Abstract
Post-transcriptional control of gene expression is one important mechanism that enables stringent and rapid modulation of cytokine, chemokines or growth factors expression, all relevant for immune or tumor cell function and communication. The RNA-binding protein KH-type splicing regulatory protein (KSRP) controls the mRNA stability of according genes by initiation of mRNA decay and inhibition of translation, and by enhancing the maturation of microRNAs. Therefore, KSRP plays a pivotal role in immune cell function and tumor progression. In this review, we summarize the current knowledge about KSRP with regard to the regulation of immunologically relevant targets, and the functional role of KSRP on immune responses and tumorigenesis. KSRP is involved in the control of myeloid hematopoiesis. Further, KSRP-mediated mRNA decay of pro-inflammatory factors is necessary to keep immune homeostasis. In case of infection, functional impairment of KSRP is important for the induction of robust immune responses. In this regard, KSRP seems to primarily dampen T helper cell 2 immune responses. In cancer, KSRP has often been associated with tumor growth and metastasis. In summary, aside of initiation of mRNA decay, the KSRP-mediated regulation of microRNA maturation seems to be especially important for its diverse biological functions, which warrants further in-depth examination.
1. Introduction
To prevent an exaggerated immune response, tight control of the expression of pro-inflammatory mediators, such as cytokines or chemokines, is necessary. Since these are important regulators of immune cell function, rapid and strong expression is required to elicit a rapid response to an invading pathogen. Nevertheless, it is also important to resolve immune responses to counteract tissue destruction and to prevent autoimmune responses. Gene expression can be controlled by transcriptional and posttranscriptional mechanisms, and the latter comprise regulation of mRNA decay and translation efficiency. The 3-untranslated region (3′-UTR) of mRNA represents an important element in the post-transcriptional regulation of inflammatory cytokines/chemokines by RNA-binding proteins (RPBs) and micro (mi)RNA. In general, RPBs exert rather a stabilizing (human antigen R, HuR) or destabilizing (Tristetraprolin, TTP/KH-type splicing protein, K(H)SRP) effect on mRNA transcript half-life, and may affect its translational efficacy (e.g., T-Cell-Restricted Intracellular Antigen-1) [1,2].
KSRP (K homology [KH]-type splicing protein, KHSRP) is a single-stranded nucleic acid-binding protein which interacts with target RNA species in nuclear and cytoplasmic cell compartments [3]. In humans, the KSRP gene is located on chromosome 19p13.3, contains 21 exons, one transcript is listed in the RefSeq database (ENST00000600480.2), and encodes the 747 amino acid (aa) KSRP protein, as investigated in the literature. Some hints from the database indicate that additional transcripts may exist, but nothing is known about their biological significance.
The murine gene (ENSMUSG00000007670) is located on chromosome 17 [4] and contains 19 exons. However, the protein sequence of KSRP (in both species 747 aa) is highly conserved between both species. KSRP was first described in 1996 by Levens laboratory and categorized as a member of the far upstream element (FUSE)-binding protein (FBP) family, and was originally named FBP2 [5]. In addition to KSRP, two other members, named FBP1 and FBP3, belong to that family. FBP1 is involved in different cellular processes by regulating transcription, splicing and translation of target genes [6], whereas the biological function of FBP3 remains largely unknown [7].
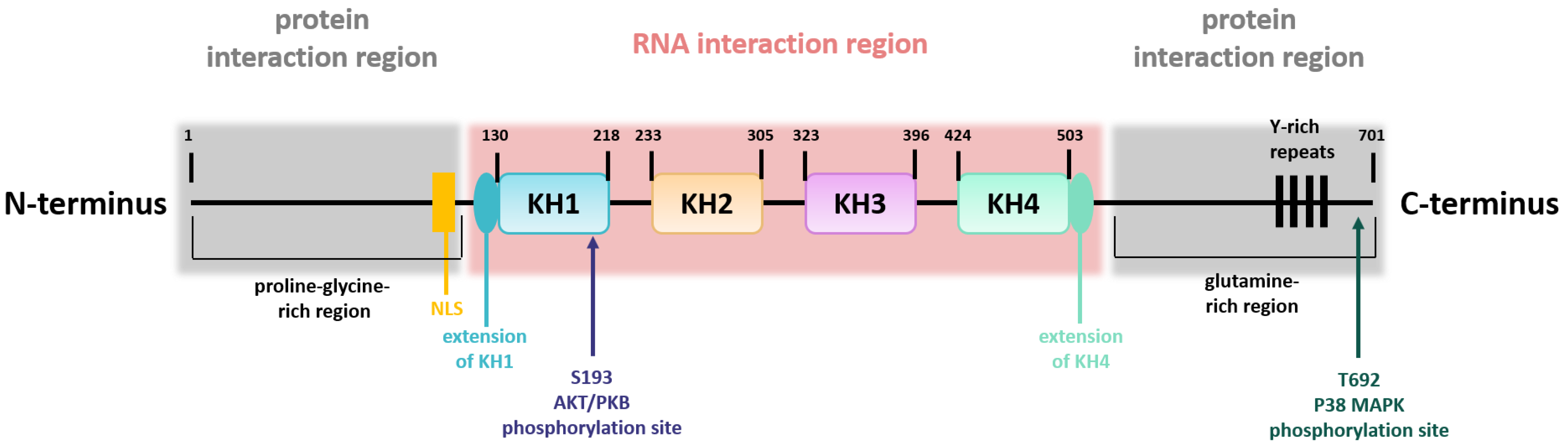
The structure of the KSRP/FBP2 protein can be divided into a central region that is flanked on either side by one additional region (Figure 1) [8]. The central region contains four KH domains mediating the interaction with single-stranded nucleic acids [9]. Whereas KH domain 1, 2, and 4 bind to a large number of sequence motifs with moderate selectivity, KH domain 3 preferably binds to G-containing sequences. In comparison with the central region, the N- and C-termini are regions with low complexity and are post-translationally modified [8]. The N-terminal part comprises a proline-glycine-rich region, whereas the C-terminal part contains a glutamine-rich region, and both contain elements for protein interaction [5,8]. Moreover, the N-terminal part contains the nuclear localization signal, and the C-terminus harbors four Y-rich repeats. Thus, the structure of KSRP is important for its flexibility in terms of target gene binding. KSRP activity is regulated by phosphorylation and other modifications as outlined below.
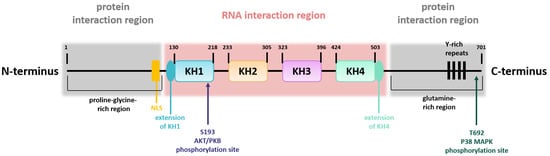
Figure 1.
KSRP structure. Schematic overview of KSRP protein structure, including the domain organization in the central region, involved in RNA interaction, and the two N- and C-terminally flanking regions, which are necessary for protein interaction. Phosphorylation sites are indicated (own illustration inspired by [10]).
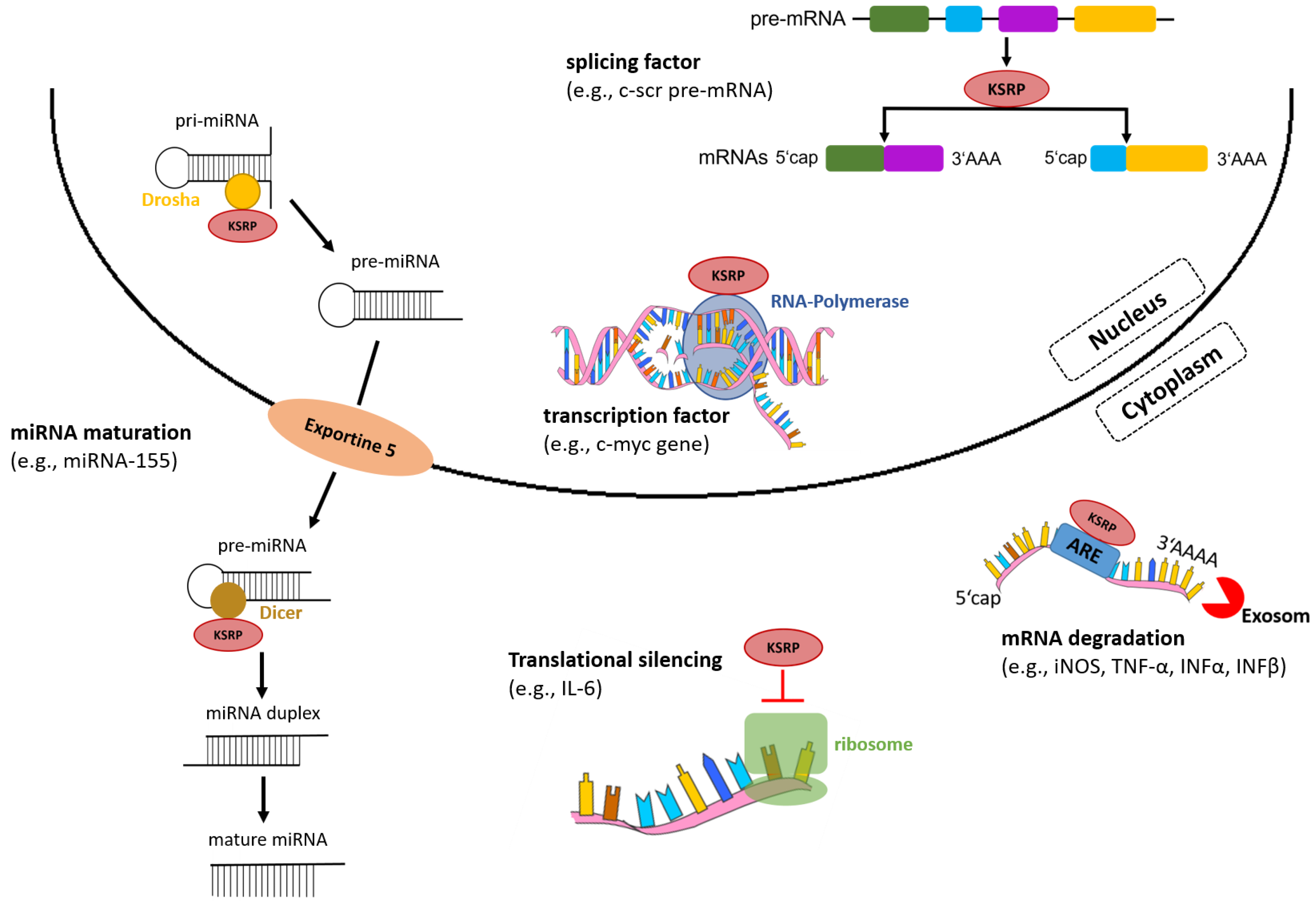
In the nucleus, KSRP acts as a transcription and splicing factor, and in the cytoplasm regulates mRNA stability by promoting its decay and translational silencing (Figure 2) [11]. In addition to regulation of gene expression via post-transcriptional mechanisms, KSRP promotes the maturation of a subset of micro (mi)RNA species, which, in turn, affect expression of multiple genes.
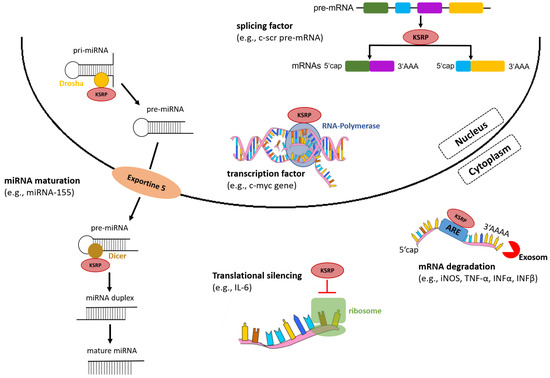
Figure 2.
KSRP regulates gene expression on various levels. In the nucleus, KSRP functions as a transcription and splicing factor, and in the cytoplasm mediates rapid decay of ARE-containing mRNAs by recruiting enzymes and silences translation of mRNAs. Moreover, KSRP promotes miRNA maturation by interacting with the ribonucleases Drosha and Dicer (own illustration inspired by [12]).
This review aims to summarize current knowledge on the multiple roles of KSRP as a regulator of gene expression, with a focus on its emerging importance on the induction and course of innate and adaptive immune responses, in addition to tumorigenesis.
2. KSRP Regulates Gene Expression on Various Levels
2.1. Gene Transcription
In 1996 KSRP was originally identified as a transcription factor of the c-myc oncogene [5]. KSRP binds to FUSE motif, and the four Y-rich regions within the C-terminus of KSRP activate c-myc transcription [8]. One year later the ability of KSRP to act as a pre-mRNA splicing regulatory protein was demonstrated. As a component of a multiprotein complex, KSRP was found to bind to an intronic splicing enhancer element of the proto-oncogene c-src and to regulate the alternative splicing process of c-scr pre-mRNA [13].
2.2. mRNA Level
KSRP plays an important role in different steps of post-transcriptional control of gene expression, including the regulation of mRNA stability and translatability. In addition to modulation of post-transcriptional target gene expression, KSRP also promotes the biogenesis of distinct micro (mi)RNA species, which, in turn, may affect expression of numerous genes as outlined in the following.
2.2.1. mRNA Stability
Many mRNAs of pro-inflammatory mediators that possess AU-rich elements (AREs) in the 3′UTR are targets of KSRP-mediated mRNA decay and are, therefore, often inherently unstable [14]. For example, it has been demonstrated that KSRP decreases stability of the mRNAs encoding tumor necrosis factor-α (TNF-α), interleukin (IL)-8, type I and III interferons (IFN) [15,16] and inducible nitric oxide synthase (iNOS) [17] by binding to ARE in their 3′-UTR. For this, KSRP recruits the exosome multiprotein complex with 3′-5′-endonucleolytic activity and other enzymes involved in mRNA decay, such as the poly (A)-specific ribonuclease (PARN) [18], the deadenylase complex consisting of poly(A) specific ribonuclease subunit 2 and 3 (PAN2/PAN3) [19], and the 5′-3′-exonuclease 1 (XRN1) [20]. Moreover, KSRP recruits mRNA decapping enzymes such as decapping mRNA 2, which activates deadenylation of poly-A-tail of mRNA, and the decapping complex consisting of decapping mRNA 1 and 2 (DCP1/DCP2) [21,22]. KH domains 3 and 4 are necessary for KSRP-mediated mRNA decay, by binding to AREs with high-affinity and interacting with mRNA decay enzymes [9]. Both KH domains act independently of each other, resulting in a broad spectrum of target mRNAs [10]. However, KH domain 3 stabilizes the interaction of KH domain 4 to target mRNAs. Thus, altogether, KSRP seems to be a central component of the ARE-mediated mRNA decay (AMD) [15].
Moreover, in 2000, Lellek and coworkers identified KSRP as a component of the apolipoprotein B mRNA editing enzyme-complex [23]. Another study identified ~100 target mRNAs of KSRP, comprising, e.g., IL-6, IL-8 and Cyclooxygenase-2, whose expression levels were upregulated in KSRP-deficient cells [24]. However, KSRP-dependent mRNA degradation could only be detected in 10% of the ~100 target mRNAs, indicative of additional modes of KSRP-mediated gene regulation.
2.2.2. Translation Efficacy
Furthermore, KSRP not only enhances mRNA degradation, but also silences mRNA translation and consequently impairs expression of, e.g., proinflammatory cytokine or chemokine genes [25]. Dhamija and coworkers compared the polysome profiles of cells with siRNA-mediated KSRP deficiency and control cells. In KSRP-deficient cells there was increased IL-6 protein. KSRP was reported to interact with ARE of IL-6 mRNA and mediate its translational silencing. However, further investigations are necessary, as to date, only Dhamjia and coworkers have identified the ability of KSRP to regulate mRNA translatability.
2.3. miRNA Biogenesis
miRNAs are small non-protein coding RNAs, which play a critical role in post-transcriptional gene regulation as constituents of RNA-induced silencing complexes (RISC) [26]. miRNA inhibit gene expression by binding to target sequences that are located most often in the 3′-UTR of mRNAs [26]. Thereby, they initiate translational repression and/or mRNA cleavage, depending on the degree of sequence homology to the target-binding site [27]. A single miRNA can target multiple transcripts and a single gene can be under the control of multiple miRNAs.
KSRP is important for proper processing of a subset of miRNAs, especially of those that contain a GC-rich stem-loop structure in the immature precursor transcript [28]. KH domain 3 binds selectively towards G-rich sequences, and KSRP interacts with ribonucleases Drosha and Dicer in nucleus and cytoplasm, respectively (Figure 2). In the nucleus, KSRP cleaves pri-miRNA into pre-miRNA. Moreover, it promotes the transport of pre-miRNA into the cytoplasm by interacting with exportine-5. In the cytoplasm, KSRP promotes maturation of pre-miRNA into mature miRNA by binding to the terminal loop of pre-miRNA and interacting with the ribonuclease Dicer [28]. Among those miRNAs whose maturation requires KSRP are miR-155 [29], let-7a [30,31] and miR-129 [32], which exert important functions in the regulation of immune processes as outlined in Table 1. This suggests that KSRP has an important function in immune cell biology that should be evaluated in detail.

Table 1.
Overview of immune cell functions mediated by miRNAs whose maturation is promoted by KSRP.
To summarize, KSRP is a versatile RNA-binding protein (RBP) which modulates gene expression at multiple levels promoted by its structural diversity. At the moment, KSRP-mediated mRNA decay and KSRP-mediated maturation of miRNAs seem to be the most important biological functions of the protein.
3. Regulation of KSRP Activity
3.1. Transcript and mRNA Level
KSRP activity is regulated at the transcriptional and post-transcriptional level. Whereas transcriptional regulation of KSRP gene expression remains largely unexplored, more information about post-transcriptional mechanisms exists. Table 2 presents a short overview about factors that regulate KSRP expression on mRNA level.

Table 2.
Summary of factors that regulate KSRP expression on the post-transcriptional level.
3.2. Protein Level
KSRP activity is also regulated by several post-translational modifications, including phosphorylation and ubiquitination, in addition to interaction with long non coding RNAs (lncRNAs).
3.2.1. Phosphorylation
KSRP contains multiple phosphorylation sites (see Figure 1) that are engaged by various kinases such as p38 mitogen-activated protein kinase (MAPK), protein kinase B (PKB) and ataxia telangiectasia mutated (ATM) kinase, thereby regulating KSRP activity [14]. Here, we only present a short overview of post-translational KSRP modifications and the functional consequences (Table 3). For detailed information see [14,49].

Table 3.
Modification of KSRP by phosphorylation.
In addition, KSRP negatively regulates the expression of prothrombin by binding to the upstream sequence element (USE) in the 3′-UTR of prothrombin mRNA [52]. Phosphorylation of KSRP via activated p38 MAPK results in dissociation of KSRP from the USE, yielding increased stabilization of prothrombin mRNA. Furthermore, the natural compound resveratrol has been demonstrated to increase KSRP activity by inhibiting threonine phosphorylation at residue 692 [53]. This, in turn, enhances the degradation of different pro-inflammatory mRNAs, which may explain some of the anti-inflammatory effects of resveratrol. Moreover, it has been described that resveratrol interferes with the transforming growth factor β (TGF-ß)-induced epithelial-to-mesenchymal transition in mammary gland cells, which depends on KSRP [54].
Phosphorylation of KSRP at its phosphorylation sites leads to reduced activity of KSRP and thus to an increased pro-inflammatory cytokine expression.
3.2.2. Ubiquitination

Table 4.
Modification of KSRP by ubiquitination.
3.2.3. Long Non-Coding RNAs
Long non-coding RNAs (lncRNA) are defined as ncRNAs longer than 200 nucleotides and are expressed in multiple cell types and tissues [61]. Some reports describe interaction of lncRNAs with KSRP that modulates KSRP-mediated mRNA decay (Table 5).

Table 5.
lncRNAs interacting with KSRP.
Altogether, KSRP activity is regulated at various levels by transcriptional and post-transcriptional mechanisms, e.g., by interaction with miRNA and other RNA-BP, in addition to post-translational mechanisms, including phosphorylation, ubiquitination and interaction with lncRNAs, in both nucleus and cytoplasm.
4. KSRP as a Regulator of Innate and Adaptive Immune Responses
PMN, monocytes/macrophages and dendritic cells (DC) recognize pathogens and are activated by pathogen-specific moieties [65] and soluble danger signals such as cytokines [66]. These innate immune cell types kill pathogens by various means, and in addition, serve as antigen presenting cells (APC) that present pathogen-derived antigens in the context of increased expression of costimulatory receptors and T cell-polarizing cytokines [67]. By this, T cells with an antigen-specific T cell receptor are activated and confer adaptive immune responses [68]. Due to its central role in AMD of pro-inflammatory mediators, KSRP was considered as an important negative regulator of inflammatory immune responses by limiting cytokine production of activated immune cells, since it promoted decay of the according mRNA, as observed in cell culture experiments [22] and when assaying primary cells isolated from KSRP−/− mice [16,69,70,71].
4.1. Innate Immune Cells
KSRP confers regulation of immune responses on several levels. As outlined above, KSRP was reported to promote granulocytic and at the same time to inhibit monocytic differentiation via processing of miR-129, and indirectly via attenuation of RUNX1 [39]. The modulation of innate immune responses by KSRP is mediated in part via regulation of type I and III interferon expression in immune and non-immune cells and modulation of retinoic acid-inducible gene (RIG-)I receptor signaling [16,72,73], in addition to other cytokines generated by innate immune cells as shown for monocytes/macrophages and PMN.
We observed that in the collagen antibody-induced arthritis (CAIA) disease, mouse model KSRP knock out (KSRP−/−) mice developed markedly lower joint inflammation compared with wild type (WT) mice, accompanied by lower expression of pro-inflammatory cytokines. In general, KSRP−/− mice were less susceptible to CAIA induction and had a much less pronounced disease severity. Myeloid cells, such as macrophages or PMN, were reduced in peripheral blood mononuclear cells isolated from KSRP−/− mice, and in LPS-stimulated spleen cells isolated from KSRP−/− mice. Since these cells are critically involved in CAIA induction [74], the lower number of myeloid cells in KSRP−/− mice may account at least in part for this phenomenon [69]. In this regard, we also showed that the frequency of apoptotic CD11b+ cells was significantly enhanced in KSRP−/− mice.
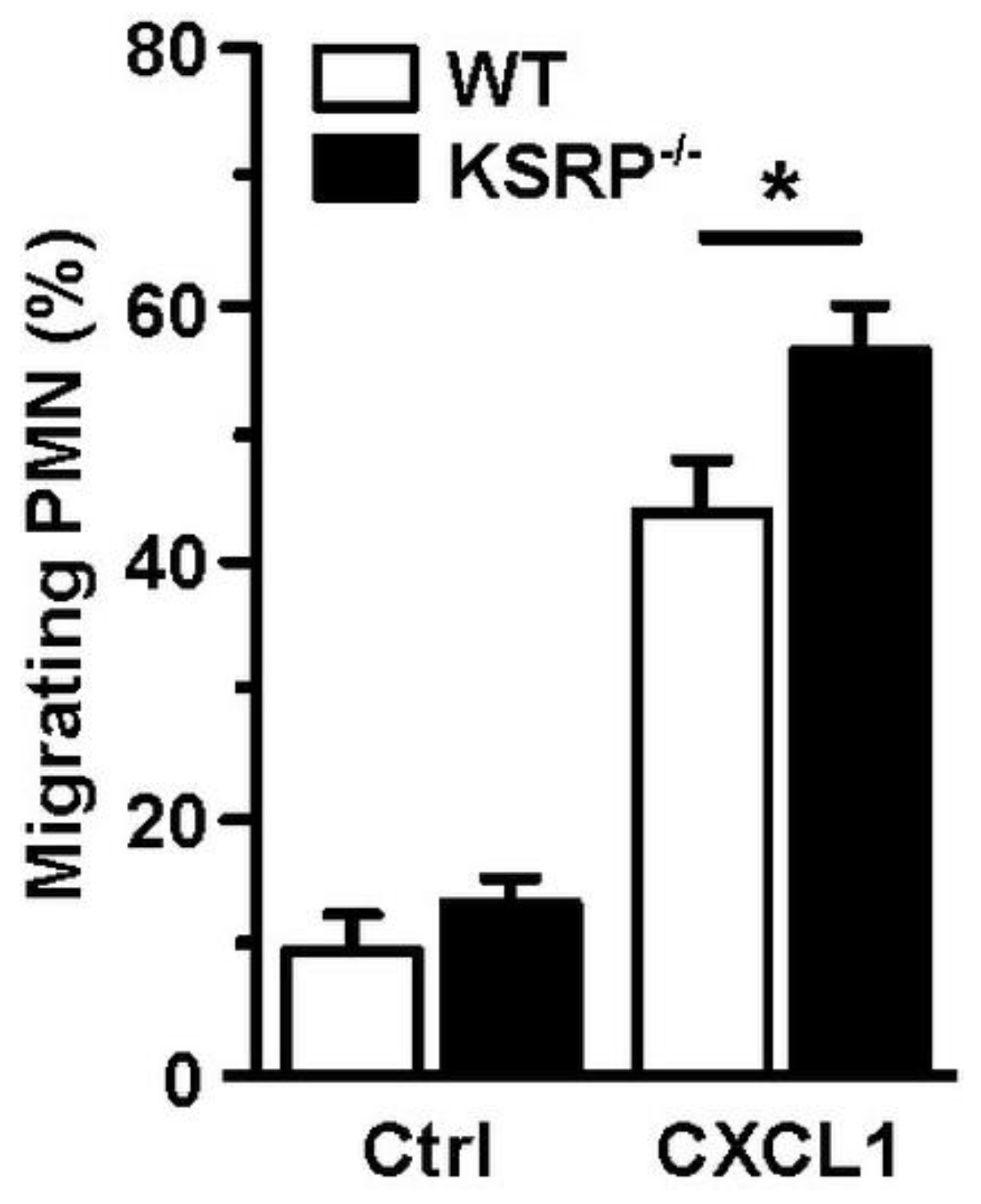
Whereas DC are the most potent type of APC [75], PMN are rather specialized in direct eradication of pathogens [76], e.g., by phagocytosis, the release of reactive oxygen species (ROS) and pathogen-binding chromatin-based extracellular traps, termed NETosis [77]. Furthermore, activated PMN secrete numerous cytokines/chemokines to attract and polarize leukocytes [78]. PMN are the first immune cell population that immigrates infected/inflamed tissue [79]. We observed that KSRP deficiency improved the migration of PMN, which suggested that KSRP may attenuate PMN migration in vivo (Figure 3).
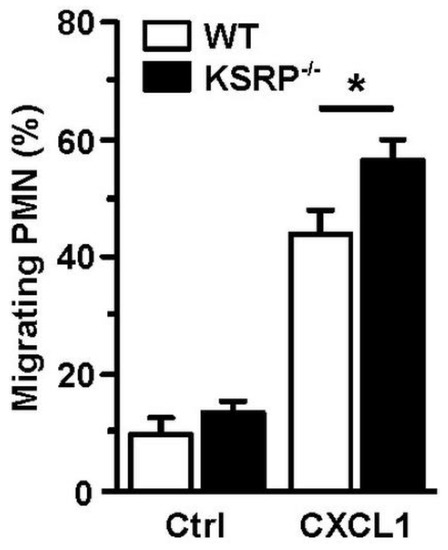
Figure 3.
KSRP coregulates PMN migration. Bone marrow cells derived from WT and KSRP−/− mice towards C-X-C motif chemokine ligand 1 (CXCL1) were assessed in a transwell (Ø 5 µm) migration assay. Bone marrow derived cells were cultivated in 24-well plates with Iscove’s Modified Dulbecco’s Medium supplemented with 5% FCS, 1% penicillin/streptavidin, 2 mM L-Glutamin and 50 µM β-Mercaptoethanol, in a final concentration of 5 × 105 cells/mL. Seeded cells were stimulated with 250 ng/mL CXCL1. Control (Ctrl): w/o chemokine. After incubation for 4 h at 37 °C and 10% CO2, the frequency of migrating Ly6G+ PMN was assessed by flow cytometric analysis (mean + SEM, n = 6; * p < 0.05).
Altogether, several lines of experimental evidence indicate that KSRP regulates/influences the differentiation and function of PMN and monocytes/macrophages.
4.2. Adaptive Immune Cells
Only limited knowledge exists about the importance of KSRP for cells of the adaptive immune system. B cells and T cells are termed adaptive immune cells since these are activated in an antigen-specific manner, and thereby are able to evoke pathogen-specific immune responses. B cells provide a variety of important functions to the adaptive immune system including antibody production, antigen presentation, and cytokine secretion [80].
In adaptive immune responses, the cytokine environment is important for the activation and differentiation of CD4+ T cells into distinct Th cell subsets (e.g., Th1, Th2, Th9 and Th17) [81]. Activated CD4+ T cells play an important regulatory role as they are not only required for full activation of CD8+ T cells [82], but also of B cells [83]. These helper functions are predominantly determined by Th-released cytokines. CD8+ T cells give rise to cytotoxic T lymphocytes (CTL) which directly recognize and kill infected (or malignant) cells that present the CTL-specific antigen via major histocompatibility complex I [84].
We demonstrated that knockdown of the KSRP protein enhanced the proliferation of polyclonally stimulated CD4+ T cells, but not of KSRP−/− CD8+ T cells. Modulation of IL-2 expression, previously reported as a KSRP target in cultures of immortalized cell lines [22], seemed not to contribute to enhanced T cell proliferation, since we were not able to detect any difference in IL-2 production on mRNA or protein level between primary KSRP−/− and WT CD4+ T cells. Another obvious finding was that upon polyclonal stimulation KSRP−/− CD4+ T cells produced higher amounts of IL-4, IL-5, IL-9, IL-10 and IL-13 as compared with WT cells. This overall change in cytokine pattern indicates that KSRP serves to inhibit Th2 polarization [70].
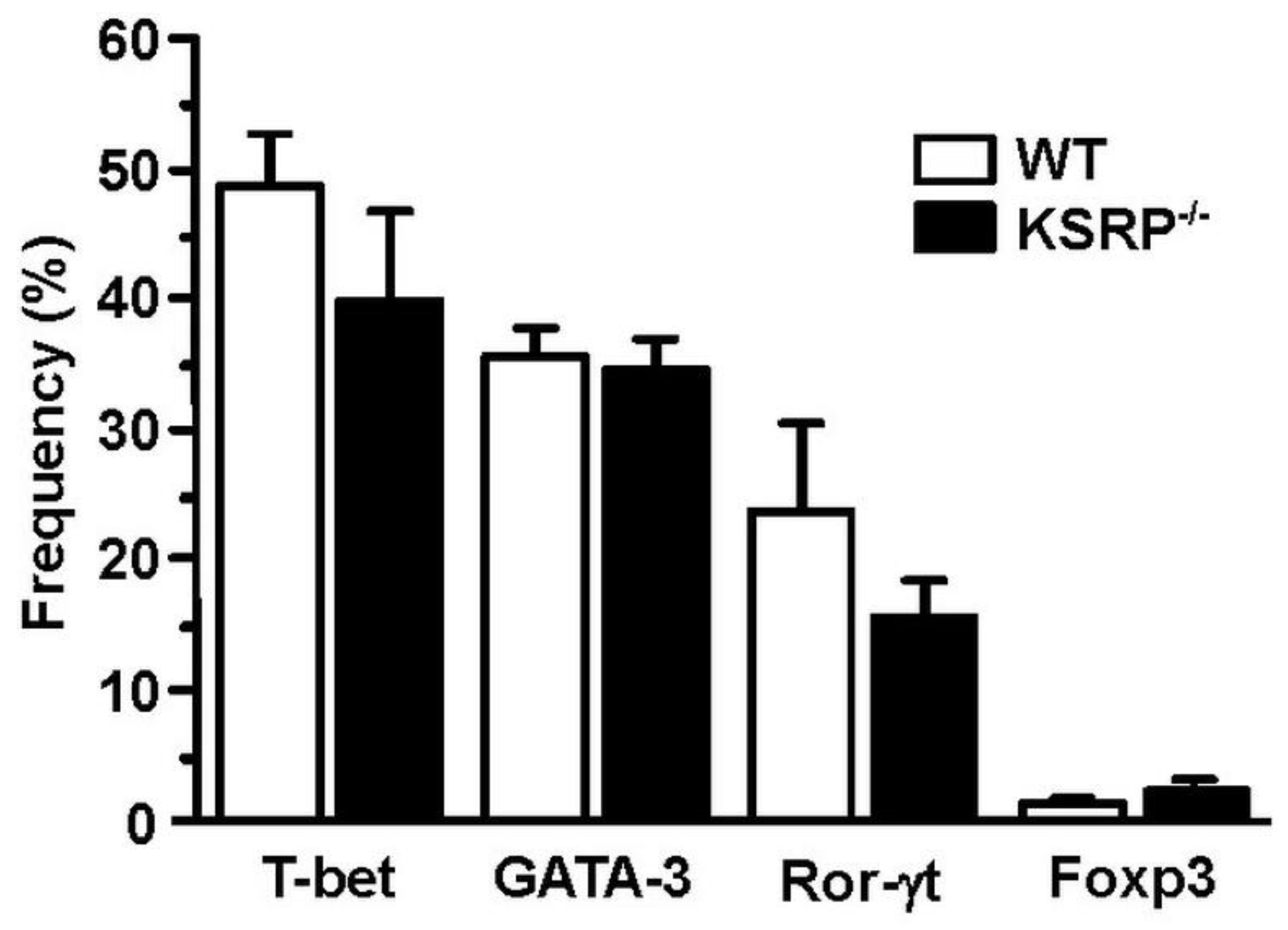
In order to identify the molecular mechanisms responsible for the altered cytokine expression and proliferation of KSRP−/− CD4+ T cells, we analyzed transcription factor expression in polyclonally stimulated CD4+ T cells. We observed no genotype-specific differences in Th2-associated GATA3 expression (Figure 4). This observation suggested that KSRP regulated Th polarization by targeting other mRNA species either directly or via miRNA regulation [85].
