β-Arrestin2 Is Critically Involved in the Differential Regulation of Phosphosignaling Pathways by Thyrotropin-Releasing Hormone and Taltirelin
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture, siRNA Inhibition and Drug Treatment
2.3. Western Blot Analysis
2.4. Protein Digestion
2.5. Phosphoproteomic Analysis by nLC-MS2
2.6. Data Analysis
3. Results
3.1. GO Enrichment Analysis of Differentially Phosphorylated Proteins
3.2. Alterations in Phosphorylation of Phosphoproteins Involved in GTPase-Mediated Signal Transduction and Protein Phosphorylation
3.2.1. Alterations in Phosphorylation of Phosphoproteins Involved in Ras GTPase-Mediated Signal Transduction Associated with the PI3K/Akt/mTOR Pathway

3.2.2. Alterations in Phosphorylation of Phosphoproteins Involved in Ras GTPase-Mediated Signal Transduction Associated with the Grb2/Sos/Ras/Raf/MEK/ERK Pathway
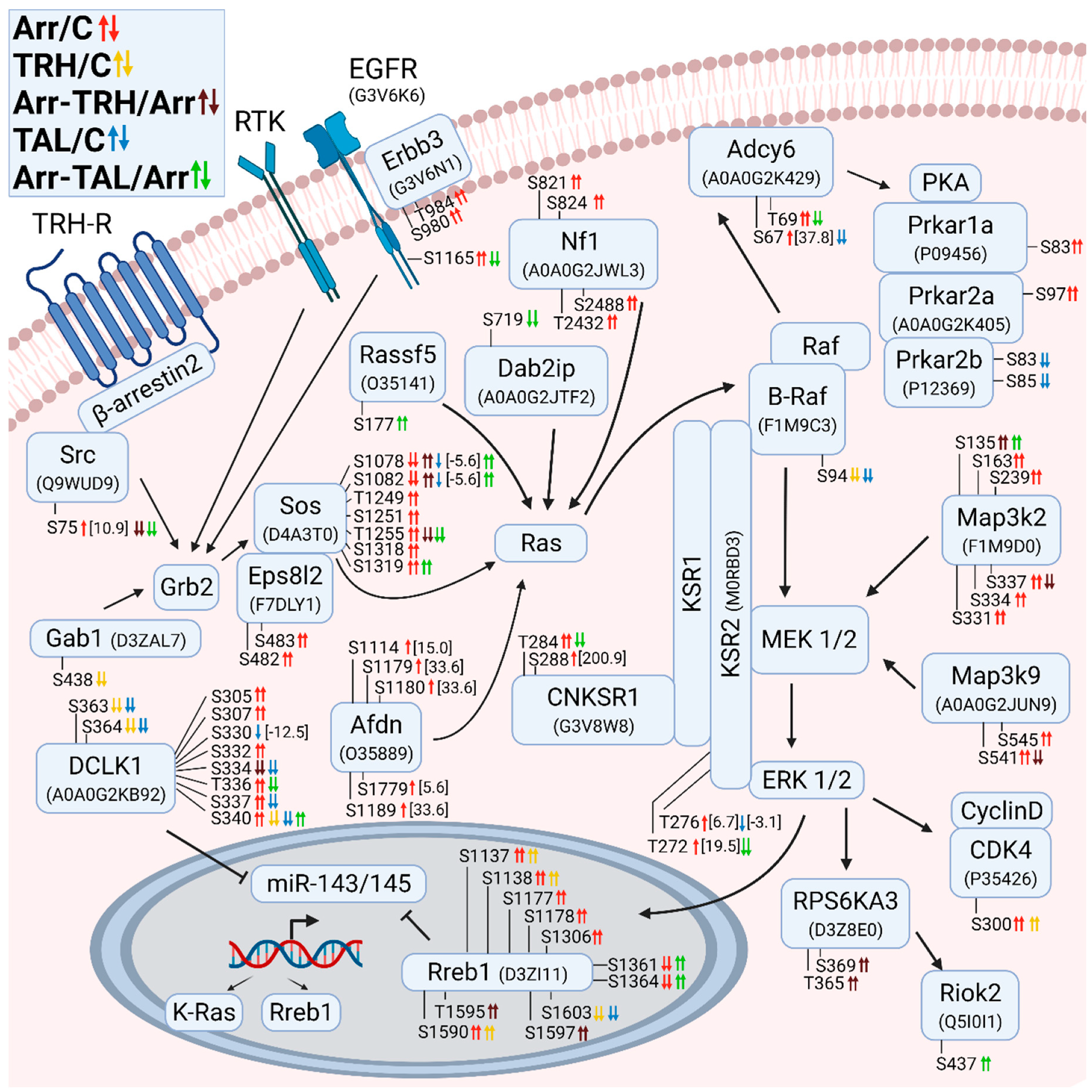
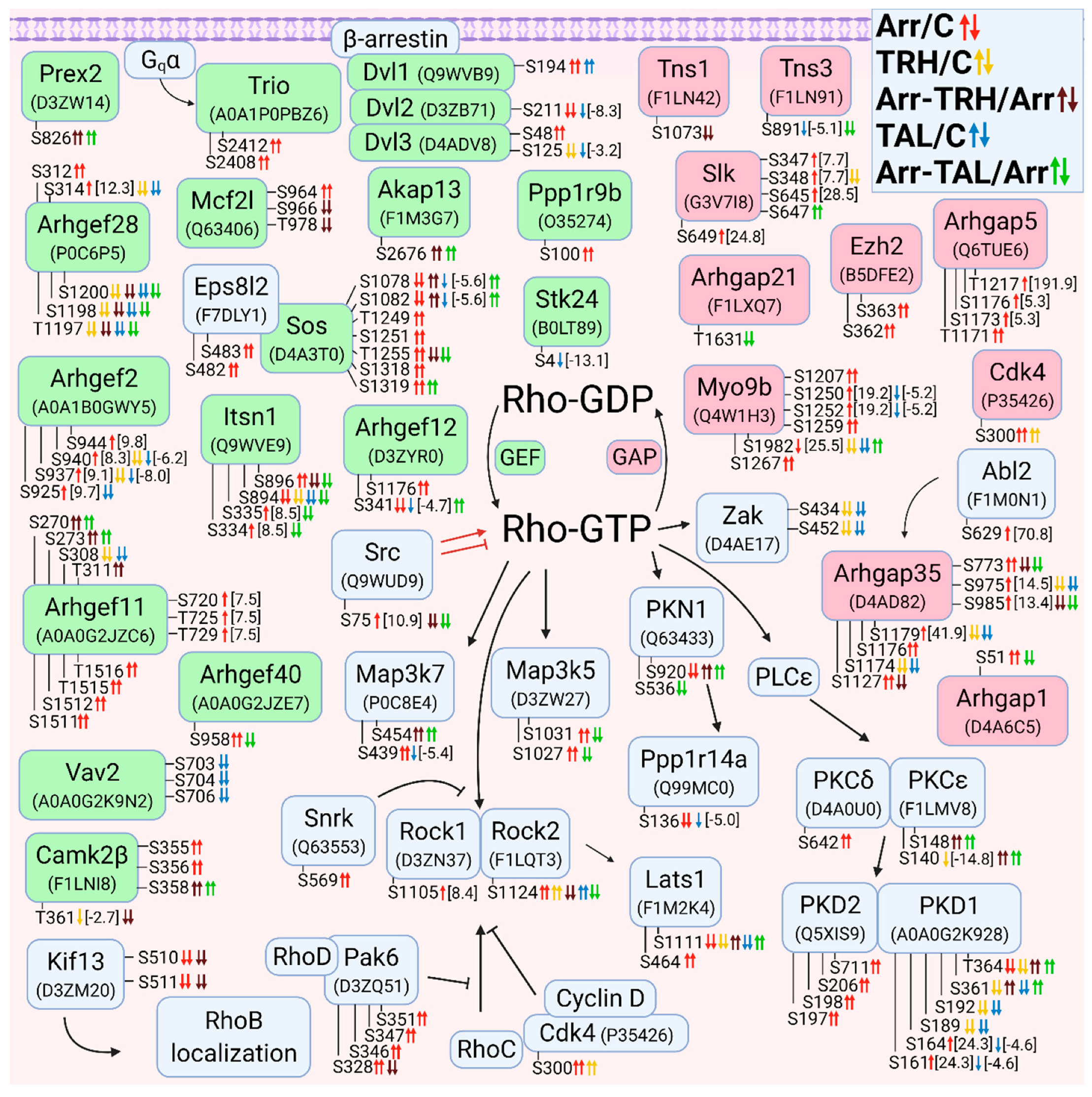
3.2.3. Alterations in Phosphorylation of Phosphoproteins Involved in Rho GTPase-Mediated Signal Transduction
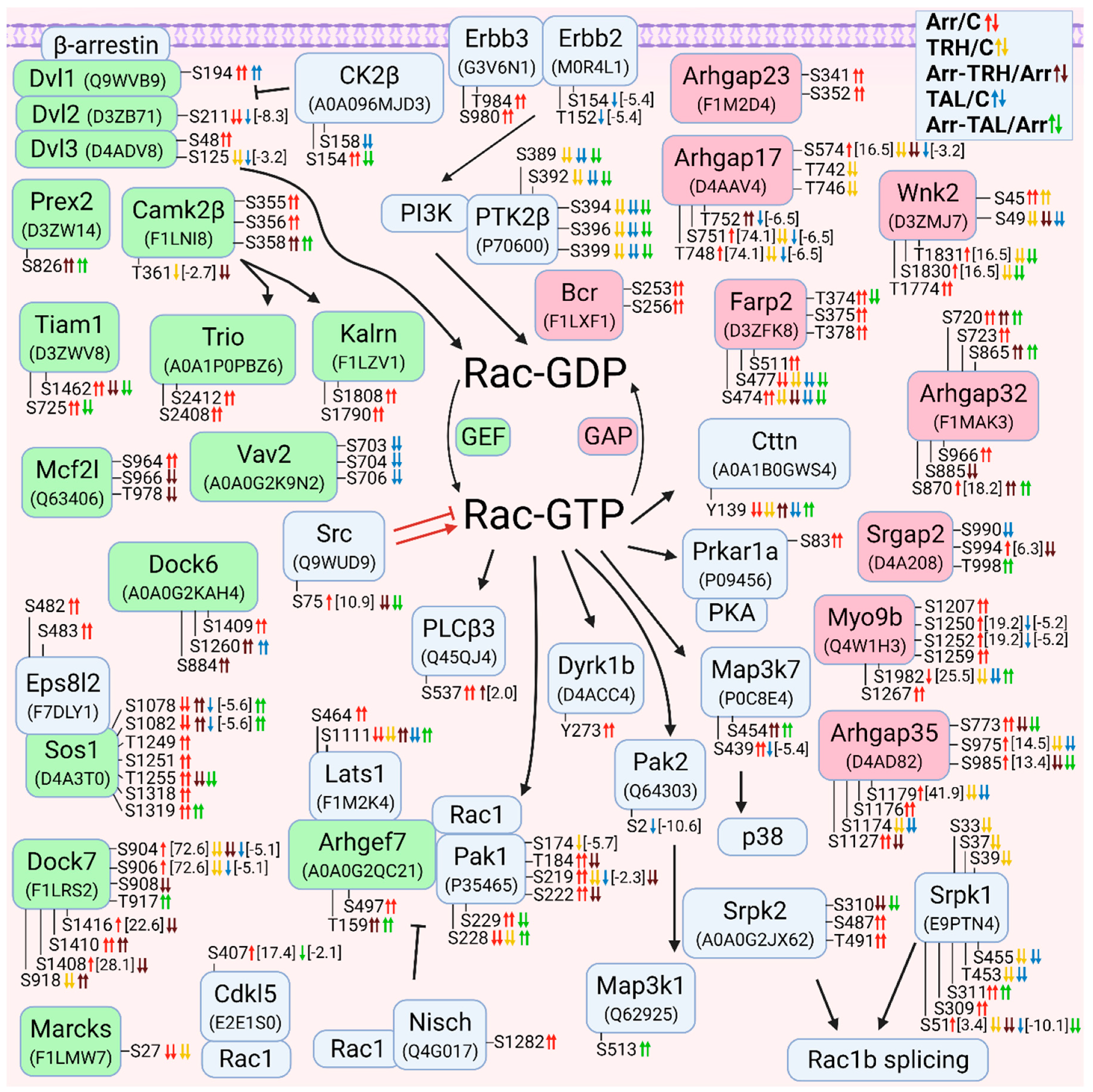
3.2.4. Alterations in Phosphorylation of Phosphoproteins Involved in Rac GTPase-Mediated Signal Transduction
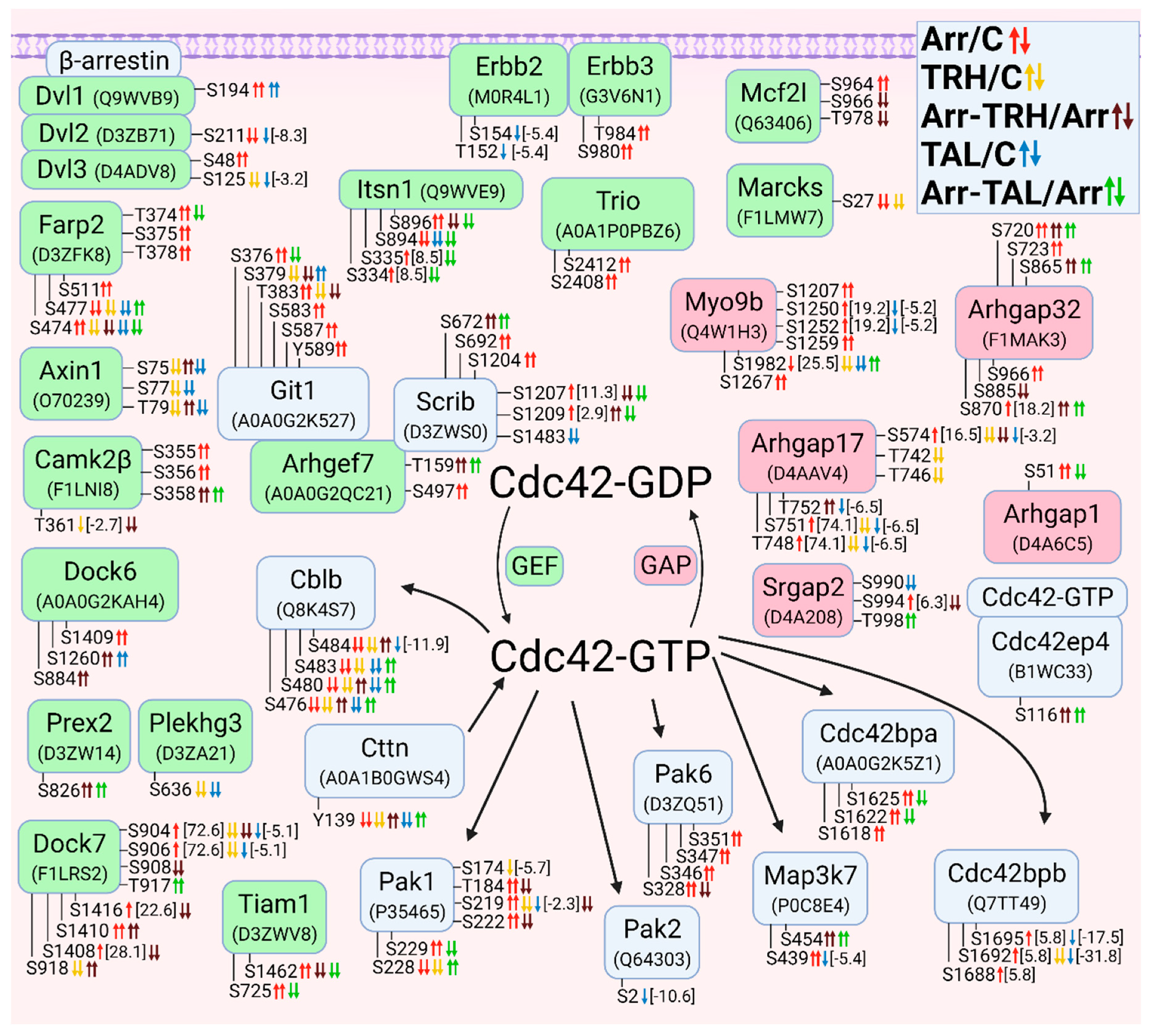
3.2.5. Alterations in Phosphorylation of Phosphoproteins Involved in Cdc42 GTPase-Mediated Signal Transduction
3.2.6. Alterations in Phosphorylation of Phosphoproteins Involved in Arf GTPase-Mediated Signal Transduction
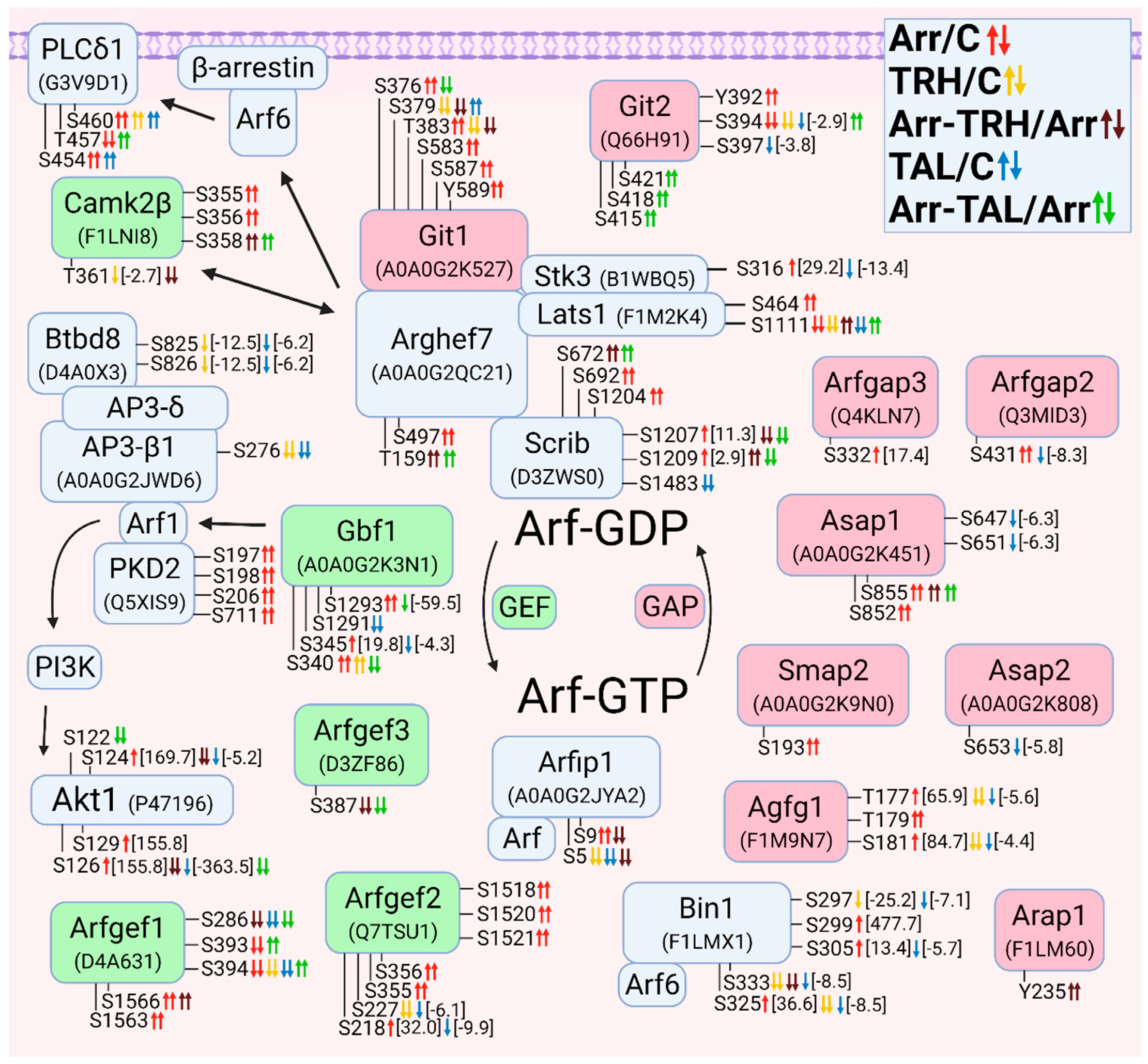
3.2.7. Alterations in Phosphorylation of Phosphoproteins Involved in Rab GTPase-Mediated Signal Transduction

3.2.8. Alterations in Phosphorylation of Phosphoproteins Involved in Ral GTPase-Mediated Signal Transduction

3.2.9. Alterations in Phosphorylation of Phosphoproteins Involved in Ran GTPase-Mediated Signal Transduction
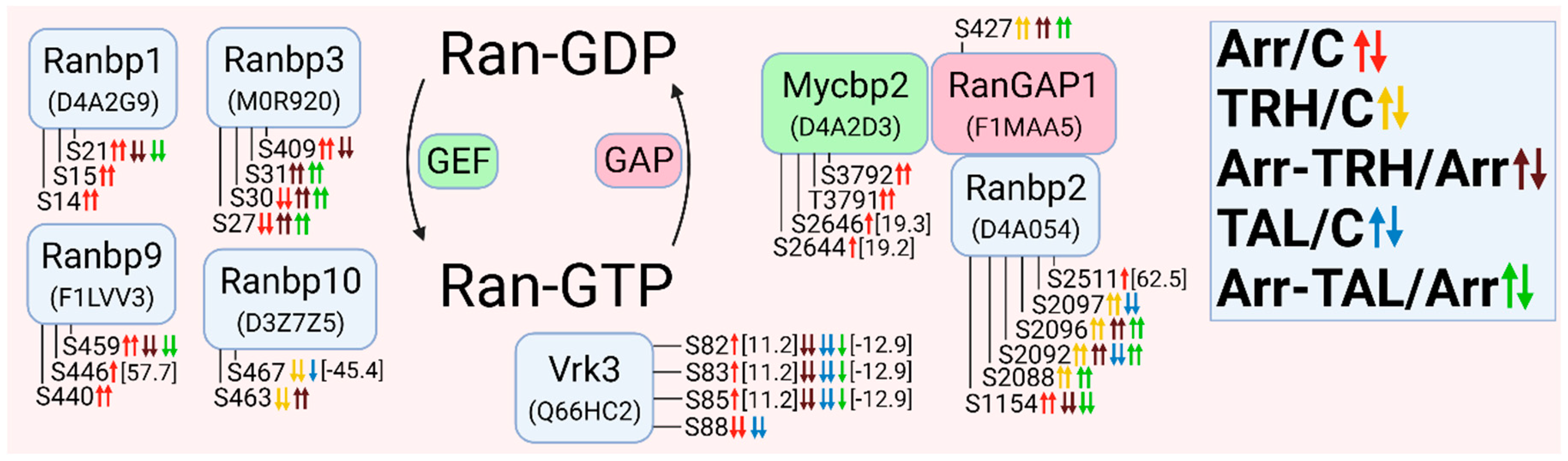
3.2.10. Alterations in Phosphorylation of Phosphoproteins Involved in Rap GTPase-Mediated Signal Transduction
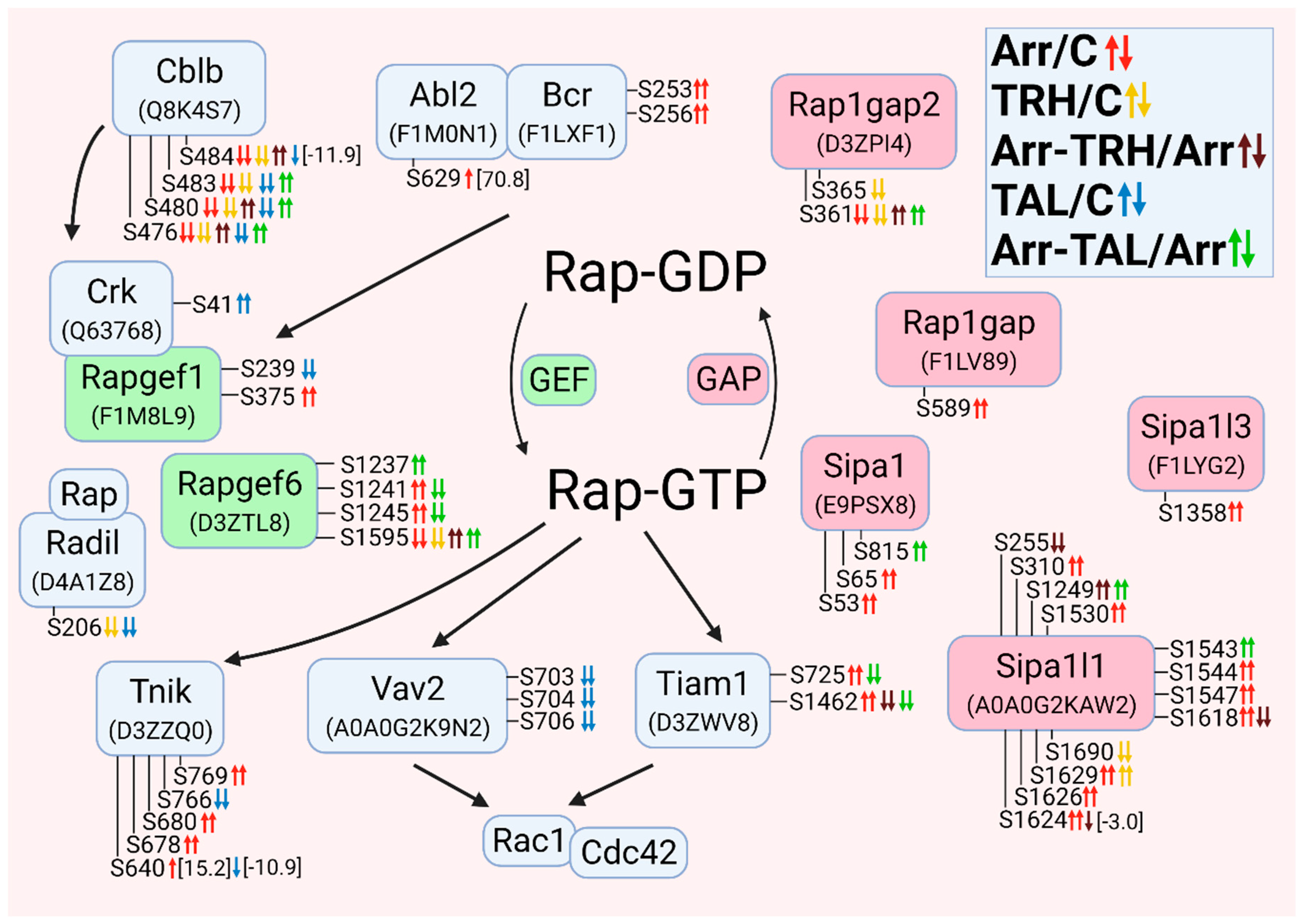
3.2.11. Alterations in Phosphorylation of Phosphoproteins Involved in the β-Catenin Signaling Pathway
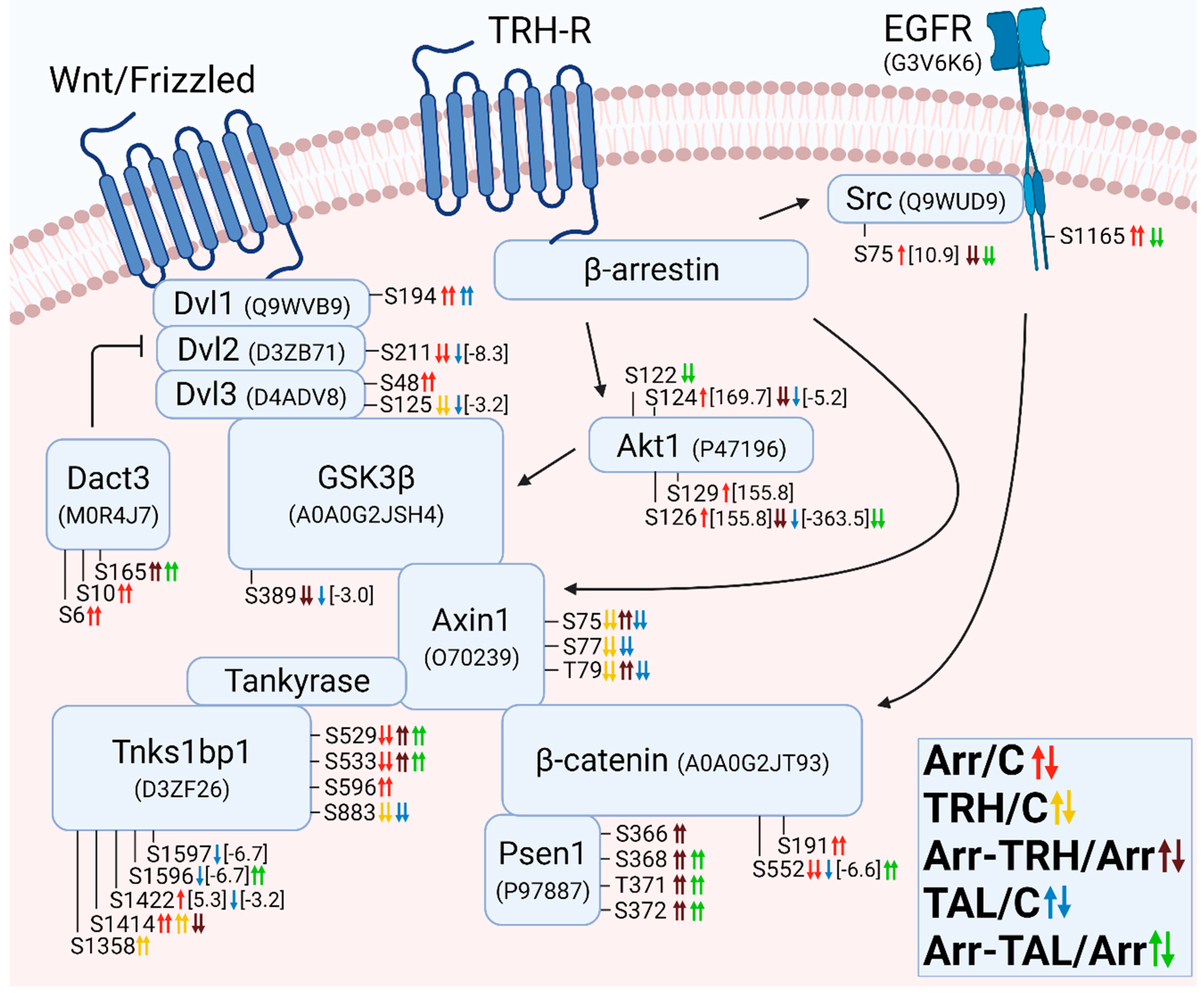
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tashjian, A.H.; Barowsky, N.J.; Jensen, D.K. Thyrotropin releasing hormone–direct evicence for stimulation of prolactin production by pituitary cells in culture. Biochem. Biophys. Res. Commun. 1971, 43, 516–523. [Google Scholar] [CrossRef]
- Kanasaki, H.; Oride, A.; Mijiddorj, T.; Kyo, S. Role of thyrotropin-releasing hormone in prolactin-producing cell models. Neuropeptides 2015, 54, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Drastichova, Z.; Bourova, L.; Hejnova, L.; Jedelsky, P.; Svoboda, P.; Novotny, J. Protein Alterations Induced by Long-Term Agonist Treatment of HEK293 Cells Expressing Thyrotropin-Releasing Hormone Receptor and G(11)alpha Protein. J. Cell. Biochem. 2010, 109, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Koo, K.B.; Suh, H.J.; Ra, K.S.; Choi, J.W. Protective Effect of Cyclo(His-Pro) on Streptozotocin-Induced Cytotoxicity and Apoptosis In Vitro. J. Microbiol. Biotechnol. 2011, 21, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Luo, J.Z.; Jackson, I. Tripeptide amide L-pyroglutamyl-histidyl-L-prolineamide (L-PHP-thyrotropin-releasing hormone, TRH) promotes insulin-producing cell proliferation. Curr. Aging Sci. 2013, 6, 8–13. [Google Scholar] [CrossRef]
- Faden, A.I.; Movsesyan, V.A.; Knoblach, S.M.; Ahmed, F.; Cernak, B. Neuroprotective effects of novel small peptides in vitro and after brain injury. Neuropharmacology 2005, 49, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Faden, A.I.; Knoblach, S.M.; Movsesyan, V.A.; Lea, P.M.; Cernak, I. Novel neuroprotective tripeptides and dipeptides. Neuroprot. Agents 2005, 1053, 472–481. [Google Scholar]
- Jaworska-Feil, L.; Jantas, D.; Leskiewicz, M.; Budziszewska, B.; Kubera, M.; Basta-Kaim, A.; Lipkowski, A.W.; Lason, W. Protective effects of TRH and its analogues against various cytotoxic agents in retinoic acid (RA)-differentiated human neuroblastoma SH-SY5Y cells. Neuropeptides 2010, 44, 495–508. [Google Scholar] [CrossRef]
- Daimon, C.M.; Chirdon, P.; Maudsley, S.; Martin, B. The role of Thyrotropin Releasing Hormone in aging and neurodegenerative diseases. Am. J. Alzheimer’s Dis. 2013, 1. [Google Scholar] [CrossRef]
- Zheng, C.; Chen, G.Q.; Tan, Y.; Zeng, W.Q.; Peng, Q.W.; Wang, J.; Cheng, C.; Yang, X.M.; Nie, S.K.; Xu, Y.; et al. TRH Analog, Taltirelin Protects Dopaminergic Neurons From Neurotoxicity of MPTP and Rotenone. Front. Cell. Neurosci. 2018, 12, 485. [Google Scholar] [CrossRef]
- Monga, V.; Meena, C.L.; Kaur, N.; Jain, R. Chemistry and biology of thyrotropin-releasing hormone (TRH) and its analogs. Curr. Med. Chem. 2008, 15, 2718–2733. [Google Scholar] [CrossRef] [PubMed]
- Fukuchi, I.; Asahi, T.; Kawashima, K.; Kawashima, Y.; Yamamura, M.; Matsuoka, Y.; Kinoshita, K. Effects of taltirelin hydrate (TA-0910), a novel thyrotropin-releasing hormone analog, on in vivo dopamine release and turnover in rat brain. Arzneimittelforschung 1998, 48, 353–359. [Google Scholar] [PubMed]
- Thirunarayanan, N.; Raaka, B.M.; Gershengorn, M.C. Taltirelin is a superagonist at the human thyrotropin-releasing hormone receptor. Front. Endocrinol. Lausanne 2012, 3, 120. [Google Scholar] [CrossRef]
- O’Dowd, B.F.; Lee, D.K.; Huang, W.; Nguyen, T.; Cheng, R.G.; Liu, Y.; Wang, B.; Gershengorn, M.C.; George, S.R. TRH-R2 exhibits similar binding and acute signaling but distinct regulation and anatomic distribution compared with TRH-R1. Mol. Endocrinol. 2000, 14, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.H.; Zupan, B.; Raaka, B.M.; Toth, M.; Gershengorn, M.C. TRH-Receptor-Type-2-Deficient Mice are Euthyroid and Exhibit Increased Depression and Reduced Anxiety Phenotypes. Neuropsychopharmacology 2009, 34, 1601–1608. [Google Scholar] [CrossRef]
- Hsieh, K.P.; Martin, T.F. Thyrotropin-releasing hormone and gonadotropin-releasing hormone receptors activate phospholipase C by coupling to the guanosine triphosphate-binding proteins Gq and G11. Mol. Endocrinol. 1992, 6, 1673–1681. [Google Scholar]
- Hinkle, P.M.; Gehret, A.U.; Jones, B.W. Desensitization, trafficking, and resensitzation of the pituitary thyrotropin-releasing hormone receptor. Front. Neurosci. 2012, 6, 180. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Yu, R.; Hinkle, P.M. Activation of MAPK by TRH requires clathrin-dependent endocytosis and PKC but not receptor interaction with beta-arrestin or receptor endocytosis. Mol. Endocrinol. 2001, 15, 1539–1548. [Google Scholar]
- Storey, N.M.; O’Bryan, J.P.; Armstrong, D.L. Rac and Rho mediate opposing hormonal regulation of the ether-a-go-go-related potassium channel. Curr. Biol. 2002, 12, 27–33. [Google Scholar] [CrossRef]
- Romano, D.; Magalon, K.; Ciampini, A.; Talet, C.; Enjalbert, A.; Gerard, C. Differential involvement of the Ras and Rap1 small GTPases in vasoactive intestinal and pituitary adenylyl cyclase activating polypeptides control of the prolactin gene. J. Biol. Chem. 2003, 278, 51386–51394. [Google Scholar] [CrossRef]
- Jones, B.W.; Hinkle, P.M. Beta-arrestin mediates desensitization and internalization but does not affect dephosphorylation of the thyrotropin-releasing hormone receptor. J. Biol. Chem. 2005, 280, 38346–38354. [Google Scholar] [CrossRef] [PubMed]
- Luttrell, L.M.; Lefkowitz, R.J. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell Sci. 2002, 115, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Mancini, A.D.; Bertrand, G.; Vivot, K.; Carpentier, E.; Tremblay, C.; Ghislain, J.; Bouvier, M.; Poitout, V. beta-Arrestin Recruitment and Biased Agonism at Free Fatty Acid Receptor 1. J. Biol. Chem. 2015, 290, 21131–21140. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Fernandez, G.; Cabezudo, S.; Garcia-Hoz, C.; Tobin, A.B.; Mayor, F.; Ribas, C. ERK5 Activation by Gq-Coupled Muscarinic Receptors Is Independent of Receptor Internalization and beta-Arrestin Recruitment. PLoS ONE 2013, 8, e84174. [Google Scholar] [CrossRef]
- Teixeira, L.B.; Parreiras-E-Silva, L.T.; Bruder-Nascimento, T.; Duarte, D.A.; Simoes, S.C.; Costa, R.M.; Rodriguez, D.Y.; Ferreira, P.A.B.; Silva, C.A.A.; Abrao, E.P.; et al. Ang-(1-7) is an endogenous beta-arrestin-biased agonist of the AT(1) receptor with protective action in cardiac hypertrophy. Sci. Rep. 2017, 7, 11903. [Google Scholar] [CrossRef]
- Luttrell, L.M.; Ferguson, S.S.G.; Daaka, Y.; Miller, W.E.; Maudsley, S.; Della Rocca, G.J.; Lin, F.T.; Kawakatsu, H.; Owada, K.; Luttrell, D.K.; et al. beta-arrestin-dependent formation of beta(2) adrenergic receptor Src protein kinase complexes. Science 1999, 283, 655–661. [Google Scholar] [CrossRef]
- Luttrell, L.M.; Roudabush, F.L.; Choy, E.W.; Miller, W.E.; Field, M.E.; Pierce, K.L.; Lefkowitz, R.J. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc. Natl. Acad. Sci. USA 2001, 98, 2449–2454. [Google Scholar] [CrossRef]
- Coffa, S.; Breitman, M.; Hanson, S.M.; Callaway, K.; Kook, S.; Dalby, K.N.; Gurevich, V.V. The Effect of Arrestin Conformation on the Recruitment of c-Raf1, MEK1, and ERK1/2 Activation. PLoS ONE 2011, 6, e28723. [Google Scholar] [CrossRef]
- Cassier, E.; Gallay, N.; Bourquard, T.; Claeysen, S.; Bockaert, J.; Crepieux, P.; Poupon, A.; Reiter, E.; Marin, P.; Vandermoere, F. Phosphorylation of beta-arrestin2 at Thr(383) by MEK underlies beta-arrestin-dependent activation of Erk1/2 by GPCRs. eLife 2017, 6, e23777. [Google Scholar] [CrossRef]
- Peterson, Y.K.; Luttrell, L.M. The Diverse Roles of Arrestin Scaffolds in G Protein-Coupled Receptor Signaling. Pharmacol. Rev. 2017, 69, 256–297. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Miranda, P.; Giráldez, T.; de la Peña, P.; Manso, D.G.; Alonso-Ron, C.; Gómez-Varela, D.; Domínguez, P.; Barros, F. Specificity of TRH receptor coupling to G-proteins for regulation of ERG K+ channels in GH3 rat anterior pituitary cells. J. Physiol. 2005, 566, 717–736. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Pan, C.; He, S.M.; Lang, B.; Gao, G.D.; Wang, X.L.; Wang, Y. The Ras Superfamily of Small GTPases in Non-neoplastic Cerebral Diseases. Front. Mol. Neurosci. 2019, 12, 121. [Google Scholar] [CrossRef]
- Ba, W.; Nadif Kasri, N. RhoGTPases at the synapse: An embarrassment of choice. Small GTPases 2017, 8, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Karginov, A.V. Phosphorylation-mediated regulation of GEFs for RhoA. Cell Adh. Migr. 2014, 8, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Shirakawa, R.; Horiuchi, H. Ral GTPases: Crucial mediators of exocytosis and tumourigenesis. J. Biochem. 2015, 157, 285–299. [Google Scholar] [CrossRef]
- Walkup, W.G.; Washburn, L.; Sweredoski, M.J.; Carlisle, H.J.; Graham, R.L.; Hess, S.; Kennedy, M.B. Phosphorylation of Synaptic GTPase-activating Protein (synGAP) by Ca2+/Calmodulin-dependent Protein Kinase II (CaMKII) and Cyclin-dependent Kinase 5 (CDK5) Alters the Ratio of Its GAP Activity toward Ras and Rap GTPases. J. Biol. Chem. 2015, 290, 4908–4927. [Google Scholar] [CrossRef]
- Humphrey, S.J.; Karayel, O.; James, D.E.; Mann, M. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat. Protoc. 2018, 13, 1897–1916. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Meth. 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. MTOR interacts with Raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef]
- Alers, S.; Loffler, A.S.; Wesselborg, S.; Stork, B. Role of AMPK-mTOR-Ulk1/2 in the Regulation of Autophagy: Cross Talk, Shortcuts, and Feedbacks. Mol. Cell. Biol. 2012, 32, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Qiao, F.; Gao, C.; Norman, B.; Optican, L.; Zelenka, P.S. Cdk5 targets active Src for ubiquitin-dependent degradation by phosphorylating Src(S75). Cell. Mol. Life Sci. 2011, 68, 3425–3436. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Ji, Q.; Li, Q. The role and mechanism of beta-arrestins in cancer invasion and metastasis. Int. J. Mol. Med. 2018, 41, 631–639. [Google Scholar] [PubMed]
- Annunziata, M.C.; Parisi, M.; Esposito, G.; Fabbrocini, G.; Ammendola, R.; Cattaneo, F. Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling. Int. J. Mol. Sci. 2020, 21, 3818. [Google Scholar] [CrossRef]
- Girardi, C.; James, P.; Zanin, S.; Pinna, L.A.; Ruzzene, M. Differential phosphorylation of Akt1 and Akt2 by protein kinase CK2 may account for isoform specific functions. Biochim. Biophys. Acta-Mol. Cell Res. 2014, 1843, 1865–1874. [Google Scholar] [CrossRef]
- Di Maira, G.; Salvi, M.; Arrigoni, G.; Marin, O.; Sarno, S.; Brustolon, F.; Pinna, L.A.; Ruzzene, M. Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell Death Differ. 2005, 12, 668–677. [Google Scholar] [CrossRef]
- Litchfield, D.W.; Bosc, D.G.; Slominski, E. The protein kinase from mitotoc human cells that phosphorylates Ser-209 on th casein kinase-II beta-subunit is P34(CDC2). Biochim. Biophys. Acta Mol. Cell Res. 1995, 1269, 69–78. [Google Scholar] [CrossRef]
- Sanders, S.S.; De Simone, F.I.; Thomas, G.M. mTORC1 Signaling Is Palmitoylation-Dependent in Hippocampal Neurons and Non-neuronal Cells and Involves Dynamic Palmitoylation of LAMTOR1 and mTOR. Front. Cell. Neurosci. 2019, 13, 115. [Google Scholar] [CrossRef]
- Li, X.D.; Wang, L.L.; Zhou, X.E.; Ke, J.Y.; De Waal, P.W.; Gu, X.; Tan, M.H.E.; Wang, D.Y.; Wu, D.H.; Xu, H.E.; et al. Structural basis of AMPK regulation by adenine nucleotides and glycogen. Cell Res. 2015, 25, 50–66. [Google Scholar] [CrossRef]
- Steinberg, G.R.; Carling, D. AMP-activated protein kinase: The current landscape for drug development. Nat. Rev. Drug Discov. 2019, 18, 527–551. [Google Scholar] [CrossRef] [PubMed]
- Ovens, A.J.; Scott, J.W.; Langendorf, C.G.; Kemp, B.E.; Oakhill, J.S.; Smiles, W.J. Post-Translational Modifications of the Energy Guardian AMP-Activated Protein Kinase. Int. J. Mol. Sci. 2021, 22, 1229. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.H.; Nousiainen, M.; Chalamalasetty, R.B.; Schafer, A.; Nigg, E.A.; Sillje, H.H.W. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 2005, 24, 2076–2086. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.P.; Moroishi, T.; Guan, K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef]
- Yamauchi, T.; Moroishi, T. Hippo Pathway in Mammalian Adaptive Immune System. Cells 2019, 8, 398. [Google Scholar] [CrossRef]
- Humbert, N.; Navaratnam, N.; Augert, A.; Da Costa, M.; Martien, S.; Wang, J.; Martinez, D.; Abbadie, C.; Carling, D.; de Launoit, Y.; et al. Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. EMBO J. 2010, 29, 376–386. [Google Scholar] [CrossRef]
- Martelli, A.M.; Evangelisti, C.; Chiarini, F.; Grimaldi, C.; McCubrey, J.A. The emerging role of the phosphatidylinositol 3-kinase/ akt/mammalian target of rapamycin signaling network in cancer stem cell biology. Cancers 2010, 2, 1576–1596. [Google Scholar] [CrossRef]
- Schoneborn, H.; Raudzus, F.; Coppey, M.; Neumann, S.; Heumann, R. Perspectives of RAS and RHEB GTPase Signaling Pathways in Regenerating Brain Neurons. Int. J. Mol. Sci. 2018, 19, 4052. [Google Scholar] [CrossRef]
- Young, K.A.; Biggins, L.; Sharpe, H.J. Protein tyrosine phosphatases in cell adhesion. Biochem. J. 2021, 478, 1061–1083. [Google Scholar] [CrossRef]
- Lai, M.C.; Chang, C.M.; Sun, H.S. Hypoxia Induces Autophagy through Translational Up-Regulation of Lysosomal Proteins in Human Colon Cancer Cells. PLoS ONE 2016, 11, e0153627. [Google Scholar] [CrossRef]
- Zhao, H.F.; Wang, J.; To, S.S.T. The phosphatidylinositol 3-kinase/Akt and c-Jun N-terminal kinase signaling in cancer: Alliance or contradiction? (Review). Int. J. Oncol. 2015, 47, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Alsaqati, M.; Heine, V.M.; Harwood, A.J. Pharmacological intervention to restore connectivity deficits of neuronal networks derived from ASD patient iPSC with a TSC2 mutation. Mol. Autism 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Goel, S.; DeCristo, M.J.; McAllister, S.S.; Zhao, J.J. CDK4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol. 2018, 28, 911–925. [Google Scholar] [CrossRef] [PubMed]
- Won, S.Y.; Park, J.J.; Shin, E.Y.; Kim, E.G. PAK4 signaling in health and disease: Defining the PAK4-CREB axis. Exp. Mol. Med. 2019, 51, 1–9. [Google Scholar] [CrossRef]
- Kucerova, L.; Kubrak, O.I.; Bengtsson, J.M.; Strnad, H.; Nylin, S.; Theopold, U.; Nassel, D.R. Slowed aging during reproductive dormancy is reflected in genome-wide transcriptome changes in Drosophila melanogaster. BMC Genom. 2016, 17, 50. [Google Scholar] [CrossRef]
- Sanchez, A.M.J.; Candau, R.B.; Csibi, A.; Pagano, A.F.; Raibon, A.; Bernardi, H. The role of AMP-activated protein kinase in the coordination of skeletal muscle turnover and energy homeostasis. Am. J. Physiol.-Cell Physiol. 2012, 303, C475–C485. [Google Scholar] [CrossRef]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef]
- Fang, C.Y.; Lai, T.C.; Hsiao, M.; Chang, Y.C. The Diverse Roles of TAO Kinases in Health and Diseases. Int. J. Mol. Sci. 2020, 21, 7463. [Google Scholar] [CrossRef]
- Spencer, J.P.E. The interactions of flavonoids within neuronal signalling pathways. Genes Nutr. 2007, 2, 257–273. [Google Scholar] [CrossRef]
- Cuesta, C.; Arevalo-Alameda, C.; Castellano, E. The Importance of Being PI3K in the RAS Signaling Network. Genes 2021, 12, 1094. [Google Scholar] [CrossRef] [PubMed]
- Beck, T.N.; Nicolas, E.; Kopp, M.C.; Golemis, E.A. Adaptors for disorders of the brain? The cancer signaling proteins NEDD9, CASS4, and PTK2B in Alzheimer’s disease. Oncoscience 2014, 1, 486–503. [Google Scholar] [CrossRef]
- Lock, L.S.; Frigault, M.M.; Saucier, C.; Park, M. Grb2-independent recruitment of Gab1 requires the C-terminal lobe and structural integrity of the met receptor kinase domain. J. Biol. Chem. 2003, 278, 30083–30090. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Hibino, K.; Yanagida, T.; Sako, Y. Switching of the positive feedback for RAS activation by a concerted function of SOS membrane association domains. Biophys. Physicobiol. 2016, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lepri, F.; De Luca, A.; Stella, L.; Rossi, C.; Baldassarre, G.; Pantaleoni, F.; Cordeddu, V.; Williams, B.J.; Dentici, M.L.; Caputo, V.; et al. SOS1 Mutations in Noonan Syndrome: Molecular Spectrum, Structural Insights on Pathogenic Effects, and Genotype-Phenotype Correlations. Hum. Mutat. 2011, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, H.; Sahmi, M.; Maisonneuve, P.; Marullo, S.A.; Thevakumaran, N.; Jin, T.; Kurinov, I.; Sicheri, F.; Therrien, M. MEK drives BRAF activation through allosteric control of KSR proteins. Nature 2018, 554, 549–553. [Google Scholar] [CrossRef]
- Dougherty, M.K.; Ritt, D.A.; Zhou, M.; Specht, S.I.; Monson, D.M.; Veenstra, T.D.; Morrison, D.K. KSR2 Is a Calcineurin Substrate that Promotes ERK Cascade Activation in Response to Calcium Signals. Mol. Cell 2009, 34, 652–662. [Google Scholar] [CrossRef]
- Nishiyama, K.; Maekawa, M.; Nakagita, T.; Nakayama, J.; Kiyoi, T.; Chosei, M.; Murakami, A.; Kamei, Y.; Takeda, H.; Takada, Y.; et al. CNKSR1 serves as a scaffold to activate an EGFR phosphatase via exclusive interaction with RhoB-GTP. Life Sci. Alliance 2021, 4, e202101095. [Google Scholar] [CrossRef]
- Cerezo, E.L.; Houles, T.; Lie, O.; Sarthou, M.K.; Audoynaud, C.; Lavoie, G.; Halladjian, M.; Cantaloube, S.; Froment, C.; Burlet-Schiltz, O.; et al. RIOK2 phosphorylation by RSK promotes synthesis of the human small ribosomal subunit. PLoS Genet. 2021, 17, e1009583. [Google Scholar] [CrossRef]
- Deng, Y.N.; Xia, Z.J.; Zhang, P.; Ejaz, S.; Liang, S.F. Transcription Factor RREB1: From Target Genes towards Biological Functions. Int. J. Biol. Sci. 2020, 16, 1463–1473. [Google Scholar] [CrossRef]
- Agulto, R.L.; Rogers, M.M.; Tan, T.C.; Ramkumar, A.; Downing, A.M.; Bodin, H.; Castro, J.; Nowakowski, D.W.; Ori-McKenney, K.M. Autoregulatory control of microtubule binding in doublecortin-like kinase 1. eLife 2021, 10, e60126. [Google Scholar] [CrossRef] [PubMed]
- Weygant, N.; Qu, D.F.; Berry, W.L.; May, R.; Chandrakesan, P.; Owen, D.B.; Sureban, S.M.; Ali, N.; Janknecht, R.; Houchen, C.W. Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Mol. Cancer 2014, 13, 103. [Google Scholar] [CrossRef]
- Patel, O.; Dai, W.W.; Mentzel, M.; Griffin, M.D.W.; Serindoux, J.; Gay, Y.; Fischer, S.; Sterle, S.; Kropp, A.; Burns, C.J.; et al. Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1. Structure 2016, 24, 1550–1561. [Google Scholar] [CrossRef]
- Sureban, S.M.; May, R.; Qu, D.F.; Weygant, N.; Chandrakesan, P.; Ali, N.; Lightfoot, S.A.; Pantazis, P.; Rao, C.V.; Postier, R.G.; et al. DCLK1 Regulates Pluripotency and Angiogenic Factors via microRNA-Dependent Mechanisms in Pancreatic Cancer. PLoS ONE 2013, 8, e73940. [Google Scholar] [CrossRef] [PubMed]
- Kent, O.A.; Fox-Talbot, K.; Halusha, M.K. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 2013, 32, 2576–2585. [Google Scholar] [CrossRef] [PubMed]
- Namba, T.; Funahashi, Y.; Nakamuta, S.; Xu, C.; Takano, T.; Kaibuchi, K. Exztacellular and intracellular signaling for neuronal polarity. Physiol. Rev. 2015, 95, 995–1024. [Google Scholar] [CrossRef]
- Llavero, F.; Arrazola Sastre, A.; Luque Montoro, M.; Martín, M.A.; Arenas, J.; Lucia, A.; Zugaza, J.L. Small GTPases of the Ras superfamily and glycogen phosphorylase regulation in T cells. Small GTPases 2021, 12, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Llavero, F.; Montoro, M.L.; Sastre, A.A.; Fernandez-Moreno, D.; Lacerda, H.M.; Parada, L.A.; Lucia, A.; Zugaza, J.L. Epidermal growth factor receptor controls glycogen phosphorylase in T cells through small GTPases of the RAS family. J. Biol. Chem. 2019, 294, 4345–4358. [Google Scholar] [CrossRef]
- Bok, S.; Shin, D.Y.; Yallowitz, A.R.; Eiseman, M.; Cung, M.; Xu, R.; Li, N.; Sun, J.; Williams, A.L.; Scott, J.E.; et al. MEKK2 mediates aberrant ERK activation in neurofibromatosis type I. Nat. Commun. 2020, 11, 5704. [Google Scholar] [CrossRef]
- Kishida, S.; Yamamoto, H.; Hino, S.; Ikeda, S.; Kishida, M.; Kikuchi, A. DIX domains of Dvl and Axin are necessary for protein interactions and their ability to regulate beta-catenin stability. Mol. Cell. Biol. 1999, 19, 4414–4422. [Google Scholar] [CrossRef]
- Bros, M.; Haas, K.; Moll, L.; Grabbe, S. RhoA as a Key Regulator of Innate and Adaptive Immunity. Cells 2019, 8, 733. [Google Scholar] [CrossRef]
- Li, Y.Y.; Shi, J.H.; Yang, J.; Ge, S.F.; Zhang, J.M.; Jia, R.B.; Fan, X.Q. Uveal melanoma: Progress in molecular biology and therapeutics. Ther. Adv. Med. Oncol. 2020, 12, 1758835920965852. [Google Scholar] [CrossRef]
- Yi, F.S.; Kong, R.R.; Ren, J.Q.; Zhu, L.; Lou, J.Z.; Wu, J.Y.; Feng, W. Noncanonical Myo9b-RhoGAP Accelerates RhoA GTP Hydrolysis by a Dual-Arginine-Finger Mechanism. J. Mol. Biol. 2016, 428, 3043–3057. [Google Scholar] [CrossRef]
- Schlessinger, K.; Hall, A.; Tolwinski, N. Wnt signaling pathways meet Rho GTPases. Genes Dev. 2009, 23, 265–277. [Google Scholar] [CrossRef]
- Cook, D.R.; Rossman, K.L.; Der, C.J. Rho guanine nucleotide exchange factors: Regulators of Rho GTPase activity in development and disease. Oncogene 2014, 33, 4021–4035. [Google Scholar] [CrossRef]
- Blangy, A. Tensins are versatile regulators of Rho GTPase signalling and cell adhesion. Biol. Cell 2017, 109, 115–126. [Google Scholar] [CrossRef]
- Gong, X.W.; Didan, Y.; Lock, J.G.; Stromblad, S. KIF13A-regulated RhoB plasma membrane localization governs membrane blebbing and blebby amoeboid cell migration. EMBO J. 2018, 37, e98994. [Google Scholar] [CrossRef] [PubMed]
- Maiwald, S.; Motazacker, M.M.; van Capelleveen, J.C.; Sivapalaratnam, S.; van der Wal, A.C.; van der Loos, C.; Kastelein, J.J.P.; Ouwehand, W.H.; Hovingh, G.K.; Trip, M.D.; et al. A rare variant in MCF2L identified using exclusion linkage in a pedigree with premature atherosclerosis. Eur. J. Hum. Genet. 2016, 24, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Akizu, N.; Martínez-Balbás, M.A. EZH2 orchestrates apicobasal polarity and neuroepithelial cell renewal. Neurogenesis 2016, 3, e1250034. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feng, X.D.; Degese, M.S.; Iglesias-Bartolome, R.; Vaque, J.P.; Molinolo, A.A.; Rodrigues, M.; Zaidi, M.R.; Ksander, B.R.; Merlino, G.; Sodhi, A.; et al. Hippo-Independent Activation of YAP by the GNAQ Uveal Melanoma Oncogene through a Trio-Regulated Rho GTPase Signaling Circuitry. Cancer Cell 2014, 25, 831–845. [Google Scholar] [CrossRef]
- Muller, P.M.; Rademacher, J.; Bagshaw, R.D.; Wortmann, C.; Barth, C.; van Unen, J.; Alp, K.M.; Giudice, G.; Eccles, R.L.; Heinrich, L.E.; et al. Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat. Cell Biol. 2020, 22, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Kim, H.J.; Han, D.C.; Lee, H.B. Effect of lovastatin on small GTP binding proteins and on TGF-beta 1 and fibronectin expression. Kidney Int. 2000, 58, S88–S92. [Google Scholar] [CrossRef][Green Version]
- Shimizu, A.; Mammoto, A.; Italiano, J.E.; Pravda, E.; Dudley, A.C.; Ingber, D.E.; Klagsbrun, M. ABL2/ARG tyrosine kinase mediates SEMA3F-induced RhoA inactivation and cytoskeleton collapse in human glioma cells. J. Biol. Chem. 2008, 283, 27230–27238. [Google Scholar] [CrossRef] [PubMed]
- Lartey, J.; Bernal, A.L. RHO protein regulation of contraction in the human uterus. Reproduction 2009, 138, 407–424. [Google Scholar] [CrossRef] [PubMed]
- Cossette, S.M.; Bhute, V.J.; Bao, X.; Harmann, L.M.; Horswill, M.A.; Sinha, I.; Gastonguay, A.; Pooya, S.; Bordas, M.; Kumar, S.N.; et al. Sucrose Nonfermenting-Related Kinase Enzyme-Mediated Rho-Associated Kinase Signaling is Responsible for Cardiac Function. Circ.-Cardiovasc. Genet. 2016, 9, 474–486. [Google Scholar] [CrossRef]
- Fokin, A.I.; Klementeva, T.S.; Nadezhdina, E.S.; Burakov, A.V. SLK/LOSK kinase regulates cell motility independently of microtubule organization and Golgi polarization. Cytoskeleton 2016, 73, 83–92. [Google Scholar] [CrossRef]
- Rangamani, P.; Levy, M.G.; Khan, S.; Oster, G. Paradoxical signaling regulates structural plasticity in dendritic spines. Proc. Natl. Acad. Sci. USA 2016, 113, E5298–E5307. [Google Scholar] [CrossRef]
- Durkin, C.H.; Leite, F.; Cordeiro, J.V.; Handa, Y.; Arakawa, Y.; Valderrama, F.; Way, M. RhoD Inhibits RhoC-ROCK-Dependent Cell Contraction via PAK6. Dev. Cell 2017, 41, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Eisler, S.A.; Curado, F.; Link, G.; Schulz, S.; Noack, M.; Steinke, M.; Olayioye, M.A.; Hausser, A. A Rho signaling network links microtubules to PKD controlled carrier transport to focal adhesions. eLife 2018, 7, e35907. [Google Scholar] [CrossRef] [PubMed]
- Forrest, M.P.; Parnell, E.; Penzes, P. Dendritic structural plasticity and neuropsychiatric disease. Nat. Rev. Neurosci. 2018, 19, 215–234. [Google Scholar] [CrossRef] [PubMed]
- Kurtzeborn, K.; Kwon, H.N.; Kuure, S. MAPK/ERK Signaling in Regulation of Renal Differentiation. Int. J. Mol. Sci. 2019, 20, 1779. [Google Scholar] [CrossRef]
- Pedraza, N.; Cemeli, T.; Monserrat, M.V.; Garí, E.; Ferrezuelo, F. Regulation of small GTPase activity by G1 cyclins. Small GTPases 2019, 10, 47–53. [Google Scholar] [CrossRef]
- Asih, P.R.; Prikas, E.; Stefanoska, K.; Tan, A.R.P.; Ahel, H.I.; Ittner, A. Functions of p38 MAP Kinases in the Central Nervous System. Front. Mol. Neurosci. 2020, 13, 570586. [Google Scholar] [CrossRef]
- Schulte, G.; Shenoy, S.K. beta-Arrestin and dishevelled coordinate biased signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 19839–19840. [Google Scholar] [CrossRef]
- Clayton, N.S.; Ridley, A.J. Targeting Rho GTPase Signaling Networks in Cancer. Front. Cell Dev. Biol. 2020, 8, 222. [Google Scholar] [CrossRef]
- Tolias, K.F.; Duman, J.G.; Um, K. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog. Neurobiol. 2011, 94, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, J.J.; Hara, M.R.; Davenport, C.L.; Kim, J.; Lefkowitz, R.J. Arrestin Development: Emerging Roles for beta-arrestins in Developmental Signaling Pathways. Dev. Cell 2009, 17, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Bryja, V.; Schambony, A.; Cajanek, L.; Dominguez, I.; Arenas, E.; Schulte, G. beta-Arrestin and casein kinase 1/2 define distinct branches of non-canonical WNT signalling pathways. EMBO Rep. 2008, 9, 1244–1250. [Google Scholar] [CrossRef] [PubMed]
- Moniz, S.; Jordan, P. Emerging roles for WNK kinases in cancer. Cell. Mol. Life Sci. 2010, 67, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Arias-Romero, L.E.; Villamar-Cruz, O.; Pacheco, A.; Kosoff, R.; Huang, M.; Muthuswamy, S.K.; Chernoff, J. A Rac-Pak signaling pathway is essential for ErbB2-mediated transformation of human breast epithelial cancer cells. Oncogene 2010, 29, 5839–5849. [Google Scholar] [CrossRef]
- Tobon, A.L.; Suresh, M.; Jin, J.; Vitriolo, A.; Pietralla, T.; Tedford, K.; Bossenz, M.; Mahnken, K.; Kiefer, F.; Testa, G.; et al. The guanine nucleotide exchange factor Arhgef7/beta Pix promotes axon formation upstream of TC10. Sci. Rep. 2018, 8, 8811. [Google Scholar] [CrossRef]
- Arash, E.H.; Song, K.M.; Song, S.; Shiban, A.; Attisano, L. Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J. 2014, 33, 2997–3011. [Google Scholar] [CrossRef]
- Fu, X.D. Both sides of the same coin: Rac1 splicing regulating by EGF signaling. Cell Res. 2017, 27, 455–456. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Yamauchi, J.; Sanbe, A.; Tanoue, A. Dock6, a Dock-C subfamily guanine nucleotide exchanger, has the dual specificity for Rac1 and Cdc42 and regulates neurite outgrowth. Exp. Cell Res. 2007, 313, 791–804. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, H.; Kim, K.; Lee, H.; Lee, S.; Lee, D. Arhgap17, a RhoGTPase activating protein, regulates mucosal and epithelial barrier function in the mouse colon. Sci. Rep. 2016, 6, 26923. [Google Scholar] [CrossRef] [PubMed]
- Okabe, T.; Nakamura, T.; Nishimura, Y.N.; Kohu, K.; Ohwada, S.; Morishita, Y.; Akiyama, T. RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the beta-catenin-N-cadherin and N-methyl-D-aspartate receptor signaling. J. Biol. Chem. 2003, 278, 9920–9927. [Google Scholar] [CrossRef] [PubMed]
- Harden, T.K.; Hicks, S.N.; Sondek, J. Phospholipase C isozymes as effectors of Ras superfamily GTPases. J. Lipid Res. 2009, 50, S243–S248. [Google Scholar] [CrossRef]
- Kichina, J.V.; Goc, A.; Al-Husein, B.; Somanath, P.R.; Kandel, E.S. PAK1 as a therapeutic target. Expert Opin. Ther. Targets 2010, 14, 703–725. [Google Scholar] [CrossRef]
- He, X.J.; Kuo, Y.C.; Rosche, T.J.; Zhang, X.W. Structural Basis for Autoinhibition of the Guanine Nucleotide Exchange Factor FARP2. Structure 2013, 21, 355–364. [Google Scholar] [CrossRef]
- Kuijl, C.; Pilli, M.; Alahari, S.K.; Janssen, H.; Khoo, P.S.; Ervin, K.E.; Calero, M.; Jonnalagadda, S.; Scheller, R.H.; Neefjes, J.; et al. Rac and Rab GTPases dual effector Nischarin regulates vesicle maturation to facilitate survival of intracellular bacteria. EMBO J. 2013, 32, 713–727. [Google Scholar] [CrossRef]
- Lopez-Guerrero, A.M.; Espinosa-Bermejo, N.; Sanchez-Lopez, I.; Macartney, T.; Pascual-Caro, C.; Orantos-Aguilera, Y.; Rodriguez-Ruiz, L.; Perez-Oliva, A.B.; Mulero, V.; Pozo-Guisado, E.; et al. RAC1-Dependent ORAI1 Translocation to the Leading Edge Supports Lamellipodia Formation and Directional Persistence. Sci. Rep. 2020, 10, 6580. [Google Scholar] [CrossRef]
- Vasco, V.R.L.L. Phosphoinositide Signal Transduction Pathway and Osteosarcoma Metastases. Jentashapir J. Cell. Mol. Biol. 2021, 12, e116225. [Google Scholar] [CrossRef]
- Zamboni, V.; Jones, R.; Umbach, A.; Ammoni, A.; Passafaro, M.; Hirsch, E.; Merlo, G.R. Rho GTPases in Intellectual Disability: From Genetics to Therapeutic Opportunities. Int. J. Mol. Sci. 2018, 19, 1821. [Google Scholar] [CrossRef]
- Wang, S.E.; Xian, B.; Guix, M.; Olivares, M.G.; Parker, J.; Chung, C.H.; Pandiella, A.; Arteaga, C.L. Transforming growth factor beta engages TACE and ErbB3 to activate phosphatidylinositol-3 kinase/Akt in ErbB2-overexpressing breast cancer and desensitizes cells to trastuzumab. Mol. Cell. Biol. 2008, 28, 5605–5620. [Google Scholar] [CrossRef]
- Manchanda, P.K.; Jones, G.N.; Lee, A.A.; Pringle, D.R.; Zhang, M.; Yu, L.; La Perle, K.M.D.; Kirschner, L.S. Rac1 is required for Prkar1a-mediated Nf2 suppression in Schwann cell tumors. Oncogene 2013, 32, 3491–3499. [Google Scholar] [CrossRef][Green Version]
- Lopez-Haber, C.; Barrio-Real, L.; Casado-Medrano, V.; Kazanietz, M.G. Heregulin/ErbB3 Signaling Enhances CXCR4-Driven Rac1 Activation and Breast Cancer Cell Motility via Hypoxia-Inducible Factor 1 alpha. Mol. Cell. Biol. 2016, 36, 2011–2026. [Google Scholar] [CrossRef] [PubMed]
- Nuche-Berenguer, B.; Ramos-Alvarez, I.; Jensen, R.T. The p21-activated kinase, PAK2, is important in the activation of numerous pancreatic acinar cell signaling cascades and in the onset of early pancreatitis events. Biochim. Biophys. Acta-Mol. Basis Dis. 2016, 1862, 1122–1136. [Google Scholar] [CrossRef]
- Mertens, A.E.; Roovers, R.C.; Collard, J.G. Regulation of Tiam1-Rac signalling. FEBS Lett. 2003, 546, 11–16. [Google Scholar] [CrossRef]
- Heo, J.; Thapar, R.; Campbell, S.L. Recognition and activation of rho GTPases by Vav1 and Vav2 guanine nucleotide exchange factors. Biochemistry 2005, 44, 6573–6585. [Google Scholar] [CrossRef] [PubMed]
- Kukimoto-Niino, M.; Tsuda, K.; Ihara, K.; Mishima-Tsumagari, C.; Honda, K.; Ohsawa, N.; Shirouzu, M. Structural Basis for the Dual Substrate Specificity of DOCK7 Guanine Nucleotide Exchange Factor. Structure 2019, 27, 741–748. [Google Scholar] [CrossRef]
- Chen, Q.A.; Zhu, Y.C.; Yu, J.; Miao, S.; Zheng, J.; Xu, L.; Zhou, Y.; Li, D.; Zhang, C.; Tao, J.; et al. CDKL5, a Protein Associated with Rett Syndrome, Regulates Neuronal Morphogenesis via Rac1 Signaling. J. Neurosci. 2010, 30, 12777–12786. [Google Scholar] [CrossRef] [PubMed]
- Welch, H.C. Regulation and function of P-Rex family Rac-GEFs. Small GTPases 2015, 6, 49–70. [Google Scholar] [CrossRef] [PubMed]
- Offenhauser, N.; Borgonovo, A.; Disanza, A.; Romano, P.; Ponzanelli, I.; Iannolo, G.; Di Fiore, P.P.; Scita, G. The eps8 family of proteins links growth factor stimulation to actin reorganization generating functional redundancy in the Ras/Rac pathway. Mol. Biol. Cell 2004, 15, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Makkar, G.; Strickland, D.K.; Blanpied, T.A.; Stumpo, D.J.; Blackshear, P.J.; Sarkar, R.; Monahan, T.S. Myristoylated Alanine-Rich Protein Kinase Substrate (MARCKS) Regulates Small GTPase Rac1 and Cdc42 Activity and Is a Critical Mediator of Vascular Smooth Muscle Cell Migration in Intimal Hyperplasia Formation. J. Am. Heart Assoc. 2015, 4, e002255. [Google Scholar] [CrossRef]
- Nola, S.; Sebbagh, M.; Marchetto, S.; Osmani, N.; Nourry, C.; Audebert, S.; Navarro, C.; Rachel, R.; Montcouquiol, M.; Sans, N.; et al. Scrib regulates PAK activity during the cell migration process. Hum. Mol. Genet. 2008, 17, 3552–3565. [Google Scholar] [CrossRef]
- Shirafuji, T.; Ueyama, T.; Yoshino, K.; Takahashi, H.; Adachi, N.; Ago, Y.; Koda, K.; Nashida, T.; Hiramatsu, N.; Matsuda, T.; et al. The Role of Pak-Interacting Exchange Factor-beta Phosphorylation at Serines 340 and 583 by PKC gamma in Dopamine Release. J. Neurosci. 2014, 34, 9268–9280. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Li, X.B.; Premont, R.T. Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes. J. Cell Sci. 2016, 129, 1963–1974. [Google Scholar] [CrossRef]
- Webb, D.J.; Mayhew, M.W.; Kovalenko, M.; Schroeder, M.J.; Jeffery, E.D.; Whitmore, L.; Shabanowitz, J.; Hunt, D.F.; Horwitz, A.F. Identification of phosphorylation sites in GIT1. J. Cell Sci. 2006, 119, 2847–2850. [Google Scholar] [CrossRef]
- Tran, C.W.; Saibil, S.D.; Le Bihan, T.; Hamilton, S.R.; Lang, K.S.; You, H.; Lin, A.E.; Garza, K.M.; Elford, A.R.; Tai, K.; et al. Glycogen Synthase Kinase-3 Modulates Cbl-b and Constrains T Cell Activation. J. Immunol. 2017, 199, 4056–4065. [Google Scholar] [CrossRef]
- Richnau, N.; Aspenstrom, P. RICH, a rho GTPase-activating protein domain-containing protein involved in signaling by Cdc42 and Rac1. J. Biol. Chem. 2001, 276, 35060–35070. [Google Scholar] [CrossRef]
- Fortin, S.P.; Ennis, M.J.; Schumacher, C.A.; Zylstra-Diegel, C.R.; Williams, B.O.; Ross, J.T.D.; Winkles, J.A.; Loftus, J.C.; Symons, M.H.; Tran, N.L. Cdc42 and the Guanine Nucleotide Exchange Factors Ect2 and Trio Mediate Fn14-Induced Migration and Invasion of Glioblastoma Cells. Mol. Cancer Res. 2012, 10, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T. Polarization and myelination in myelinating glia. ISRN Neurol. 2012, 2012, 769412. [Google Scholar] [CrossRef]
- Chen, Y.; Liang, Z.Y.; Fei, E.K.; Chen, Y.W.; Zhou, X.P.; Fang, W.Q.; Fu, W.Y.; Fu, A.K.Y.; Ip, N.Y. Axin Regulates Dendritic Spine Morphogenesis through Cdc42-Dependent Signaling. PLoS ONE 2015, 10, e0133115. [Google Scholar] [CrossRef] [PubMed]
- Brudvig, J.J.; Cain, J.T.; Sears, R.M.; Schmidt-Grimminger, G.G.; Wittchen, E.S.; Adler, K.B.; Ghashghaei, H.T.; Weimer, J.M. MARCKS regulates neuritogenesis and interacts with a CDC42 signaling network. Sci. Rep. 2018, 8, 13278. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, A.J.; Calvo, F. The Borg family of Cdc42 effector proteins Cdc42EP1-5. Biochem. Soc. Trans. 2016, 44, 1709–1716. [Google Scholar] [CrossRef]
- Boissier, P.; Huynh-Do, U. The guanine nucleotide exchange factor Tiam1: A Janus-faced molecule in cellular signaling. Cell Signal. 2014, 26, 483–491. [Google Scholar] [CrossRef]
- Lai, F.P.L.; Szczodrak, M.; Oelkers, J.M.; Ladwein, M.; Acconcia, F.; Benesch, S.; Auinger, S.; Faix, J.; Small, J.V.; Polo, S.; et al. Cortactin Promotes Migration and Platelet-derived Growth Factor-induced Actin Reorganization by Signaling to Rho-GTPases. Mol. Biol. Cell 2009, 20, 3209–3223. [Google Scholar] [CrossRef]
- Eiseler, T.; Wille, C.; Koehler, C.; Illing, A.; Seufferlein, T. Protein Kinase D2 Assembles a Multiprotein Complex at the Trans-Golgi Network to Regulate Matrix Metalloproteinase Secretion. J. Biol. Chem. 2016, 291, 462–477. [Google Scholar] [CrossRef]
- Boulay, P.L.; Cotton, M.; Melancon, P.; Claing, A. ADP-ribosylation Factor 1 Controls the Activation of the Phosphatidylinositol 3-Kinase Pathway to Regulate Epidermal Growth Factor-dependent Growth and Migration of Breast Cancer Cells. J. Biol. Chem. 2008, 283, 36425–36434. [Google Scholar] [CrossRef]
- Schoppe, J.; Schubert, E.; Apelbaum, A.; Yavavli, E.; Birkholz, O.; Stephanowitz, H.; Han, Y.P.; Perz, A.; Hofnagel, O.; Liu, F.; et al. Flexible open conformation of the AP-3 complex explains its role in cargo recruitment at the Golgi. J. Biol. Chem. 2021, 297, 101334. [Google Scholar] [CrossRef]
- Piccini, A.; Castroflorio, E.; Valente, P.; Guarnieri, F.C.; Aprile, D.; Michetti, C.; Bramini, M.; Giansante, G.; Pinto, B.; Savardi, A.; et al. APache Is an AP2-Interacting Protein Involved in Synaptic Vesicle Trafficking and Neuronal Development. Cell Rep. 2017, 21, 3596–3611. [Google Scholar] [CrossRef] [PubMed]
- Schürmann, B.; Bermingham, D.P.; Kopeikina, K.J.; Myczek, K.; Yoon, S.; Horan, K.E.; Kelly, C.J.; Martin-de-Saavedra, M.D.; Forrest, M.P.; Fawcett-Patel, J.M.; et al. A novel role for the late-onset Alzheimer’s disease (LOAD)-associated protein Bin1 in regulating postsynaptic trafficking and glutamatergic signaling. Mol. Psychiatry 2020, 25, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Monetta, P.; Slavin, F.; Romero, N.; Alvarez, C. Rab1b interacts with GBF1 and modulates both ARR dynamics and COPI association. Mol. Biol. Cell 2007, 18, 2400–2410. [Google Scholar] [CrossRef] [PubMed]
- Sztul, E.; Chen, P.W.; Casanova, J.E.; Cherfils, J.; Decks, J.B.; Lambright, D.G.; Lee, F.J.S.; Randazzo, P.A.; Santy, L.C.; Schurmann, A.; et al. ARF GTPases and their GEFs and GAPs: Concepts and challenges. Mol. Biol. Cell 2019, 30, 1249–1271. [Google Scholar] [CrossRef] [PubMed]
- Villarroel-Campos, D.; Bronfman, F.C.; Gonzalez-Billault, C. Rab GTPase Signaling in Neurite Outgrowth and Axon Specification. Cytoskeleton 2016, 73, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Xiaofeng, D.; Shuanglin, X. Endocytosis and human innate immunity. J. Immunol. Sci. 2018, 2, 65–70. [Google Scholar]
- Lohr, N.L. Collateral development: The quest continues. Circ. Res. 2014, 114, 591–593. [Google Scholar] [CrossRef]
- Cezanne, A.; Lauer, J.; Solomatina, A.; Sbalzarini, I.F.; Zerial, M. A non-linear system patterns Rab5 GTPase on the membrane. eLife 2020, 9, e54434. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, K.; Lv, X.L.; Lan, Y.G.; Hu, S.Y.; Shi, J.C.; Guan, J.Y.; Yang, Y.W.; Lu, H.J.; He, H.B.; et al. Ulk1 Governs Nerve Growth Factor/TrkA Signaling by Mediating Rab5 GTPase Activation in Porcine Hemagglutinating Encephalomyelitis Virus-Induced Neurodegenerative Disorders. J. Virol. 2018, 92, e00325-18. [Google Scholar] [CrossRef]
- Lyon, A.M.; Dutta, S.; Boguth, C.A.; Skiniotis, G.; Tesmer, J.J.G. Full-length G alpha(q)-phospholipase C-beta 3 structure reveals interfaces of the C-terminal coiled-coil domain. Nat. Struct. Mol. Biol. 2013, 20, 355–362. [Google Scholar] [CrossRef]
- van den Eshof, B.L.; Hoogendijk, A.J.; Simpson, P.J.; van Alphen, F.P.J.; Zanivan, S.; Mertens, K.; Meijer, A.B.; van den Biggelaar, M. Paradigm of Biased PAR1 (Protease-Activated Receptor-1) Activation and Inhibition in Endothelial Cells Dissected by Phosphoproteomics. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1891–1902. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, E.; Christoforidis, S.; Uttenweiler-Joseph, S.; Miaczynska, M.; Dewitte, F.; Wilm, M.; Hoflack, B.; Zerial, M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 2000, 151, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Eathiraj, S.; Pan, X.J.; Ritacco, C.; Lambright, D.G. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature 2005, 436, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.D.; Donaldson, J.G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2009, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Linford, A.; Yoshimura, S.; Bastos, R.N.; Langemeyer, L.; Gerondopoulos, A.; Rigden, D.J.; Barr, F.A. Rab14 and Its Exchange Factor FAM116 Link Endocytic Recycling and Adherens Junction Stability in Migrating Cells. Dev. Cell 2012, 22, 952–966. [Google Scholar] [CrossRef]
- Chaineau, M.; Ioannou, M.S.; McPherson, P.S. Rab35: GEFs, GAPs and Effectors. Traffic 2013, 14, 1109–1117. [Google Scholar] [CrossRef]
- Fuchs, E.; Haas, A.K.; Spooner, R.A.; Yoshimura, S.I.; Lord, J.M.; Barr, F.A. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 2007, 177, 1133–1143. [Google Scholar] [CrossRef]
- Gallo, L.I.; Liao, Y.; Ruiz, W.G.; Clayton, D.R.; Li, M.; Liu, Y.J.; Jiang, Y.; Fukuda, M.; Apodaca, G.; Yin, X.M. TBC1D9B functions as a GTPase-activating protein for Rab11a in polarized MDCK cells. Mol. Biol. Cell 2014, 25, 3779–3797. [Google Scholar] [CrossRef]
- Spearman, P. Viral interactions with host cell Rab GTPases. Small GTPases 2018, 9, 192–201. [Google Scholar] [CrossRef]
- Tzeng, H.T.; Wang, Y.C. Rab-mediated vesicle trafficking in cancer. J. Biomed. Sci. 2016, 23, 70. [Google Scholar] [CrossRef]
- Elbaz-Alon, Y.; Guo, Y.T.; Segev, N.; Harel, M.; Quinnell, D.E.; Geiger, T.; Avinoam, O.; Li, D.; Nunnari, J. PDZD8 interacts with Protrudin and Rab7 at ER-late endosome membrane contact sites associated with mitochondria. Nat. Commun. 2020, 11, 3645. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Fukuda, M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013, 14, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Nassari, S.; Del Olmo, T.; Jean, S. Rabs in Signaling and Embryonic Development. Int. J. Mol. Sci. 2020, 21, 1064. [Google Scholar] [CrossRef] [PubMed]
- Zachari, M.; Ganley, I.G. The mammalian ULK1 complex and autophagy initiation. Signal. Mech. Autophagy 2017, 61, 585–596. [Google Scholar]
- Wang, C.Y.; Wang, H.F.; Zhang, D.Y.; Luo, W.W.; Liu, R.L.; Xu, D.Q.; Diao, L.; Liao, L.J.; Liu, Z.X. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy. Nat. Commun. 2018, 9, 3492. [Google Scholar] [CrossRef]
- Marat, A.L.; Dokainish, H.; McPherson, P.S. DENN Domain Proteins: Regulators of Rab GTPases. J. Biol. Chem. 2011, 286, 13791–13800. [Google Scholar] [CrossRef]
- Kiral, F.R.; Kohrs, F.E.; Jin, E.J.; Hiesinger, P.R. Rab GTPases and Membrane Trafficking in Neurodegeneration. Curr. Biol. 2018, 28, R471–R486. [Google Scholar] [CrossRef]
- Ghelfi, E.; Grondin, Y.; Millet, E.J.; Bartos, A.; Bortoni, M.; dos Santos, C.O.G.; Trevino-Villarreal, H.J.; Sepulveda, R.; Rogers, R. In vitro gentamicin exposure alters caveolae protein profile in cochlear spiral ligament pericytes. Proteome Sci. 2018, 16, 7. [Google Scholar] [CrossRef]
- Morgan, N.E.; Cutrona, M.B.; Simpson, J.C. Multitasking Rab Proteins in Autophagy and Membrane Trafficking: A Focus on Rab33b. Int. J. Mol. Sci. 2019, 20, 3916. [Google Scholar] [CrossRef]
- Takahashi, T.; Minami, S.; Tsuchiya, Y.; Tajima, K.; Sakai, N.; Suga, K.; Hisanaga, S.; Ohbayashi, N.; Fukuda, M.; Kawahara, H. Cytoplasmic control of Rab family small GTPases through BAG6. EMBO Rep. 2019, 20, e46794. [Google Scholar] [CrossRef]
- Ceresa, B.P. Regulation of EGFR endocytic trafficking by rab proteins. Histol. Histopathol. 2006, 21, 987–993. [Google Scholar]
- Hermle, T.; Schneider, R.; Schapiro, D.; Braun, D.A.; van der Ven, A.T.; Warejko, J.K.; Daga, A.; Widmeier, E.; Nakayama, M.; Jobst-Schwan, T.; et al. GAPVD1 and ANKFY1 Mutations Implicate RAB5 Regulation in Nephrotic Syndrome. J. Am. Soc. Nephrol. 2018, 29, 2123–2138. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Henriques, M.; Anton, V. Mitochondrial Surveillance by Cdc48/p97: MAD vs. Membrane Fusion. Int. J. Mol. Sci. 2020, 21, 6841. [Google Scholar] [CrossRef] [PubMed]
- D’Aloia, A.; Berruti, G.; Costa, B.; Schiller, C.; Ambrosini, R.; Pastori, V.; Martegani, E.; Ceriani, M. RalGPS2 is involved in tunneling nanotubes formation in 5637 bladder cancer cells. Exp. Cell Res. 2018, 362, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Rebhun, J.F.; Chen, H.S.; Quilliam, L.A. Identification and characterization of a new family of guanine nucleotide exchange factors for the Ras-related GTPase Ral. J. Biol. Chem. 2000, 275, 13406–13410. [Google Scholar] [CrossRef]
- Cornish, J.; Owen, D.; Mott, H.R. RLIP76: A Structural and Functional Triumvirate. Cancers 2021, 13, 2206. [Google Scholar] [CrossRef]
- Herlevsen, M.C.; Theodorescu, D. Mass spectroscopic phosphoprotein mapping of Ral binding protein 1 (RalBP1/Rip1/RLIP76). Biochem. Biophys. Res. Commun. 2007, 362, 56–62. [Google Scholar] [CrossRef][Green Version]
- Koo, T.H.; Eipper, B.A.; Donaldson, J.G. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 2007, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Osmani, N.; Peglion, F.; Chavrier, P.; Etienne-Manneville, S. Cdc42 localization and cell polarity depend on membrane traffic. J. Cell Biol. 2010, 191, 1261–1269. [Google Scholar] [CrossRef]
- Conner, S.D.; Schmid, S.L. Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J. Cell Biol. 2002, 156, 921–929. [Google Scholar] [CrossRef]
- Lee, D.W.; Wu, X.F.; Eisenberg, E.; Greene, L.E. Recruitment dynamics of GAK and auxilin to clathrin-coated pits during endocytosis. J. Cell Sci. 2006, 119, 3502–3512. [Google Scholar] [CrossRef] [PubMed]
- Ricotta, D.; Conner, S.D.; Schmid, S.L.; von Figura, K.; Honing, S. Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 2002, 156, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Jullien-Flores, V.; Mahe, Y.; Mirey, G.; Leprince, C.; Meunier-Bisceuil, B.; Sorkin, A.; Camonis, J.H. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: Involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 2000, 113, 2837–2844. [Google Scholar] [CrossRef] [PubMed]
- Fillatre, J.; Delacour, D.; Van Hove, L.; Bagarre, T.; Houssin, N.; Soulika, M.; Veitia, R.A.; Moreau, J. Dynamics of the subcellular localization of RalBP1/RLIP through the cell cycle: The role of targeting signals and of protein-protein interactions. FASEB J. 2012, 26, 2164–2174. [Google Scholar] [CrossRef] [PubMed]
- Personnic, N.; Lakisic, G.; Gouin, E.; Rousseau, A.; Gautreau, A.; Cossart, P.; Bierne, H. A role for Ral GTPase-activating protein subunit beta in mitotic regulation. FEBS J. 2014, 281, 2977–2989. [Google Scholar] [CrossRef]
- Matunis, M.J.; Wu, J.A.; Blobel, G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 1998, 140, 499–509. [Google Scholar] [CrossRef]
- Dorr, A.; Pierre, S.; Zhang, D.D.; Henke, M.; Holland, S.; Scholich, K. MYCBP2 Is a Guanosine Exchange Factor for Ran Protein and Determines Its Localization in Neurons of Dorsal Root Ganglia. J. Biol. Chem. 2015, 290, 25620–25635. [Google Scholar] [CrossRef]
- Yoon, S.O.; Shin, S.; Liu, Y.; Ballif, B.A.; Woo, M.S.; Gygi, S.P.; Blenis, J. Ran-binding protein 3 phosphorylation links the Ras and PI3-kinase pathways to nucleocytoplasmic transport. Mol. Cell 2008, 29, 362–375. [Google Scholar] [CrossRef]
- Bischoff, F.R.; Klebe, C.; Kretschmer, J.; Wittinghofer, A.; Ponstingl, H. RanGAP1 induced GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 1994, 91, 2587–2591. [Google Scholar] [CrossRef]
- He, Y.J.; Yang, Z.G.; Zhao, C.S.; Xiao, Z.H.; Gong, Y.; Li, Y.Y.; Chen, Y.Q.; Du, Y.T.; Feng, D.Y.; Altman, A.; et al. T-cell receptor (TCR) signaling promotes the assembly of RanBP2/RanGAP1-SUMO1/Ubc9 nuclear pore subcomplex via PKC-theta-mediated phosphorylation of RanGAP1. eLife 2021, 10, e67123. [Google Scholar] [CrossRef]
- Sanz-Garcia, M.; Lopez-Sanchez, I.; Lazo, P.A. Proteomics Identification of Nuclear Ran GTPase as an Inhibitor of Human VRK1 and VRK2 (Vaccinia-related Kinase) Activities. Mol. Cell. Prot. 2008, 7, 2199–2214. [Google Scholar] [CrossRef] [PubMed]
- Radha, V.; Mitra, A.; Dayma, K.; Sasikumar, K. Signalling to actin: Role of C3G, a multitasking guanine-nucleotide-exchange factor. Biosci. Rep. 2011, 31, 231–244. [Google Scholar] [CrossRef]
- Chen, X.; Shibata, A.C.E.; Hendi, A.; Kurashina, M.; Fortes, E.; Weilinger, N.L.; MacVicar, B.A.; Murakoshi, H.; Mizumoto, K. Rap2 and TNIK control Plexin-dependent tiled synaptic innervation in C. elegans. eLife 2018, 7, e38801. [Google Scholar] [CrossRef]
- Smolen, G.A.; Schott, B.J.; Stewart, R.A.; Diederichs, S.; Muir, B.; Provencher, H.L.; Look, A.T.; Sgroi, D.C.; Peterson, R.T.; Haber, D.A. A Rap GTPase interactor, RADIL, mediates migration of neural crest precursors. Gen. Dev. 2007, 21, 2131–2136. [Google Scholar] [CrossRef] [PubMed]
- Sequera, C.; Manzano, S.; Guerrero, C.; Porras, A. How Rap and its GEFs control liver physiology and cancer development. C3G alterations in human hepatocarcinoma. Hepatic Oncol. 2018, 5, HEP05. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Pan, D.X.; Welch, H.C.E. Small GTPases and their guanine-nucleotide exchange factors and GTPase-activating proteins in neutrophil recruitment. Curr. Opin. Hematol. 2016, 23, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Schultess, J.; Danielewski, O.; Smolenski, A.P. Rap1GAP2 is a new GTPase-activating protein of Rap1 expressed in human platelets. Blood 2005, 105, 3185–3192. [Google Scholar] [CrossRef]
- Park, Y.G.; Zhao, X.H.; Lesueur, F.; Lowy, D.R.; Lancaster, M.; Pharoah, P.; Qian, X.L.; Hunter, K.W. Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat. Genet. 2005, 37, 1055–1062. [Google Scholar] [CrossRef]
- Rosano, L.; Cianfrocca, R.; Masi, S.; Spinella, F.; Di Castro, V.; Biroccio, A.; Salvati, E.; Nicotra, M.R.; Natali, P.G.; Bagnato, A. beta-Arrestin links endothelin A receptor to beta-catenin signaling to induce ovarian cancer cell invasion and metastasis. Proc. Natl. Acad. Sci. USA 2009, 106, 2806–2811. [Google Scholar] [CrossRef]
- Eisemann, T.; McCauley, M.; Langelier, M.F.; Gupta, K.; Roy, S.; Van Duyne, G.D.; Pascal, J.M. Tankyrase-1 Ankyrin Repeats Form an Adaptable Binding Platform for Targets of ADP-Ribose Modification. Structure 2016, 24, 1679–1692. [Google Scholar] [CrossRef]
- Murayama, M.; Tanaka, S.; Palacino, J.; Murayama, O.; Honda, T.; Sun, X.Y.; Yasutake, K.; Nihonmatsu, N.; Wolozin, B.; Takashima, A. Direct association of presenilin-1 with beta-catenin. FEBS Lett. 1998, 433, 73–77. [Google Scholar] [CrossRef]
- Lee, G.; Goretsky, T.; Managlia, E.; Dirisina, R.; Singh, A.P.; Brown, J.B.; May, R.; Yang, G.Y.; Ragheb, J.W.; Evers, B.M.; et al. Phosphoinositide 3-Kinase Signaling Mediates beta-Catenin Activation in Intestinal Epithelial Stem and Progenitor Cells in Colitis. Gastroenterology 2010, 139, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Tan, J.; Li, J.S.; Kivimaee, S.; Yang, X.J.; Zhuang, L.; Lee, P.L.; Chan, M.T.W.; Stanton, L.W.; Liu, E.T.; et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 2008, 13, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Sastre-Perona, A.; Riesco-Eizaguirre, G.; Zaballos, M.A.; Santisteban, P. beta-catenin signaling is required for RAS-driven thyroid cancer through PI3K activation. Oncotarget 2016, 7, 49435–49449. [Google Scholar] [CrossRef]
- Mariotti, L.; Pollock, K.; Guettler, S. Regulation of Wnt/beta-catenin signalling by tankyrase-dependent poly(ADP-ribosyl) ation and scaffolding. Br. J. Pharmacol. 2017, 174, 4611–4636. [Google Scholar] [CrossRef]
- Fang, D.X.; Hawke, D.; Zheng, Y.H.; Xia, Y.; Meisenhelder, J.; Nika, H.; Mills, G.B.; Kobayashi, R.; Hunter, T.; Lu, Z.M. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J. Biol. Chem. 2007, 282, 11221–11229. [Google Scholar] [CrossRef]
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 2015, 519, 106–109. [Google Scholar] [CrossRef]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef]
- van Gastel, J.; Hendrickx, J.O.; Leysen, H.; Santos-Otte, P.; Luttrell, L.M.; Martin, B.; Maudsley, S. beta-Arrestin Based Receptor Signaling Paradigms: Potential Therapeutic Targets for Complex Age-Related Disorders. Front. Pharmacol. 2018, 9, 1369. [Google Scholar] [CrossRef]
- Jones, B.W.; Hinkle, P.M. Arrestin binds to different phosphorylated regions of the thyrotropin-releasing hormone receptor with distinct functional consequences. Mol. Pharmacol. 2008, 74, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Oride, A.; Kanasaki, H.; Mutiara, S.; Purwana, I.N.; Miyazaki, K. Activation of extracellular signal-regulated kinase (ERK) and induction of mitogen-activated protein kinase phosphatase 1 (MKP-1) by perifused thyrotropin-releasing hormone (TRH) stimulation in rat pituitary GH3 cells. Mol. Cell. Endocrinol. 2008, 296, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Sibilski, C.; Mueller, T.; Kollipara, L.; Zahedi, R.P.; Rapp, U.R.; Rudel, T.; Baljuls, A. Tyr(728) in the Kinase Domain of the Murine Kinase Suppressor of RAS 1 Regulates Binding and Activation of the Mitogen-activated Protein Kinase Kinase. J. Biol. Chem. 2013, 288, 35237–35252. [Google Scholar] [CrossRef] [PubMed]
- McKay, M.M.; Ritt, D.A.; Morrison, D.K. Signaling dynamics of the KSR1 scaffold complex. Proc. Natl. Acad. Sci. USA 2009, 106, 11022–11027. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.C.; Wang, X.H.; Tan, T.H. MAP4K Family Kinases in Immunity and Inflammation. Adv. Immunol. 2016, 129, 277–314. [Google Scholar] [PubMed]
- Gurevich, V.V.; Gurevich, E.V. Arrestin-mediated signaling: Is there a controversy? World J. Biol. Chem. 2018, 9, 25–35. [Google Scholar] [CrossRef]
- Bourquard, T.; Landomiel, F.; Reiter, E.; Crepieux, P.; Ritchie, D.W.; Aze, J.; Poupon, A. Unraveling the molecular architecture of a G protein-coupled receptor/beta-arrestin/Erk module complex. Sci. Rep. 2015, 5, 10760. [Google Scholar] [CrossRef]
- Claing, A. beta-Arrestins: Modulators of Small GTPase Activation and Function. Mol. Biol. Arrestins 2013, 118, 149–174. [Google Scholar]
- Du, R.W.; Du, R.H.; Bu, W.G. beta-arrestin 2 mediates the anti-inflammatory effects of fluoxetine in lipopolysaccharide-stimulated microglial cells. J. Neuroimmune Pharmacol. 2014, 9, 582–590. [Google Scholar] [CrossRef]
- Arakaki, A.K.S.; Pan, W.A.; Wedegaertner, H.; Roca-Mercado, I.; Chinn, L.; Gujral, T.S.; Trejo, J.A. α-Arrestin ARRDC3 tumor suppressor function is linked to GPCR-induced TAZ activation and breast cancer metastasis. J. Cell Sci. 2021, 134, jcs254888. [Google Scholar] [CrossRef]
- Wang, Y.H.; Jue, S.F.; Maurer, R.A. Thyrotropin-releasing hormone stimulates phosphorylation of the epidermal growth factor receptor in GH(3) pituitary cells. Mol. Endocrinol. 2000, 14, 1328–1337. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, L.G.; Yano, N.; Luo, J.Z.Q. The molecular mechanism of EGF receptor activation in pancreatic beta-cells by thyrotropin-releasing hormone. Am. J. Physiol.-Endocrinol. Metabol. 2006, 290, E889–E899. [Google Scholar] [CrossRef] [PubMed]
- Collazos, A.; Diouf, B.; Guerineau, N.C.; Quittau-Prevostel, C.; Peter, M.; Coudane, F.; Hollande, F.; Joubert, D. A spatiotemporally coordinated cascade of protein kinase C activation controls isoform-selective translocation. Mol. Cell. Biol. 2006, 26, 2247–2261. [Google Scholar] [CrossRef] [PubMed]
- Lampe, M.; Vassilopoulos, S.; Merrifield, C. Clathrin coated pits, plaques and adhesion. J. Struct. Biol. 2016, 196, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Thirunarayanan, N.; Nir, E.A.; Raaka, B.M.; Gershengorn, M.C. Thyrotropin-Releasing Hormone Receptor Type 1 (TRH-R1), not TRH-R2, Primarily Mediates Taltirelin Actions in the CNS of Mice. Neuropsychopharmacology 2013, 38, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Cue, C.; Rueda, N. Signalling Pathways Implicated in Alzheimer’s Disease Neurodegeneration in Individuals with and without Down Syndrome. Int. J. Mol. Sci. 2020, 21, 6906. [Google Scholar] [CrossRef] [PubMed]
- Cherubini, M.; Wade-Martins, R. Convergent pathways in Parkinson’s disease. Cell Tissue Res. 2018, 373, 79–90. [Google Scholar] [CrossRef]
- Bohush, A.; Niewiadomska, G.; Filipek, A. Role of Mitogen Activated Protein Kinase Signaling in Parkinson’s Disease. Int. J. Mol. Sci. 2018, 19, 2973. [Google Scholar] [CrossRef]
- Fawdar, S.; Trotter, E.W.; Li, Y.Y.; Stephenson, N.L.; Hanke, F.; Marusiak, A.A.; Edwards, Z.C.; Ientile, S.; Waszkowycz, B.; Miller, C.J.; et al. Targeted genetic dependency screen facilitates identification of actionable mutations in FGFR4, MAP3K9, and PAK5 in lung cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 12426–12431. [Google Scholar] [CrossRef]
- Kahle, M.P.; Cuevas, B.D. Interaction with the Paxillin LD1 Motif Relieves MEKK2 Auto-inhibition. J. Mol. Signal. 2015, 10, 4. [Google Scholar] [CrossRef][Green Version]
- Stark, M.S.; Woods, S.L.; Gartside, M.G.; Bonazzi, V.F.; Dutton-Regester, K.; Aoude, L.G.; Chow, D.; Sereduk, C.; Niemi, N.M.; Tang, N.Y.; et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat. Genet. 2012, 44, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, H.; Opoku-Ansah, J.; Pastorino, S.; Renganathan, H.; Matter, M.L.; Ramos, J.W. ERK MAP kinase is targeted to RSK2 by the phosphoprotein PEA-15. Proc. Natl. Acad. Sci. USA 2007, 104, 19837–19842. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, E.; Hu, H.; Yuzwa, S.A.; Hernandez-Miranda, L.R.; Kraemer, N.; Ninnemann, O.; Musante, L.; Boltshauser, E.; Schindler, D.; Hubner, A.; et al. Homozygous ARHGEF2 mutation causes intellectual disability and midbrain-hindbrain malformation. PLoS Genet. 2017, 13, e1006746. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Cieply, B.; Yang, Y.Q.; Peart, N.; Glaser, C.; Chan, P.; Carstens, R.P. Esrp1-Regulated Splicing of Arhgef11 Isoforms Is Required for Epithelial Tight Junction Integrity. Cell Rep. 2018, 25, 2417–2430. [Google Scholar] [CrossRef] [PubMed]
- Rykx, A.; De Kimpe, L.; Mikhalap, S.; Vantus, T.; Seufferlein, T.; Vandenheede, J.R.; Van Lint, J. Protein kinase D: A family affair. FEBS Lett. 2003, 546, 81–86. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drastichova, Z.; Trubacova, R.; Novotny, J. β-Arrestin2 Is Critically Involved in the Differential Regulation of Phosphosignaling Pathways by Thyrotropin-Releasing Hormone and Taltirelin. Cells 2022, 11, 1473. https://doi.org/10.3390/cells11091473
Drastichova Z, Trubacova R, Novotny J. β-Arrestin2 Is Critically Involved in the Differential Regulation of Phosphosignaling Pathways by Thyrotropin-Releasing Hormone and Taltirelin. Cells. 2022; 11(9):1473. https://doi.org/10.3390/cells11091473
Chicago/Turabian StyleDrastichova, Zdenka, Radka Trubacova, and Jiri Novotny. 2022. "β-Arrestin2 Is Critically Involved in the Differential Regulation of Phosphosignaling Pathways by Thyrotropin-Releasing Hormone and Taltirelin" Cells 11, no. 9: 1473. https://doi.org/10.3390/cells11091473
APA StyleDrastichova, Z., Trubacova, R., & Novotny, J. (2022). β-Arrestin2 Is Critically Involved in the Differential Regulation of Phosphosignaling Pathways by Thyrotropin-Releasing Hormone and Taltirelin. Cells, 11(9), 1473. https://doi.org/10.3390/cells11091473





