 3T3-L1 adipocytes. PLoS ONE 2015, 10, e0145850. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Cooper, D.E.; Cluntun, A.A.; Warmoes, M.O.; Zhao, S.; Reid, M.A.; Liu, J.; Lund, P.J.; Lopes, M.; Garcia, B.A.; et al. Acetate production from glucose and coupling to mitochondrial metabolism in mammals. Cell 2018, 175, 502–513. [Google Scholar] [CrossRef] [Green Version]
- Meiser, J.; Tumanov, S.; Maddocks, O.; Labuschagne, C.F.; Athineos, D.; Van Den Broek, N.; Mackay, G.M.; Gottlieb, E.; Blyth, K.; Vousden, K.; et al. Serine one-carbon catabolism with formate overflow. Sci. Adv. 2016, 2, e1601273. [Google Scholar] [CrossRef] [Green Version]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate metabolism in health and disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef]
- Langenbach, F.; Handschel, J. Effects of dexamethasone, ascorbic acid and β-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Res. Ther. 2013, 4, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäck, L.M.; Noack, S.; Winkler, R.; Wißmann, G.; Behrens, P.; Wellmann, M.; Jagodzinski, M.; Krettek, C.; Hoffmann, A. The phosphate source influences gene expression and quality of mineralization during in vitro osteogenic differentiation of human mesenchymal stem cells. PLoS ONE 2013, 8, e65943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, S.J.; Stewart, A.J.; Sadler, P.J.; Farquharson, C. Human PHOSPHO1 exhibits high specific phosphoethanolamine and phosphocholine phosphatase activities. Biochem. J. 2004, 382, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, M.C.; Simão, A.M.S.; Narisawa, S.; Huesa, C.; McKee, M.D.; Farquharson, C.; Millán, J.L. Loss of skeletal mineralization by the simultaneous ablation of PHOSPHO1 and alkaline phosphatase function: A unified model of the mechanisms of initiation of skeletal calcification. J. Bone Miner. Res. 2011, 26, 286–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
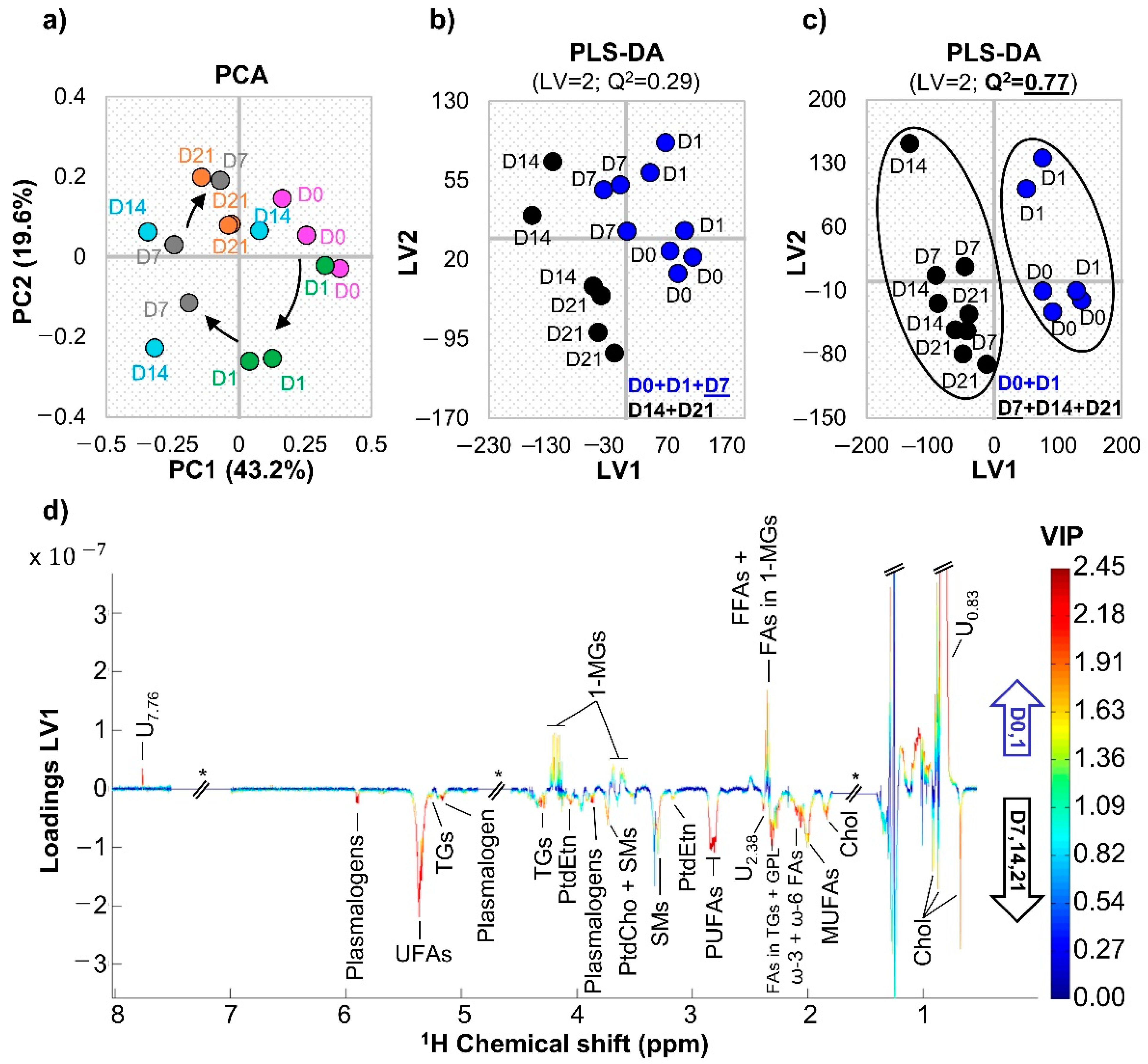
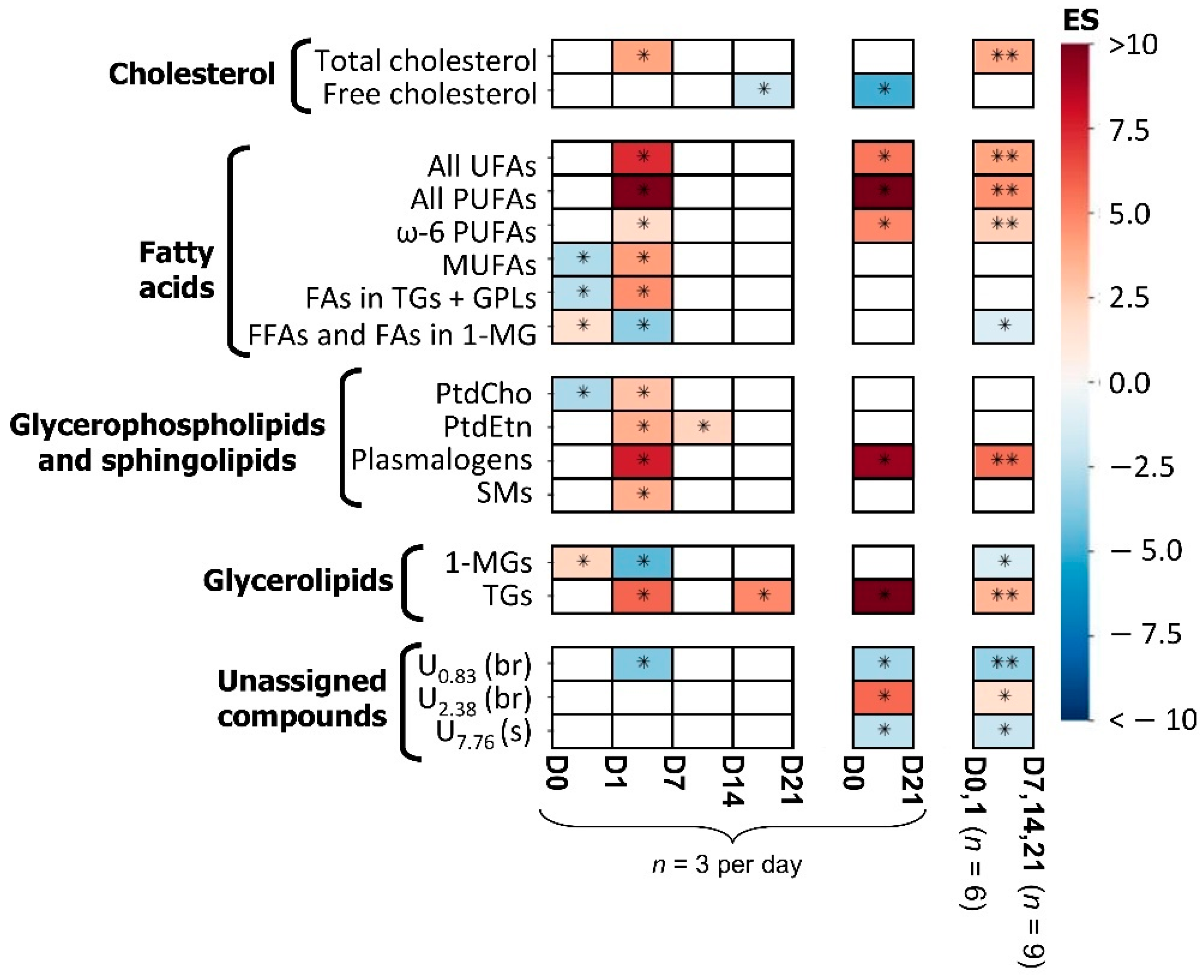


| Metabolite | δ 1H (Multiplicity) a | D0 + D1 vs. D7 + D14 + D21 | ||
|---|---|---|---|---|
| Effect Size (ES Error %) | p-Values | FDR Adjusted p-Values | ||
| Cholesterol | ||||
| Total cholesterol | 0.68 (s) | 3.8 (44.8%) | 0.001 | 0.004 |
| Fatty acids | ||||
| All UFAs | 5.35 (m) | 4.0 (44.0%) | 0.001 | 0.004 |
| All PUFAs | 2.82 (m) | 4.6 (42.3%) | 0.001 | 0.004 |
| ω-6 PUFAs | 2.06 (m) | 2.5 (55.1%) | 0.003 | 0.008 |
| FFAs + FAs in 1-MGs | 2.35 (t) | −1.2 (90.9%) | 0.045 | 0.048 |
| Glycerophospholipids | ||||
| Plasmalogens | 5.90 (d) | 5.6 (40.3%) | 0.001 | 0.004 |
| Glycerolipids | ||||
| 1-MGs | 4.18 (ddd) | −1.3 (84%) | 0.025 | 0.033 |
| TGs | 4.29 (dd) | 11.9 (58.2%) | 0.001 | 0.004 |
| Unassigned compounds | ||||
| U0.83 | 0.83 (br) | −3.0 (78.3%) | 0.001 | 0.004 |
| U2.38 | 2.38 (m) | 1.8 (66.8%) | 0.013 | 0.023 |
| U7.76 | 7.76 (s) | −2.3 (89.1%) | 0.013 | 0.023 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bispo, D.S.C.; Michálková, L.; Correia, M.; Jesus, C.S.H.; Duarte, I.F.; Goodfellow, B.J.; Oliveira, M.B.; Mano, J.F.; Gil, A.M. Endo- and Exometabolome Crosstalk in Mesenchymal Stem Cells Undergoing Osteogenic Differentiation. Cells 2022, 11, 1257. https://doi.org/10.3390/cells11081257
Bispo DSC, Michálková L, Correia M, Jesus CSH, Duarte IF, Goodfellow BJ, Oliveira MB, Mano JF, Gil AM. Endo- and Exometabolome Crosstalk in Mesenchymal Stem Cells Undergoing Osteogenic Differentiation. Cells. 2022; 11(8):1257. https://doi.org/10.3390/cells11081257
Chicago/Turabian StyleBispo, Daniela S. C., Lenka Michálková, Marlene Correia, Catarina S. H. Jesus, Iola F. Duarte, Brian J. Goodfellow, Mariana B. Oliveira, João F. Mano, and Ana M. Gil. 2022. "Endo- and Exometabolome Crosstalk in Mesenchymal Stem Cells Undergoing Osteogenic Differentiation" Cells 11, no. 8: 1257. https://doi.org/10.3390/cells11081257
APA StyleBispo, D. S. C., Michálková, L., Correia, M., Jesus, C. S. H., Duarte, I. F., Goodfellow, B. J., Oliveira, M. B., Mano, J. F., & Gil, A. M. (2022). Endo- and Exometabolome Crosstalk in Mesenchymal Stem Cells Undergoing Osteogenic Differentiation. Cells, 11(8), 1257. https://doi.org/10.3390/cells11081257