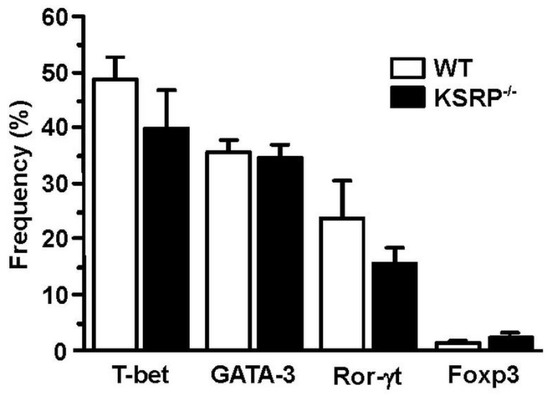
Figure 4.
Expression of GATA-3 in stimulated CD4+ T cells is not affected by KSRP deficiency. Splenic CD4+ T cells (WT, KSRP−/−) were isolated by magnetic bead separation, and were polyclonally stimulated with agonistic CD3- (1 µg/mL) and CD28- (2 µg/mL) specific antibodies for 72 h. Transcription factor expression was delineated by intracellular flow cytometric analysis. Data denote the frequencies of transcription factor-positive CD4+ T cell (mean + SEM, n = 4).
All cytokine mRNA species whose expression is differentially regulated by KSRP in CD4+ T cells contain ARE in their 3′-UTR, but whereas responsiveness of IL-10 and IL-13 mRNA for rapid degradation by other ARE-binding RBP such as TTP [86], AUF1 [87] and HuR [88] has been documented, much less is known about mechanisms of post-transcriptional regulation of IL-5 [89] and IL-9 [90] mRNA expression. We detected direct binding of KSRP to the IL-10 and IL-13 mRNA 3′-UTR, respectively, in pull down experiments, but not to the IL-5 and IL-9 mRNA 3′-UTR [70]. Further, we observed no direct effect of KSRP on the decay of either mRNA monitored. However, polyclonally stimulated KSRP−/− CD4+ T cells displayed increased expression of IL-4 on mRNA and protein level as compared with the corresponding WT control, which may be explained by a longer IL-4 mRNA half-life [70]. Therefore, our data suggested that KSRP is a negative regulator of IL-4 expression. IL-4 on one hand is a master regulator of Th2 polarization [91], and on the other hand constitutes the prototypic Th2-associated cytokine [92]. Further studies are necessary to elucidate by which mechanisms KSRP modulates Th2 polarization on a molecular level.
In summary, KSRP is an important negative regulator of pro-inflammatory mediators by using its several functions, and is involved in immune response.
4.3. KSRP as a Negative Modulator of Immune Responses in Infection
In the case of immune responses in consequence to infections, the host innate immune system plays a significant role in the elimination of pathogen infection [93]. Danger receptors such as Toll-like receptors (TLR) play an essential role in the activation of innate immunity by recognizing specific patterns of microbial components and activating downstream intracellular signaling pathways such as nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) [94]. On one hand, the resulting expression of, e.g., pro-inflammatory cytokines needs to be fast and robust to establish a swift immune response to an invading pathogen [95]. On the other hand, cytokine expression has to be controlled strictly to avoid an excessive immune response resulting in a cytokine storm [96], extensive tissue destruction [97] or autoimmune reactions [98]. Due to its central role in AMD of mRNA species encoding pro-inflammatory mediators, KSRP is considered as an important negative regulator of inflammatory immune responses [15,22].
In this regard, KSRP was demonstrated to inhibit the activation of the retinoic acid-inducible gene (RIG-)I receptor, which is involved in antiviral defense mechanisms [73]. In the absence of KSRP RIG-I receptor induced antiviral signaling was enhanced and accordingly viral replication was reduced.
Type I interferons (IFN-α and IFN-β) play crucial roles in the innate immune response against viral infection [99,100]. Lin and colleagues detected in cells derived from KSRP−/− mice that type I interferons were upregulated, which implied that KSRP plays a crucial role in maintaining low basal IFN-α/β expression levels in the absence of stimuli [72]. