The Autophagy Process in Cervical Carcinogenesis: Role of Non-Coding-RNAs, Molecular Mechanisms, and Therapeutic Targets
Abstract
:1. Introduction
2. Autophagy in Premalignant Cervical Lesions and Cervical Cancer
3. The Role of HPV E5, E6, and E7 Oncoproteins in the Autophagy Process in Cervical Cancer
4. Function of ncRNAs in the Regulation of Autophagy in Cervical Cancer Cells
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Akt | RAC-alpha serine/threonine-protein kinase |
| AMPK | AMP-activated protein kinase |
| ATF2 | Activating transcription factor 2 |
| ATF4, 6 | Activating transcription factors 4, 6 (tax-responsive enhancer element B67) |
| ATGs1-12 | Autophagy-related genes 1-12 |
| Bax | Bcl-2-associated X protein |
| Bcl-2 | B-cell lymphoma 2 |
| Bcl-XL | B-cell lymphoma-extra large |
| Becn1 | Beclin-1 |
| CD95 | Fas receptor |
| CDKs | Cyclin-dependent kinase |
| c-FLIP | CASP8 and FADD-like apoptosis regulator |
| CHK1 | Checkpoint kinase-1 |
| CircRNAs | Circular RNAs |
| CPT | Camptothecin |
| DAPK | Death-associated protein kinase |
| DRAM | Damage-regulated autophagy modulators |
| ER | Endoplasmic reticulum |
| EGFR | Epidermal growth factor receptor |
| FLICE | Inhibitory protein (c-FLIP) |
| EMT | Epithelial–mesenchymal transition |
| Erk | Extracellular signal-regulated kinases |
| FAK | Focal adhesion kinase |
| GSK3B | Glycogen synthase kinase 3 beta |
| HCPT | Hydroxycamptothecin |
| HDLEC | Human dermal lymphatic endothelial cells |
| HIF | Hypoxia-inducible transcription factor |
| H-SIL | High-grade squamous intraepithelial lesion |
| HOTAIR | HOX transcript antisense RNA |
| HPV | Human Papilloma Virus |
| HPV E5 | Human Papilloma Virus E5 oncoprotein |
| HPV E6 | Human Papilloma Virus E6 oncoprotein |
| HPV E7 | Human Papilloma Virus E7 oncoprotein |
| Hr-HPV | High-risk HPV |
| IRE1 | Inositol-requiring enzyme 1 |
| KGFR | Keratinocyte growth factor receptor |
| LAMP1 | Lysosomal-associated membrane protein 1 |
| LC3-II | Proteins 1A/1B light chain 3B |
| L-SIL | Low-grade squamous intraepithelial lesion |
| LncRNAs | Long noncoding RNAs |
| Lr-HPV | Low risk HPV |
| Lmnb1 | Lamin-B1 |
| MCF7 | Michigan Cancer Foundation-7 cell line |
| MG132 | Potent, reversible, and cell-permeable proteasome inhibitors |
| MIR-G-1 | GRSF1-RNA immunoprecipitation RIP-deep sequencing |
| mRNA | Messenger RNA |
| mTOR | Mammalian target of rapamycin kinase |
| nc-RNAs | Non-coding RNAs |
| PLD1 | Phospholipase D |
| PERK | Eukaryotic translation initiation factor 2-alpha kinase 3 |
| PI3K | Phosphoinositide 3-kinase |
| PIK3C3 | Phosphatidylinositol 3-kinase catalytic subunit type 3 |
| PTEN | Phosphatase and tensin homolog |
| P53 | p53 protein |
| pRb | Retinoblastoma protein |
| PSAP | Prosaposin |
| RHEB | Ras homolog enriched in brain |
| Rictor | Rapamycin-insensitive companion of mammalian target of rapamycin |
| RNAi | RNA interference |
| ROS | Receptor tyrosine kinase |
| RPS6KB2 | Ribosomal protein S6 kinase beta-2 |
| RXRA | Transcription factor retinoic X receptor alpha |
| siRNAs | Small interference RNAs |
| SNPs | Single-nucleotide polymorphisms |
| SQSTM1 | Sequestosome-1 |
| STX12 | Syntaxin-12 |
| TAP1 | Transporter associated with antigen processing 1 |
| TCA | Tricarboxylic acid cycle |
| TCGA | The Cancer Genome Atlas |
| THBS2 | Thrombospondin 2 |
| THP | Tetrahydropalmatine |
| TIMP2 | Tissue inhibitor of metalloproteinases 2 |
| TMED5 | Transmembrane emp24 domain-containing protein 5 |
| TSC1, TSC2 | Tuberous sclerosis complex 1, 2 |
| TPP1 | Tripeptidyl peptidase I |
| UBC9 | SUMO-conjugating enzyme UBC9 |
| ULK1 | Like autophagy activating kinase |
| UPR | Unfolded protein response |
| VEGFA | Vascular endothelial growth factor A |
| Wnt7B | Protein Wnt-7b |
References
- Levine, B.; Kroemer, G. Biological functions of autophagy genes: A disease perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariosa, A.R.; Lahiri, V.; Lei, Y.; Yang, Y.; Yin, Z.; Zhang, Z.; Klionsky, D.J. A perspective on the role of autophagy in cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166262. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Rivera, A.K.; Cruz-Gregorio, A.; Briones-Herrera, A.; Pedraza-Chaverri, J. Regulation of autophagy by high- and low-risk human papillomaviruses. Rev. Med. Virol. 2021, 31, e2169. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Chandravati. Autophagy in cervical cancer: An emerging therapeutic target. Asian Pac. J. Cancer Prev. 2012, 13, 4867–4871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lum, J.J.; Bauer, D.E.; Kong, M.; Harris, M.H.; Li, C.; Lindsten, T.; Thompson, C.B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005, 120, 237–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryter, S.W.; Bhatia, D.; Choi, M.E. Autophagy: A lysosome-dependent process with implications in cellular redox homeostasis and human disease. Antioxid. Redox Signal. 2019, 30, 138–159. [Google Scholar] [CrossRef]
- Crighton, D.; Wilkinson, S.; O’Prey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 126, 121–134. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.A.; Park, M.N.; Rahman, M.H.; Rashid, M.M.; Islam, R.; Uddin, M.J.; Hannan, M.A.; Kim, B. p53 Modulation of autophagy signaling in cancer therapies: Perspectives mechanism and therapeutic targets. Front. Cell Dev. Biol. 2022, 10, 761080. [Google Scholar] [CrossRef]
- Tur, M.K.; Daramola, A.K.; Gattenlöhner, S.; Herling, M.; Chetty, S.; Barth, S. Restoration of DAP kinase tumor suppressor function: A therapeutic strategy to selectively induce apoptosis in cancer cells using immunokinase fusion proteins. Biomedicines 2017, 5, 59. [Google Scholar] [CrossRef] [Green Version]
- Mei, Y.; Glover, K.; Su, M.; Sinha, S.C. Conformational flexibility of BECN1: Essential to its key role in autophagy and beyond. Protein Sci. 2016, 25, 1767–1785. [Google Scholar] [CrossRef] [Green Version]
- Loopik, D.L.; Bentley, H.A.; Eijgenraam, M.N.; IntHout, J.; Bekkers, R.L.M.; Bentley, J.R. The natural history of cervical intraepithelial neoplasia grades 1, 2, and 3: A systematic review and meta-analysis. J. Low. Genit. Tract Dis. 2021, 25, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, S.R.; Le, T.; Lockhart, A.; Sanusi, A.; Dal Santo, L.; Davis, M.; McKinney, D.A.; Brown, M.; Poole, C.; Willame, C.; et al. Patterns of persistent HPV infection after treatment for cervical intraepithelial neoplasia (CIN): A systematic review. Int. J. Cancer 2017, 141, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Pirš, B.; Škof, E.; Smrkolj, V.; Smrkolj, Š. Overview of immune checkpoint inhibitors in gynecological cancer treatment. Cancers 2022, 14, 631. [Google Scholar] [CrossRef] [PubMed]
- Gheit, T. Mucosal and cutaneous human papillomavirus infections and cancer biology. Front. Oncol. 2019, 9, 355. [Google Scholar] [CrossRef] [Green Version]
- Haręża, D.A.; Wilczyński, J.R.; Paradowska, E. Human papillomaviruses as infectious agents in gynecological cancers. oncogenic properties of viral proteins. Int. J. Mol. Sci. 2022, 23, 1818. [Google Scholar] [CrossRef]
- Estêvão, D.; Costa, N.R.; Gil da Costa, R.M.; Medeiros, R. Hallmarks of HPV carcinogenesis: The role of E6, E7 and E5 oncoproteins in cellular malignancy. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 153–162. [Google Scholar] [CrossRef]
- Jose, L.; Gilson, T.; Androphy, E.J.; DeSmet, M. Regulation of the human papillomavirus lifecyle through post-translational modifications of the viral E2 protein. Pathogens 2021, 10, 793. [Google Scholar] [CrossRef]
- Da Silva, M.L.R.; De Albuquerque, B.H.D.R.; Allyrio, T.A.M.F.; De Almeida, V.D.; Cobucci, R.N.O.; Bezerra, F.L.; Andrade, V.S.; Lanza, D.C.F.; De Azevedo, J.C.V.; De Araújo, J.M.G.; et al. The role of HPV-induced epigenetic changes in cervical carcinogenesis (Review). Biomed. Rep. 2021, 15, 60. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, J.; Zhu, X.; Yuan, H. MiR-126 reverses drug resistance to TRAIL through inhibiting the expression of c-FLIP in cervical cancer. Gene 2017, 627, 420–427. [Google Scholar] [CrossRef]
- Vonsky, M.; Shabaeva, M.; Runov, A.; Lebedeva, N.; Chowdhury, S.; Palefsky, J.M.; Isaguliants, M. Carcinogenesis associated with human papillomavirus infection. mechanisms and potential for immunotherapy. Biochemistry 2019, 84, 782–799. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, M.; Li, X.; Yin, S.; Wang, B. An overview of novel agents for cervical cancer treatment by inducing apoptosis: Emerging drugs ongoing clinical trials and preclinical studies. Front. Med. 2021, 8, 682366. [Google Scholar] [CrossRef] [PubMed]
- Lagunas-Martínez, A.; Madrid-Marina, V.; Gariglio, P. Modulation of apoptosis by early human papillomavirus proteins in cervical cancer. Biochim. Biophys. Acta 2010, 1805, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Ravanan, P.; Srikumar, I.F.; Talwar, P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017, 188, 53–67. [Google Scholar] [CrossRef]
- Ambrosio, S.; Majello, B. Autophagy roles in genome maintenance. Cancers 2020, 12, 1793. [Google Scholar] [CrossRef]
- Bata, N.; Cosford, N.D.P. Cell survival and cell death at the intersection of autophagy and apoptosis: Implications for current and future cancer therapeutics. ACS Pharmacol. Transl. Sci. 2021, 4, 1728–1746. [Google Scholar] [CrossRef]
- Ziegler, D.V.; Huber, K.; Fajas, L. The intricate interplay between cell cycle regulators and autophagy in cancer. Cancers 2021, 14, 153. [Google Scholar] [CrossRef]
- Raj, S.; Chandel, V.; Kumar, A.; Kesari, K.K.; Asthana, S.; Ruokolainen, J.; Kamal, M.A.; Kumar, D. Molecular mechanisms of interplay between autophagy and metabolism in cancer. Life Sci. 2020, 259, 118184. [Google Scholar] [CrossRef]
- Jiang, M.; Ju, M.; Bu, W.; Chen, K.; Li, L.; Li, M.; Chen, X.; Gu, H. HPV infection downregulates the expression of autophagy-related genes in condylomata acuminata. Dermatology 2019, 235, 418–425. [Google Scholar] [CrossRef]
- Zhang, B.; Song, Y.; Sun, S.; Han, R.; Hua, C.; van der Veen, S.; Cheng, H. Human papillomavirus 11 early protein E6 activates autophagy by repressing AKT/mTOR and Erk/mTOR. J. Virol. 2019, 93, e00172-19. [Google Scholar] [CrossRef] [Green Version]
- Mattoscio, D.; Casadio, C.; Miccolo, C.; Maffini, F.; Raimondi, A.; Tacchetti, C.; Gheit, T.; Tagliabue, M.; Galimberti, V.E.; De Lorenzi, F.; et al. Autophagy regulates UBC9 levels during viral mediated tumorigenesis. PLoS Pathog. 2017, 13, e1006262. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Tan, J.; Miao, Y.; Li, M.; Zhang, Q. Crosstalk of autophagy and apoptosis: Involvement of the dual role of autophagy under ER stress. J. Cell Physiol. 2017, 232, 2977–2984. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Wu, Y.S.; Li, D.Y.; Tang, J.X.; Liu, H.F. Autophagy in viral infection and pathogenesis. Front. Cell Dev. Biol. 2021, 9, 766142. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Yang, G.F.; Huang, Y.H.; Huang, Q.W.; Gao, J.; Zhao, X.D.; Huang, L.M.; Chen, H.L. Reduced expression of autophagy markers correlates with high-risk human papillomavirus infection in human cervical squamous cell carcinoma. Oncol. Lett. 2014, 8, 1492–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Gong, Z.; Zhang, L.; Zhao, C.; Zhao, X.; Gu, X.; Chen, H. Autophagy knocked down by high-risk HPV infection and uterine cervical carcinogenesis. Int. J. Clin. Exp. Med. 2015, 8, 10304–10314. [Google Scholar] [PubMed]
- Griffin, L.M.; Cicchini, L.; Pyeon, D. Human papillomavirus infection is inhibited by host autophagy in primary human keratinocytes. Virology 2013, 437, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surviladze, Z.; Sterk, R.T.; DeHaro, S.A.; Ozbun, M.A. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J. Virol. 2013, 87, 2508–2517. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.J.; Wu, L.X.; Wang, W.; Ye, Y.Y.; Yang, J.; Chen, H.; Yang, Q.F.; Zhang, X.; Wang, B.; Chen, W.X. Nucleotide variation in ATG4A and susceptibility to cervical cancer in Southwestern Chinese women. Oncol. Lett. 2018, 15, 2992–3000. [Google Scholar] [CrossRef]
- Ilahi, N.E.; Bhatti, A. Impact of HPV E5 on viral life cycle via EGFR signaling. Microb. Pathog. 2020, 139, 103923. [Google Scholar] [CrossRef]
- Bergner, S.; Halec, G.; Schmitt, M.; Aubin, F.; Alonso, A.; Auvinen, E. Individual and complementary effects of human papillomavirus oncogenes on epithelial cell proliferation and differentiation. Cells Tissues Organs 2016, 201, 97–108. [Google Scholar] [CrossRef]
- Belleudi, F.; Nanni, M.; Raffa, S.; Torrisi, M.R. HPV16 E5 deregulates the autophagic process in human keratinocytes. Oncotarget 2015, 6, 9370–9386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roperto, S.; Russo, V.; De Falco, F.; Urraro, C.; Maiolino, P.; Del Piero, F.; Roperto, F. Bovine papillomavirus E5 oncoprotein expression and its association with an interactor network in aggresome-autophagy pathway. Vet. Microbiol. 2019, 233, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Lagunas-Martínez, A.; García-Villa, E.; Arellano-Gaytán, M.; Contreras-Ochoa, C.O.; Dimas-González, J.; López-Arellano, M.E.; Madrid-Marina, V.; Gariglio, P. MG132 plus apoptosis antigen-1 (APO-1) antibody cooperate to restore p53 activity inducing autophagy and p53-dependent apoptosis in HPV16 E6-expressing keratinocytes. Apoptosis 2017, 22, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.C.; Hung, Y.C.; Lin, T.Y.; Chang, H.W.; Chiang, I.P.; Chen, Y.Y.; Chow, K.C. Human papillomavirus infection and expression of ATPase family AAA domain containing 3A, a novel anti-autophagy factor, in uterine cervical cancer. Int. J. Mol. Med. 2011, 28, 689–696. [Google Scholar] [CrossRef]
- Lang, L.; Loveless, R.; Teng, Y. Emerging links between control of mitochondrial protein ATAD3A and cancer. Int. J. Mol. Sci. 2020, 21, 7917. [Google Scholar] [CrossRef]
- Hanning, J.E.; Saini, H.K.; Murray, M.J.; Caffarel, M.M.; van Dongen, S.; Ward, D.; Barker, E.M.; Scarpini, C.G.; Groves, I.J.; Stanley, M.A.; et al. Depletion of HPV16 early genes induces autophagy and senescence in a cervical carcinogenesis model, regardless of viral physical state. J. Pathol. 2013, 231, 354–366. [Google Scholar] [CrossRef]
- Tingting, C.; Shizhou, Y.; Songfa, Z.; Junfen, X.; Weiguo, L.; Xiaodong, C.; Xing, X. Human papillomavirus 16E6/E7 activates autophagy via Atg9B and LAMP1 in cervical cancer cells. Cancer Med. 2019, 8, 4404–4416. [Google Scholar] [CrossRef]
- Akgül, B.; Kirschberg, M.; Storey, A.; Hufbauer, M. Human papillomavirus type 8 oncoproteins E6 and E7 cooperate in downregulation of the cellular checkpoint kinase-1. Int. J. Cancer 2019, 145, 797–806. [Google Scholar] [CrossRef]
- Conrady, M.C.; Suarez, I.; Gogl, G.; Frecot, D.I.; Bonhoure, A.; Kostmann, C.; Cousi-do-Siah, A.; Mitschler, A.; Lim, J.; Masson, M.; et al. Structure of high-risk papillomavirus 31 E6 oncogenic protein and characterization of E6/E6AP/p53 complex formation. J. Virol. 2020, 95, e00730-20. [Google Scholar] [CrossRef]
- Vliet-Gregg, P.A.; Robinson, K.L.; Levan, J.; Matsumoto, L.R.; Katzenellenbogen, R.A. NFX1-123 is highly expressed in cervical cancer and increases growth and te-lomerase activity in HPV 16E6 expressing cells. Cancer Lett. 2019, 449, 106–113. [Google Scholar] [CrossRef]
- Graham, S.V.; Faizo, A.A.A. Control of human papillomavirus gene expression by alternative splicing. Virus Res. 2017, 231, 83–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales-Garcia, V.; Contreras-Paredes, A.; Martinez-Abundis, E.; Gomez-Crisostomo, N.P.; Lizano, M.; Hernandez-Landero, F.; de la Cruz-Hernandez, E. The high-risk HPV E6 proteins modify the activity of the eIF4E protein via the MEK/ERK and AKT/PKB pathways. FEBS Open Bio. 2020, 10, 2541–2552. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Nor Rashid, N.; Mohamad Razif, M.F.; Othman, S. Proteasomal deg-radation of p130 facilitate cell cycle deregulation and impairment of cellular differen-tiation in high-risk Human Papillomavirus 16 and 18 E7 transfected cells. Mol. Biol. Rep. 2021, 48, 5121–5133. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhao, Y.; Li, Q.; Ci, X.; Ye, X.; Chen, G.; Tu, Q.; Feng, W.; Jiang, P.; Zhu, S.; et al. Sustained expression of HPV16 E7 oncopro-tein promotes p-AKT (Ser473)/p-Src (Tyr527) signalling to drive precancerous lesions to invasive cervical cancer. Carcinogenesis 2022, 43, bgac010. [Google Scholar] [CrossRef]
- Feng, Q.; Wang, J.; Cui, N.; Liu, X.; Wang, H. Autophagy-related long non-coding RNA signature for potential prognostic biomarkers of patients with cervical cancer: A study based on public databases. Ann. Transl. Med. 2021, 9, 1668. [Google Scholar] [CrossRef]
- Wan, G.; Xie, W.; Liu, Z.; Xu, W.; Lao, Y.; Huang, N.; Cui, K.; Liao, M.; He, J.; Jiang, Y.; et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy 2014, 10, 70–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, N.; Wu, J.; Qiu, W.; Lyu, Q.; He, J.; Xie, W.; Xu, N.; Zhang, Y. MiR-15a and miR-16 induce autophagy and enhance chemosensitivity of Camptothecin. Cancer Biol. Ther. 2015, 16, 941–948. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Chen, G.; Hu, M.; Huang, J.; Li, B.; Zhou, L.; Hong, L. Has-miR-30a regulates autophagic activity in cervical cancer upon hydroxycamptothecin exposure. Biomed. Pharmacother. 2015, 75, 67–74. [Google Scholar] [CrossRef]
- Wu, Y.; Ni, Z.; Yan, X.; Dai, X.; Hu, C.; Zheng, Y.; He, F.; Lian, J. Targeting the MIR34C-5p-ATG4B-autophagy axis enhances the sensitivity of cervical cancer cells to pirarubicin. Autophagy 2016, 12, 1105–1117. [Google Scholar] [CrossRef] [Green Version]
- Peralta-Zaragoza, O.; Deas, J.; Meneses-Acosta, A.; De-la-O-Gómez, F.; Fernández-Tilapa, G.; Gómez-Cerón, C.; Benítez-Boijseauneau, O.; Burguete-García, A.; Torres-Poveda, K.; Bermúdez-Morales, V.H.; et al. Relevance of miR-21 in regulation of tumor suppressor gene PTEN in human cervical cancer cells. BMC Cancer 2016, 16, 215–232. [Google Scholar] [CrossRef] [Green Version]
- Fang, W.; Shu, S.; Yongmei, L.; Endong, Z.; Lirong, Y.; Bei, S. miR-224-3p inhibits autophagy in cervical cancer cells by targeting FIP200. Sci. Rep. 2016, 6, 33229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, R.; Yang, Z.; Xu, G.; Yu, S. miR-338 modulates proliferation and autophagy by PI3K/AKT/mTOR signaling pathway in cervical cancer. Biomed. Pharmacother. 2018, 105, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, Z.; Yang, X.; Li, T.; Liu, M.; Tang, H. Corrigendum to “miR-346 functions as a pro-survival factor under ER stress by activating mitophagy”. Cancer Lett. 2018, 413, 69–81, Erratum in Cancer Lett. 2020, 493, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Zhou, C.; Han, S.; Hou, X.; Kang, S.; Zhang, Y. MicroRNA-378 enhances migration and invasion in cervical cancer by directly targeting autophagy-related protein 12. Mol. Med. Rep. 2018, 17, 6319–6326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Dong, J.; Luo, R.; Zhou, X.; Wang, J.; Chen, F. MicroRNA-20a regulates cell proliferation, apoptosis and autophagy by targeting thrombospondin 2 in cervical cancer. Eur. J. Pharmacol. 2019, 844, 102–109. [Google Scholar] [CrossRef]
- Li, N.; Guo, X.; Liu, L.; Wang, L.; Cheng, R. Molecular mechanism of miR-204 regulates proliferation, apoptosis and autophagy of cervical cancer cells by targeting ATF2. Artif Cells Nanomed Biotechnol. 2019, 47, 2529–2535. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.H.; Du, X.; Lin, H.; Wang, P.C.; Li, M. Paclitaxel inhibits the progression of cervical cancer by inhibiting autophagy via lncRNARP11-381N20.2. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3010–3017. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, M.; Huang, Y.; Zhang, J.; Yin, L. Promotion of cell autophagy and apoptosis in cervical cancer by inhibition of long noncoding RNA LINC00511 via transcription factor RXRA-regulated PLD1. J. Cell Physiol. 2020, 235, 6592–6604. [Google Scholar] [CrossRef]
- Guo, J.; Chen, M.; Ai, G.; Mao, W.; Li, H.; Zhou, J. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed. Pharmacother. 2019, 115, 108957. [Google Scholar] [CrossRef]
- Revathidevi, S.; Murugan, A.K.; Nakaoka, H.; Inoue, I.; Munirajan, A.K. APOBEC: A molecular driver in cervical cancer pathogenesis. Cancer Lett. 2021, 496, 104–116. [Google Scholar] [CrossRef]
- Kari, I.; Syrjänen, S.; Johansson, B.; Peri, P.; He, B.; Roizman, B.; Hukkanen, V. Antisense RNA directed to the human papillomavirus type 16 E7 mRNA from herpes simplex virus type 1 derived vectors is expressed in CaSki cells and downregulates E7 mRNA. Virol. J. 2007, 4, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisarska, J.; Baldy-Chudzik, K. MicroRNA-based fingerprinting of cervical lesions and cancer. J. Clin. Med. 2020, 9, 3668. [Google Scholar] [CrossRef] [PubMed]
- Salazar-León, J.; Reyes-Román, F.; Meneses-Acosta, A.; Merchant, H.; Lagunas-Martínez, A.; Meda-Monzón, E.; Pita-López, M.L.; Gómez-Cerón, C.; Bermúdez-Morales, V.H.; Madrid-Marina, V.; et al. Silencing of HPV16 E6 and E7 oncogenic activities by small interference RNA induces autophagy and apoptosis in human cervical cancer cells. J. Nucleic Acids Investig. 2011, 2, 59–69. [Google Scholar] [CrossRef]
- Cargnello, M.; Tcherkezian, J.; Roux, P.P. The expanding role of mTOR in cancer cell growth and proliferation. Mutagenesis 2015, 30, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korkmaz, G.; le Sage, C.; Tekirdag, K.A.; Agami, R.; Gozuacik, D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 2012, 8, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Shan, C.; Chen, X.; Cai, H.; Hao, X.; Li, J.; Zhang, Y.; Gao, J.V.; Zhou, Z.; Li, X.; Liu, C.; et al. The emerging roles of autophagy-related microRNAs in cancer. Int. J. Biol. Sci. 2021, 17, 134–150. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fu, H.; Sun, F.; Zhang, H.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008, 36, 5391–5404. [Google Scholar] [CrossRef]
- Wei, Y.; Li, C.; Zhang, Y.; He, H.; Zhang, G.; Hao, X.; Liu, H.; Wang, H.; Tian, W. Hydroxycamptothecin mediates antiproliferative effects through apoptosis and autophagy in A549 cells. Oncol. Lett. 2018, 15, 6322–6328. [Google Scholar] [CrossRef]
- Zhao, S.; Yao, D.; Chen, J.; Ding, N.; Ren, F. MiR-20a promotes cervical cancer proliferation and metastasis in vitro and in vivo. PLoS ONE 2015, 10, e0120905. [Google Scholar] [CrossRef]
- Cui, X.; Wang, X.; Zhou, X.; Jia, J.; Chen, H.; Zhao, W. miR-106a Regulates cell prolif-eration and autophagy by targeting LKB1 in HPV-16-associated cervical cancer. Mol. Cancer Res. 2020, 18, 1129–1141. [Google Scholar] [CrossRef]
- Wang, F.; Shan, S.; Huo, Y.; Xie, Z.; Fang, Y.; Qi, Z.; Chen, F.; Li, Y.; Sun, B. MiR-155-5p in-hibits PDK1 and promotes autophagy via the mTOR pathway in cervical cancer. Int. J. Biochem. Cell Biol. 2018, 99, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lu, D.; Zheng, X.; Xu, J.; Li, Z.; Deng, L.; Hu, Y. Dysregulation of autopha-gy-associated microRNAs in condyloma acuminatum. Infect. Genet. Evol. 2021, 93, 104878. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.H.; Cui, Y.H.; Wang, T.; Luo, Y. Long non-coding RNA HOTAIR in cervical cancer: Molecular marker, mechanistic insight, and therapeutic target. Adv. Clin. Chem. 2020, 97, 117–140. [Google Scholar] [CrossRef] [PubMed]
 ) indicate inhibition and black arrows (
) indicate inhibition and black arrows (  ) indicate activation. Red arrows (
) indicate activation. Red arrows (  ) represent increase in mRNA.
) represent increase in mRNA.
 ) indicate inhibition and black arrows (
) indicate inhibition and black arrows (  ) indicate activation. Red arrows (
) indicate activation. Red arrows (  ) represent increase in mRNA.
) represent increase in mRNA.
 ) indicate inhibition black arrows (
) indicate inhibition black arrows (  ) indicate activation. Red arrows represent increase (
) indicate activation. Red arrows represent increase (  ) or decrease (
) or decrease (  ) in mRNA or cellular proteins, respectively. See the text for more details.
) in mRNA or cellular proteins, respectively. See the text for more details.
 ) indicate inhibition black arrows (
) indicate inhibition black arrows (  ) indicate activation. Red arrows represent increase (
) indicate activation. Red arrows represent increase (  ) or decrease (
) or decrease (  ) in mRNA or cellular proteins, respectively. See the text for more details.
) in mRNA or cellular proteins, respectively. See the text for more details.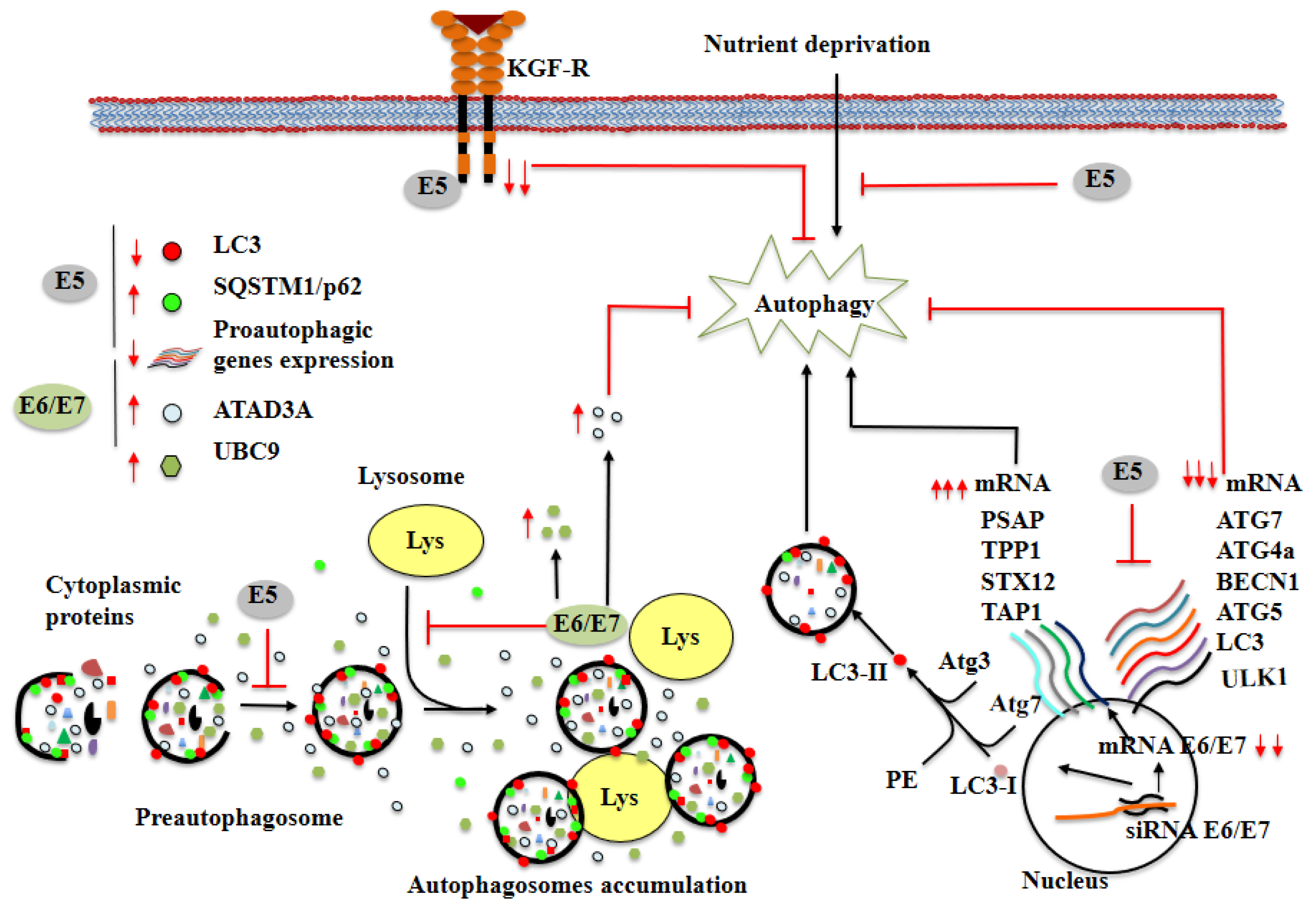
| NcRNA | Target Genes | Biologic Effects | Reference |
|---|---|---|---|
| miR-155 | RHEB, Rictor, RPS6KB2 | Hypoxia-induced miR155 induces autophagy. Knocking down endogenous miR155 alleviates hypoxia-induced autophagy. The members of the mTOR pathway, RHEB, RICTOR, and RPS6KB2; are direct targets of miR155. | [56] |
| miR-15a/miR-16 | Rictor | MiR-15a and miR-16 are potent inducers of autophagy. Rictor, a component of mTORC2 complex, is directly targeted by miR-15a/miR-16. | [57] |
| miR-30a | LC3II, Beclin-1 | The decreased expression of miR-30a is involved in HCPT-induced autophagy in HeLa cells. MiR-30a directly target to Beclin-1. | [58] |
| miR-34c-5p | ATG4B | THP triggers downregulation of miR-34c-5p, associated with upregulation of ATG4B and autophagy induction. Overexpression of miR-34c-5p decreases the level of ATG4B and attenuates autophagy, accompanied by enhanced cell death and apoptosis in THP-treated cervical cancer cells. | [59] |
| miR-21 | PTEN | There is an inverse correlation between miR-21 expression and PTEN mRNA level, as well as PTEN protein expression, in cervical cancer cells. Tumor cells exhibit reduced cell proliferation along with autophagy and apoptosis induction. | [60] |
| miR-224-3p | FIP200 | MiR-224-3p regulates autophagy in cervical cancer tissues and cell lines. The overexpression of miR-224-3p inhibits autophagy in HPV-infected cells, while knocking down endogenous miR-224-3p increases autophagy activity. MiR-224-3p inhibits the expression of the FIP200 gene. | [61] |
| miR-338 | p-mTOR, p-p70S6 | Levels of miR-338 are decreased in cervical cancer tissues and cells, and negatively correlate with the protein level of ATF2. Inhibition of miR-338 expression decreases the expression of p-mTOR and p-p70S6, thus miR-338 decreases autophagy in cervical cancer cells by activating mTOR-signaling pathway. | [62] |
| miR-346 | GSK3B | MiR-346 induced under ER stress modulates autophagic flux in HeLa cells. MiR-346 activates autophagy by interrupting the association between BCL2 and BECN1 in a GSK3B-dependent manner under ER stress. | [63] |
| miR-378 | ATG12 | ATG12 gene is a direct target of miR-378 and its expression is downregulated by miR-378 in cervical cancer cells. Thus miR-378 has a potential role in autophagy. | [64] |
| miR-20a | THBS2 | The inhibition of miR-20a results in reduced proliferation, increased apoptosis and downregulated autophagic activity in cervical cancer cells. Thrombospondin 2 (THBS2) is a direct target of miR-20a. | [65] |
| miR-204 | Bcl-2, LC3I-II, Bax, Caspase-3 | The overexpression of miR-204 reduces protein expression of Bcl-2 and LC3I/II and increases protein expression of Bax and Caspase-3 in cervical cancer cells. MiR-204 regulates the expression of ATF2. | [66] |
| lncRNA RP11-381N20.2 | Atg7, LC3A/B-II | The expression of RP11-381N20.2 is negatively correlated with the treatment time and dose of paclitaxel in cervical cancer cells. Paclitaxel combined with RP11-381N20.2 increases apoptosis of cervical cancer cell. | [67] |
| lncRNA LINC00511 | RXRA | LINC00511 influences the occurrence of cervical cancer by upregulating PLD1 expression via recruiting transcription factor RXRA. SiRNA-LINC00511, siRNA-RXRA or siRNA-PLD1 trigger repression of proliferation and promotion of autophagy and apoptosis of cervical cancer cells. | [68] |
| circRNA 0023404 | miR-5047, VEGFA | Hsa_circ_0023404 knockdown attenuates invasion of cervical cancer cells and lymphatic vessel formation of HDLEC cells. Hsa_circ_0023404 knockdown and miR-5047 downregulate the expression levels of VEGFA. Autophagy-associated genes (Beclin1 and p62) are dysregulated in hsa_circ_0023404 depleted and overexpressed in HeLa cells. | [69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lagunas-Martínez, A.; Madrid-Marina, V.; Gómez-Cerón, C.; Deas, J.; Peralta-Zaragoza, O. The Autophagy Process in Cervical Carcinogenesis: Role of Non-Coding-RNAs, Molecular Mechanisms, and Therapeutic Targets. Cells 2022, 11, 1323. https://doi.org/10.3390/cells11081323
Lagunas-Martínez A, Madrid-Marina V, Gómez-Cerón C, Deas J, Peralta-Zaragoza O. The Autophagy Process in Cervical Carcinogenesis: Role of Non-Coding-RNAs, Molecular Mechanisms, and Therapeutic Targets. Cells. 2022; 11(8):1323. https://doi.org/10.3390/cells11081323
Chicago/Turabian StyleLagunas-Martínez, Alfredo, Vicente Madrid-Marina, Claudia Gómez-Cerón, Jessica Deas, and Oscar Peralta-Zaragoza. 2022. "The Autophagy Process in Cervical Carcinogenesis: Role of Non-Coding-RNAs, Molecular Mechanisms, and Therapeutic Targets" Cells 11, no. 8: 1323. https://doi.org/10.3390/cells11081323
APA StyleLagunas-Martínez, A., Madrid-Marina, V., Gómez-Cerón, C., Deas, J., & Peralta-Zaragoza, O. (2022). The Autophagy Process in Cervical Carcinogenesis: Role of Non-Coding-RNAs, Molecular Mechanisms, and Therapeutic Targets. Cells, 11(8), 1323. https://doi.org/10.3390/cells11081323






