Hypoxia-Induced circRNAs in Human Diseases: From Mechanisms to Potential Applications
Abstract
:1. Introduction
2. circRNAs Function as Novel Regulators in Hypoxia
2.1. Basic Features and Functions of circRNAs
2.2. circRNAs Are New Players for Hypoxic Response
2.3. Emerging Roles of circRNAs in Pathological Responses to Hypoxia
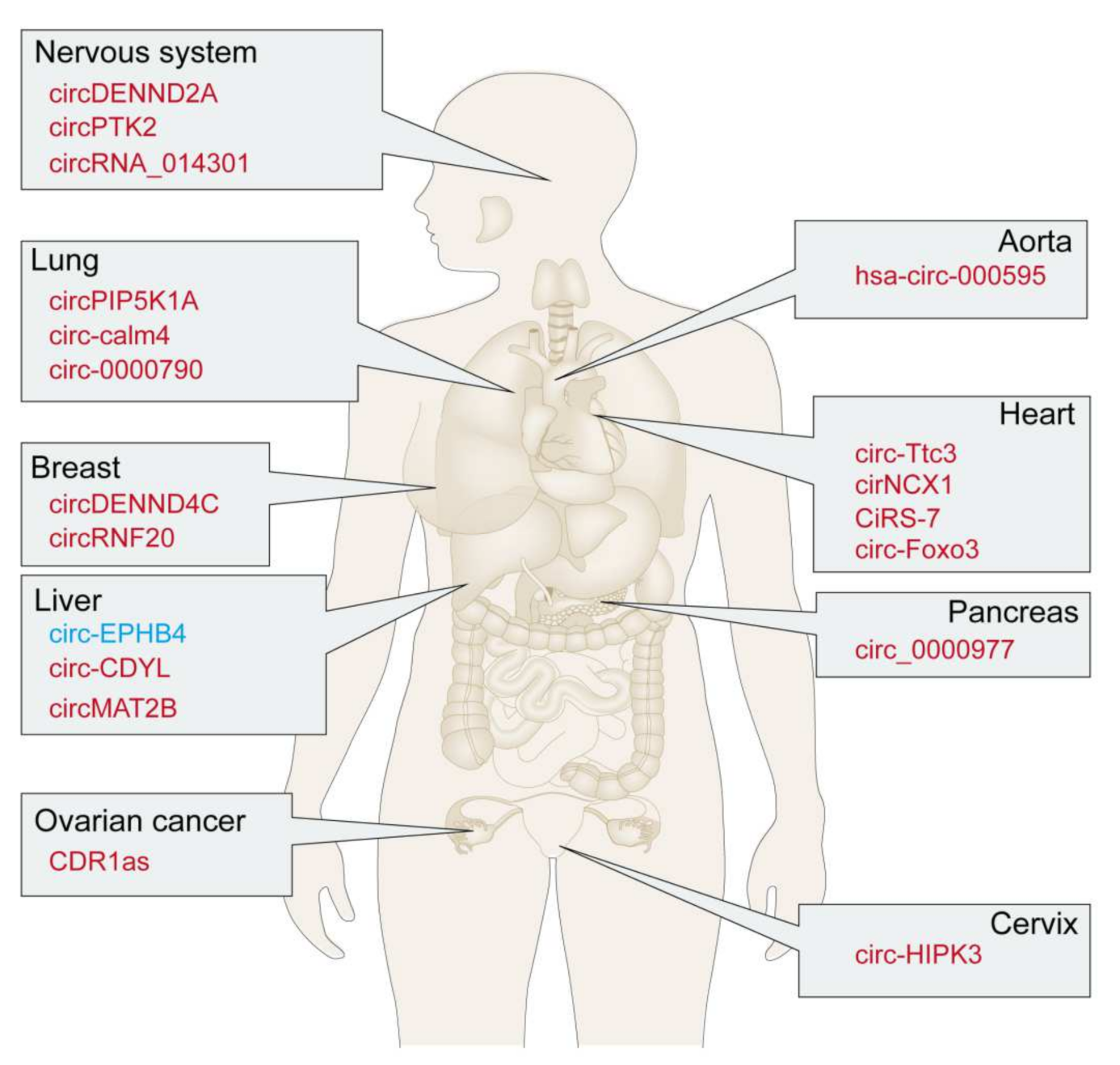
3. Functions of circRNAs in Hypoxic Microenvironments
3.1. Hypoxic circRNAs in Cancer Progression
3.2. Hypoxic circRNAs Regulate Therapeutic Resistance
3.3. Hypoxic circRNAs Regulate Angiogenesis
3.4. Hypoxic circRNAs Influence Energy Metabolism
3.5. Other Regulations by Hypoxic circRNAs
4. Potential Clinical Applications of circRNAs in Human Diseases
5. Concluding Remarks and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev Mol. Cell. Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Leng, K.; Yao, Y.; Kang, P.; Liao, G.; Han, Y.; Shi, G.; Ji, D.; Huang, P.; Zheng, W.; et al. A Circular RNA, Cholangiocarcinoma-Associated Circular RNA 1, Contributes to Cholangiocarcinoma Progression, Induces Angiogenesis, and Disrupts Vascular Endothelial Barriers. Hepatology 2021, 73, 1419–1435. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.G.; Li, J.; Xu, Y.; Li, W.; Fang, S.X.; Zhang, Q.; Xin, H.W.; Ma, Z. Long non-coding RNAs and circular RNAs in tumor angiogenesis: From mechanisms to clinical significance. Mol. Ther. Oncolytics 2021, 22, 336–354. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Li, J.; Hu, J.; Wang, L.; Huang, J.R.; Sethi, G.; Ma, Z. Circular RNAs in cell cycle regulation: Mechanisms to clinical significance. Cell Prolif. 2021, 54, e13143. [Google Scholar] [CrossRef]
- Choudhry, H.; Harris, A.L. Advances in Hypoxia-Inducible Factor Biology. Cell. Metab. 2018, 27, 281–298. [Google Scholar] [CrossRef]
- Shih, J.W.; Kung, H.J. Long non-coding RNA and tumor hypoxia: New players ushered toward an old arena. J. Biomed. Sci. 2017, 24, 53. [Google Scholar] [CrossRef]
- Li, Y.; Patel, S.P.; Roszik, J.; Qin, Y. Hypoxia-Driven Immunosuppressive Metabolites in the Tumor Microenvironment: New Approaches for Combinational Immunotherapy. Front. Immunol. 2018, 9, 1591. [Google Scholar] [CrossRef] [Green Version]
- Rankin, E.B.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Li, S.; Du, W.; Ke, Q.; Cai, J.; Yang, J. Hypoxia regulates CD44 expression via hypoxia-inducible factor-1alpha in human gastric cancer cells. Oncol. Lett. 2017, 13, 967–972. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Wang, L.Z.; Cheng, J.T.; Lam, W.S.T.; Ma, X.; Xiang, X.; Wong, A.L.; Goh, B.C.; Gong, Q.; Sethi, G.; et al. Targeting Hypoxia-Inducible Factor-1-Mediated Metastasis for Cancer Therapy. Antioxid. Redox Signal. 2021, 34, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Zou, D.; Sun, Y.; Dai, Y. Hypoxia-associated circDENND2A promotes glioma aggressiveness by sponging miR-625-5p. Cell. Mol. Biol. Lett. 2019, 24, 24. [Google Scholar] [CrossRef] [PubMed]
- Dang, R.Y.; Liu, F.L.; Li, Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem. Biophys. Res. Commun. 2017, 490, 104–110. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Ma, J.; Zhou, W.; Cao, B.; Zhou, X.; Zhang, H.; Zhao, Q.; Hong, L.; Fan, D. Reciprocal regulations between miRNAs and HIF-1alpha in human cancers. Cell. Mol. Life Sci. 2019, 76, 453–471. [Google Scholar] [CrossRef]
- Wang, W.; Han, Y.; Jo, H.A.; Lee, J.; Song, Y.S. Non-coding RNAs shuttled via exosomes reshape the hypoxic tumor microenvironment. J. Hematol. Oncol. 2020, 13, 67. [Google Scholar] [CrossRef]
- Choudhry, H.; Harris, A.L.; McIntyre, A. The tumour hypoxia induced non-coding transcriptome. Mol. Aspects Med. 2016, 47–48, 35–53. [Google Scholar] [CrossRef]
- Kumar, A.; Deep, G. Exosomes in hypoxia-induced remodeling of the tumor microenvironment. Cancer Lett. 2020, 488, 1–8. [Google Scholar] [CrossRef]
- Hsu, M.T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Li, X.; Zhang, P.; Wang, J.; Zhou, Y.; Chen, M. Circular RNA: An emerging key player in RNA world. Brief Bioinform. 2017, 18, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013, 51, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Filonov, G.S.; Noto, J.J.; Schmidt, C.A.; Hatkevich, T.L.; Wen, Y.; Jaffrey, S.R.; Matera, A.G. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 2015, 21, 1554–1565. [Google Scholar] [CrossRef] [Green Version]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Zhao, G.; Yan, X.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z.; et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018, 19, 218. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell. Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017, 66, 22–37.e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, T.; Nikolopoulou, E.A.; Xu, Q.; Qu, A. Hypoxia inducible factor as a therapeutic target for atherosclerosis. Pharmacol. Ther. 2018, 183, 22–33. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol 2020, 21, 268–283. [Google Scholar] [CrossRef]
- Gunton, J.E. Hypoxia-inducible factors and diabetes. J. Clin. Invest. 2020, 130, 5063–5073. [Google Scholar] [CrossRef]
- Pral, L.P.; Fachi, J.L.; Correa, R.O.; Colonna, M.; Vinolo, M.A.R. Hypoxia and HIF-1 as key regulators of gut microbiota and host interactions. Trends Immunol. 2021, 42, 604–621. [Google Scholar] [CrossRef]
- Gkagkalidis, K.; Kampantais, S.; Dimitriadis, G.; Gourvas, V.; Kapoukranidou, D.; Mironidou-Tzouveleki, M. Expression of HIF-2a in clear-cell renal cell carcinoma independently predicts overall survival. Med. Mol. Morphol. 2020, 53, 229–237. [Google Scholar] [CrossRef]
- Boddy, J.L.; Fox, S.B.; Han, C.; Campo, L.; Turley, H.; Kanga, S.; Malone, P.R.; Harris, A.L. The androgen receptor is significantly associated with vascular endothelial growth factor and hypoxia sensing via hypoxia-inducible factors HIF-1a, HIF-2a, and the prolyl hydroxylases in human prostate cancer. Clin. Cancer Res. 2005, 11, 7658–7663. [Google Scholar] [CrossRef] [Green Version]
- McGettrick, A.F.; O’Neill, L.A.J. The Role of HIF in Immunity and Inflammation. Cell. Metab. 2020, 32, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Luo, Q.; Song, Y.; Yang, F.; Wang, Y.; Jin, M.; Zhang, D. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1alpha regulation. J. Cell. Biochem. 2019, 120, 19019–19030. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Du, B.; Zhan, Y.; Wang, K.; Wang, X.; Chen, B.; Wei, X.; Xiao, J. Antitumor effects of circ-EPHB4 in hepatocellular carcinoma via inhibition of HIF-1alpha. Mol. Carcinog. 2019, 58, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, Y.; Meng, Q.; Cui, L.; Xu, C. Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am. J. Transl. Res. 2019, 11, 6838–6849. [Google Scholar]
- Liang, G.; Liu, Z.; Tan, L.; Su, A.N.; Jiang, W.G.; Gong, C. HIF1alpha-associated circDENND4C Promotes Proliferation of Breast Cancer Cells in Hypoxic Environment. Anticancer Res. 2017, 37, 4337–4343. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Chen, X.; Liang, C.; Ling, Y.; Yang, X.; Ye, X.; Zhang, H.; Yang, P.; Cui, X.; Ren, Y.; et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology 2020, 71, 130–147. [Google Scholar] [CrossRef]
- Qian, W.; Huang, T.; Feng, W. Circular RNA HIPK3 Promotes EMT of Cervical Cancer Through Sponging miR-338-3p to Up-Regulate HIF-1alpha. Cancer Manag. Res. 2020, 12, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Ji, Y. CircC6orf132 Facilitates Proliferation, Migration, Invasion, and Glycolysis of Gastric Cancer Cells Under Hypoxia by Acting on the miR-873-5p/PRKAA1 Axis. Front. Genet. 2021, 12, 636392. [Google Scholar] [CrossRef]
- Xu, L.; Liao, W.L.; Lu, Q.J.; Zhang, P.; Zhu, J.; Jiang, G.N. Hypoxic tumor-derived exosomal circular RNA SETDB1 promotes invasive growth and EMT via the miR-7/Sp1 axis in lung adenocarcinoma. Mol. Ther. Nucleic Acids 2021, 23, 1078–1092. [Google Scholar] [CrossRef]
- Feng, D.; Xu, Y.; Hu, J.; Zhang, S.; Li, M.; Xu, L. A novel circular RNA, hsa-circ-0000211, promotes lung adenocarcinoma migration and invasion through sponging of hsa-miR-622 and modulating HIF1-alpha expression. Biochem. Biophys. Res. Commun. 2020, 521, 395–401. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, F.; Xiong, Y.; Wang, N.; Gu, Y.; Qiu, X. CircZFR functions as a sponge of miR-578 to promote breast cancer progression by regulating HIF1A expression. Cancer Cell. Int. 2020, 20, 400. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wang, X.; Li, C.; Zhang, H.; Liu, Y.; Han, D.; Li, Y.; Li, Z.; Luo, D.; Zhang, N.; et al. CircHIF1A regulated by FUS accelerates triple-negative breast cancer progression by modulating NFIB expression and translocation. Oncogene 2021, 40, 2756–2771. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Mao, M.; Jiang, J.; Zhu, D.; Li, P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019, 12, 3869–3879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Liu, Y.; Gao, R.; Xiu, Z.; Sun, T. Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721 cell proliferation, vasculogenic mimicry, and radioresistance. Cell. Signal. 2019, 60, 122–135. [Google Scholar] [CrossRef]
- Joseph, N.A.; Chiou, S.H.; Lung, Z.; Yang, C.L.; Lin, T.Y.; Chang, H.W.; Sun, H.S.; Gupta, S.K.; Yen, L.; Wang, S.D.; et al. The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J. Hematol. Oncol. 2018, 11, 74. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Yang, W.; Jiang, N.; Shi, J.; Chen, L.; Zhong, G.; Bi, J.; Dong, W.; Wang, Q.; Wang, C.; et al. Hypoxia-elevated circELP3 contributes to bladder cancer progression and cisplatin resistance. Int. J. Biol. Sci. 2019, 15, 441–452. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhao, Y.; Chen, Q.; Zhu, S.; Niu, Y.; Ye, Z.; Hu, P.; Chen, D.; Xu, P.; Chen, J.; et al. Hypoxic exosomal HIF-1alpha-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene 2021, 40, 5505–5517. [Google Scholar] [CrossRef]
- Ou, Z.L.; Luo, Z.; Wei, W.; Liang, S.; Gao, T.L.; Lu, Y.B. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: Role of circ_0000977/miR-153 axis. RNA Biol. 2019, 16, 1592–1603. [Google Scholar] [CrossRef]
- Boeckel, J.N.; Jae, N.; Heumuller, A.W.; Chen, W.; Boon, R.A.; Stellos, K.; Zeiher, A.M.; John, D.; Uchida, S.; Dimmeler, S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ. Res. 2015, 117, 884–890. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Y.; Wang, L.; Ren, Y.X.; Pang, Z.; Liu, Y.; Sun, X.D.; Tu, J.; Zhi, Z.; Qin, Y.; Sun, L.N.; et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1alpha translation. Mol. Cancer 2020, 19, 164. [Google Scholar] [CrossRef]
- Zhou, R.M.; Shi, L.J.; Shan, K.; Sun, Y.N.; Wang, S.S.; Zhang, S.J.; Li, X.M.; Jiang, Q.; Yan, B.; Zhao, C. Circular RNA-ZBTB44 regulates the development of choroidal neovascularization. Theranostics 2020, 10, 3293–3307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Sun, W.; Han, J.; Cheng, S.; Yu, P.; Shen, L.; Fan, M.; Tong, H.; Zhang, H.; Chen, J.; et al. The circular RNA hsa_circ_0007623 acts as a sponge of microRNA-297 and promotes cardiac repair. Biochem. Biophys. Res. Commun. 2020, 523, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.Q.; Kong, P.; Li, C.L.; Sun, H.X.; Li, W.W.; Yu, Y.; Nie, L.; Zhao, L.L.; Miao, S.B.; Li, X.K.; et al. Smooth muscle SIRT1 reprograms endothelial cells to suppress angiogenesis after ischemia. Theranostics 2020, 10, 1197–1212. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yao, M.D.; Li, C.P.; Shan, K.; Yang, H.; Wang, J.J.; Liu, B.; Li, X.M.; Yao, J.; Jiang, Q.; et al. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics 2017, 7, 2863–2877. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, R.; Liu, W.; Wang, Z.; Rong, J.; Long, X.; Liu, Z.; Ge, J.; Shi, B. Exosomal circHIPK3 Released from Hypoxia-Pretreated Cardiomyocytes Regulates Oxidative Damage in Cardiac Microvascular Endothelial Cells via the miR-29a/IGF-1 Pathway. Oxid. Med. Cell. Longev. 2019, 2019, 7954657. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pan, X.; Zhu, D.; Deng, Z.; Jiang, R.; Wang, X. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. Hepatology 2019, 70, 1298–1316. [Google Scholar] [CrossRef]
- Cao, L.; Wang, M.; Dong, Y.; Xu, B.; Chen, J.; Ding, Y.; Qiu, S.; Li, L.; Karamfilova Zaharieva, E.; Zhou, X.; et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1alpha/HK2. Cell. Death Dis. 2020, 11, 145. [Google Scholar] [CrossRef]
- Li, M.; Ding, W.; Tariq, M.A.; Chang, W.; Zhang, X.; Xu, W.; Hou, L.; Wang, Y.; Wang, J. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 2018, 8, 5855–5869. [Google Scholar] [CrossRef]
- Geng, H.H.; Li, R.; Su, Y.M.; Xiao, J.; Pan, M.; Cai, X.X.; Ji, X.P. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS ONE 2016, 11, e0151753. [Google Scholar] [CrossRef]
- Cai, L.; Qi, B.; Wu, X.; Peng, S.; Zhou, G.; Wei, Y.; Xu, J.; Chen, S.; Liu, S. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J. Mol. Cell. Cardiol. 2019, 130, 10–22. [Google Scholar] [CrossRef]
- Zheng, C.; Niu, H.; Li, M.; Zhang, H.; Yang, Z.; Tian, L.; Wu, Z.; Li, D.; Chen, X. Cyclic RNA hsacirc000595 regulates apoptosis of aortic smooth muscle cells. Mol. Med. Rep. 2015, 12, 6656–6662. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, Y.; Qi, J.; Yu, X.; Ren, H.; Zhao, X.; Xin, W.; He, S.; Zheng, X.; Ma, C.; et al. Circ-calm4 Serves as an miR-337-3p Sponge to Regulate Myo10 (Myosin 10) and Promote Pulmonary Artery Smooth Muscle Proliferation. Hypertension 2020, 75, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liang, H.; Meng, X.; Shen, L.; Guan, Z.; Hei, B.; Yu, H.; Qi, S.; Wen, X. mmu_circ_0000790 Is Involved in Pulmonary Vascular Remodeling in Mice with HPH via MicroRNA-374c-Mediated FOXC1. Mol. Ther. Nucleic Acids 2020, 20, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Z.; Gao, J.; Liao, Q. Circular RNA circPTK2 regulates oxygen-glucose deprivation-activated microglia-induced hippocampal neuronal apoptosis via miR-29b-SOCS-1-JAK2/STAT3-IL-1beta signaling. Int. J. Biol. Macromol. 2019, 129, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, Q.; Liu, N.; Zhao, J.; Yu, J.; Tao, S. Circular RNA circ-Ttc3 protects HaCaT cells from hypoxic injury by downregulation of miR-449a. IUBMB Life 2020, 72, 505–514. [Google Scholar] [CrossRef]
- Lu, S.; Yang, X.; Wang, C.; Chen, S.; Lu, S.; Yan, W.; Xiong, K.; Liu, F.; Yan, J. Current status and potential role of circular RNAs in neurological disorders. J. Neurochem. 2019, 150, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- Azzouzi, H.E.; Leptidis, S.; Doevendans, P.A.; De Windt, L.J. HypoxamiRs: Regulators of cardiac hypoxia and energy metabolism. Trends Endocrinol. Metab. 2015, 26, 502–508. [Google Scholar] [CrossRef]
- Aufiero, S.; Reckman, Y.J.; Pinto, Y.M.; Creemers, E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019, 16, 503–514. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factors in physiology and medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhu, M.C.; Kalionis, B.; Wu, J.Z.; Wang, L.L.; Ge, H.Y.; Chen, C.C.; Tang, X.D.; Song, Y.L.; He, H.; et al. Characteristics of circular RNA expression in lung tissues from mice with hypoxiainduced pulmonary hypertension. Int. J. Mol. Med. 2018, 42, 1353–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, L.; An, G.; Wang, S.; Chen, X.; Liu, Y.; Liu, Z.; Ma, Q.; Wang, J. Circular RNA Expression Profiling and Selection of Key Circular RNAs in the Hypothalamus of Heat-Acclimated Rats. Front. Physiol. 2019, 10, 1112. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhao, R.; Huang, L.; Mo, S.; Yu, Z.; Jiang, L.; Qiao, L. Circular RNA Expression in the Brain of a Neonatal Rat Model of Periventricular White Matter Damage. Cell. Physiol. Biochem. 2018, 49, 2264–2276. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, H.; Fan, Z.; Zhao, R.; Xia, Z. Circular RNA expression profiles in neonatal rats following hypoxic-ischemic brain damage. Int. J. Mol. Med. 2019, 43, 1699–1708. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, L.; Yu, B.; Jiao, L.; Han, Z.; Zhao, H.; Li, G.; Ma, Y.; Luo, Y. Circular RNA profiling of neutrophil transcriptome provides insights into asymptomatic Moyamoya disease. Brain Res. 2019, 1719, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Rao, H.; Chen, W.; Luo, X.; Tong, C.; Qi, H. Profiles of circular RNAs in human placenta and their potential roles related to preeclampsia. Biol. Reprod. 2018, 98, 705–712. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Ren, C.; Xiao, Y.; Xia, X.; Fang, X. Expression Profile Analysis of Circular RNAs in Ovarian Endometriosis by Microarray and Bioinformatics. Med Sci Monit 2018, 24, 9240–9250. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, X.; Gao, W.; Hu, H.; Wang, X.; Hao, D. lncRNA/circRNAmiRNAmRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep 2019, 20, 3160–3174. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Hansen, T.B.; Veno, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Xiang, X.; Li, S.; Xie, P.; Gong, Q.; Goh, B.C.; Wang, L. Targeting hypoxia-inducible factor-1, for cancer treatment: Recent advances in developing small-molecule inhibitors from natural compounds. Semin. Cancer Biol. 2022, 80, 379–390. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [Green Version]
- Wong, B.W.; Marsch, E.; Treps, L.; Baes, M.; Carmeliet, P. Endothelial cell metabolism in health and disease: Impact of hypoxia. EMBO J. 2017, 36, 2187–2203. [Google Scholar] [CrossRef] [PubMed]
- Herbert, S.P.; Stainier, D.Y. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat Rev Mol Cell. Biol. 2011, 12, 551–564. [Google Scholar] [CrossRef] [Green Version]
- Cho, W.C.; Jour, G.; Aung, P.P. Role of angiogenesis in melanoma progression: Update on key angiogenic mechanisms and other associated components. Semin. Cancer Biol. 2019, 59, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Ho, Q.T.; Kuo, C.J. Vascular endothelial growth factor: Biology and therapeutic applications. Int. J. Biochem. Cell. Biol. 2007, 39, 1349–1357. [Google Scholar] [CrossRef] [Green Version]
- Bourhis, M.; Palle, J.; Galy-Fauroux, I.; Terme, M. Direct and Indirect Modulation of T Cells by VEGF-A Counteracted by Anti-Angiogenic Treatment. Front. Immunol. 2021, 12, 616837. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Zhang, L.F.; Zhang, H.W.; Hu, S.; Lu, M.H.; Liang, S.; Li, B.; Li, Y.; Li, D.; Wang, E.D.; et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012, 31, 1985–1998. [Google Scholar] [CrossRef] [Green Version]
- Ren, S.; Liu, J.; Feng, Y.; Li, Z.; He, L.; Li, L.; Cao, X.; Wang, Z.; Zhang, Y. Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. J. Exp. Clin. Cancer Res. 2019, 38, 388. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Qiu, J.; Wang, S.; Yang, Y.; Guo, M.; Wang, D.; Luo, Q.; Xu, L. Comprehensive circular RNA profiling identifies CircFAM120A as a new biomarker of hypoxic lung adenocarcinoma. Ann. Transl. Med. 2019, 7, 442. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, J.; Zou, C.; Xie, X.; Wang, Y.; Wang, B.; Zhao, Z.; Tu, J.; Wang, X.; Li, H.; et al. Microarray Expression Profile and Functional Analysis of Circular RNAs in Osteosarcoma. Cell. Physiol. Biochem. 2017, 43, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Di Liddo, A.; de Oliveira Freitas Machado, C.; Fischer, S.; Ebersberger, S.; Heumuller, A.W.; Weigand, J.E.; Muller-McNicoll, M.; Zarnack, K. A combined computational pipeline to detect circular RNAs in human cancer cells under hypoxic stress. J. Mol. Cell. Biol. 2019, 11, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, K.; Tan, S.; Xin, J.; Yuan, Q.; Xu, H.; Xu, X.; Liang, Q.; Christiani, D.C.; Wang, M.; et al. Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Mol. Cancer 2021, 20, 13. [Google Scholar] [CrossRef]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 79. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Boutros, M.; Ahringer, J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 2008, 9, 554–566. [Google Scholar] [CrossRef]
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes. Dev. 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell. 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal. Transduct. Target. Ther. 2021, 6, 185. [Google Scholar] [CrossRef]
- Sanghani, N.S.; Haase, V.H. Hypoxia-Inducible Factor Activators in Renal Anemia: Current Clinical Experience. Adv. Chronic Kidney Dis. 2019, 26, 253–266. [Google Scholar] [CrossRef] [PubMed]


| CircRNA | Interacting Partner | Targets/Pathways | Functions | Diseases/Cells/Organs | Reference |
|---|---|---|---|---|---|
| Cancer progression | |||||
| Circ-CDYL | miR-328-3p | HIF1AN | Promotes stem-like characteristics and tumor growth in vitro and in vivo | HCC | [46] |
| CircPIP5K1A | miR-600 | HIF-1α | Promotes non-small cell lung cancer (NSCLC) proliferation and metastasis in vitro and in vivo | NSCLC | [42] |
| Circ-HIPK3 | miR-338-3p | HIF-1α | Promotes EMT of cervical cancer (CC) in vitro | CC | [47] |
| CircC6orf132 | miR-873-5p | Protein kinase AMP-activated alpha 1 catalytic subunit (PRKAA1) | Promotes gastric cancer proliferation, migration, invasion and glycolysis under hypoxic conditions in vitro and in vivo | Gastric cancer | [48] |
| CircSETDB1 | miR-7 | Specificity protein 1 (Sp1) | Promotes invasive growth and EMT in vitro and in vivo | Lung adenocarcinoma (LUAD) | [49] |
| hsa-circ-0000211 | miR-622 | HIF-1α | Promotes lung adenocarcinoma migration and invasion in vitro | LUAD | [50] |
| CircDENND2A | miR-625-5p | Promotes glioma aggressiveness in vitro | Glioma | [14] | |
| CircDENND4C | Promotes the proliferation of breast cancer cells under hypoxia in vitro | Breast cancer | [45] | ||
| CircZFR | miR-578 | HIF1A | Promotes breast cancer progression in vitro and in vivo | Breast cancer | [51] |
| circHIF1A | NFIB and FUS | Promotes TNBC growth and metastasis in vitro and in vivo | Breast cancer | [52] | |
| Circ-EPHB4 | HIF-1α and PI3K-AKT pathways | Inhibits tumorigenesis, tumor development, and metastasis in vitro and in vivo | HCC | [43] | |
| CDR1as | miR-135b-5p | HIF1AN | Suppresses ovarian cancer progression in vitro | Ovarian cancer | [53] |
| Therapeutic resistance | |||||
| cZNF292 | Wnt/β-catenin pathway | Promotes hypoxic human hepatoma SMMC7721 cell proliferation, vasculogenic mimicry, and radioresistance in vitro and in vivo | Hepatoma | [54] | |
| CirRNA CCDC66 | Increases EMT and drug resistance of LADC cells in vitro | LUAD | [55] | ||
| CircELP3 | Contributes to bladder cancer progression and cisplatin resistance in vitro and in vivo | Bladder cancer | [56] | ||
| CircZNF91 | Sirtuin1 (SIRT1) and HIF-1α | Facilitates glycolysis and gemcitabine chemoresistance of recipient PC cells in vitro and in vivo | Pancreatic cancer | [57] | |
| Circ_0000977 | miR-153 | HIF1 and ADAM10 | Modulates HIF1A-mediated immune escape of PC cells in vitro | Pancreatic cancer | [58] |
| Angiogenesis | |||||
| cZNF292 | Induces tube formation and spheroid sprouting of endothelial cells in vitro | Endothelial cells | [59] | ||
| Circ-Erbin | miR-125a-5p and miR-138-5p | 4E binding protein 1(4EBP-1) | Facilitates the proliferation, migration, and metastasis of colorectal cancer (CRC) in vitro and in vivo | CRC | [60] |
| cZBTB44 | miR-578 | VEGFA/VCAM1 | Induces endothelial cell viability, proliferation, migration, and tube formation in vitro and in vivo | Choroidal neovascularization (CNV) | [61] |
| hsa_circ_0007623 | miR-297 | VEGFA | Promotes cardiac repair after acute myocardial ischemia and protects cardiac function in vitro and in vivo | Heart | [62] |
| cZFP609 | HIF-1α | VEGFA | Inhibits VEGFA expression and endothelial angiogenic functions in vitro and in vivo | Vascular smooth muscle cells (VSMCs) | [63] |
| cZNF609 | miR-615-5p | MEF2A | Decreases endothelial cell migration and tube formation in vitro and in vivo | Vascular dysfunction | [64] |
| hsa_circ_0010729 | miR-186 | HIF-1α | Regulates vascular endothelial cell proliferation and apoptosis in vitro | Human umbilical vein endothelial cells (HUVECs) | [15] |
| CircHIPK3 | miR-29a | IGF-1 | Decrease in oxidative stress-induced CMVECs dysfunction in vitro and in vivo | Cardiac microvascular endothelial cells (CMVECs) | [65] |
| Energy metabolism | |||||
| CircMAT2B | MiR-338-3p | PKM2 | Promotes HCC progression by enhanced glycolysis in vitro and in vivo | HCC | [66] |
| circRNF20 | miR-487a | HIF-1α/HK2 | Promotes the proliferation and aerobic glycolysis in vitro and in vivo | Breast cancer | [67] |
| Other regulation | |||||
| CircNCX1 | miR-133a-3p | Cell death-inducing protein (CDIP1) | Promotes cardiomyocyte apoptosis in vitro and in vivo | Cardiomyocyte apoptosis | [68] |
| Cdr1as | miR-7a | PARP and SP1 | Increases the cardiac infarct size in vitro and in vivo | Myocardial infarction (MI) | [69] |
| Circ-Ttc3 | miR-15b | Arl2 | In cardiomyocytes counteracted hypoxia-induced ATP depletion and apoptotic death in vitro and in vivo | MI | [70] |
| Circ-Foxo3 | ID-1, E2F1, FAK, and HIF1α | Promotes cardiac senescence in vitro and in vivo | Heart | [16] | |
| hsa-circ-000595 | miR-19a | Increases the apoptotic rate of human aortic smooth muscle cells in vitro | Aortic smooth muscle cells | [71] | |
| Circ-calm4 | miR-337-3p | Myo10 (myosin 10) | Promotes pulmonary artery smooth muscle proliferation in vitro and in vivo | Pulmonary hypertension | [72] |
| mmu_circ_0000790 | miR-374c | Forkhead transcription factor 1 (FOXC1) | Induces proliferation and inhibits apoptosis of hypoxic PASMCs in vitro and in vivo | Hypoxic pulmonary hypertension (HPH) | [73] |
| CircPTK2 | miR-29b | SOCS-1-JAK2/STAT3-IL-1β | Regulates oxygen-glucose deprivation-activated microglia-induced hippocampal neuronal apoptosis in vitro and in vivo | Microglia | [74] |
| Circ-Ttc3 | miR-449a | NF-κB and PI3K/AKT pathways | Alleviates hypoxic injury in vitro | HaCaT cells | [75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Q.; Yang, J.; Goh, R.M.W.-J.; You, M.; Wang, L.; Ma, Z. Hypoxia-Induced circRNAs in Human Diseases: From Mechanisms to Potential Applications. Cells 2022, 11, 1381. https://doi.org/10.3390/cells11091381
Huang Q, Yang J, Goh RMW-J, You M, Wang L, Ma Z. Hypoxia-Induced circRNAs in Human Diseases: From Mechanisms to Potential Applications. Cells. 2022; 11(9):1381. https://doi.org/10.3390/cells11091381
Chicago/Turabian StyleHuang, Qi, Juan Yang, Robby Miguel Wen-Jing Goh, Mingliang You, Lingzhi Wang, and Zhaowu Ma. 2022. "Hypoxia-Induced circRNAs in Human Diseases: From Mechanisms to Potential Applications" Cells 11, no. 9: 1381. https://doi.org/10.3390/cells11091381
APA StyleHuang, Q., Yang, J., Goh, R. M. W.-J., You, M., Wang, L., & Ma, Z. (2022). Hypoxia-Induced circRNAs in Human Diseases: From Mechanisms to Potential Applications. Cells, 11(9), 1381. https://doi.org/10.3390/cells11091381







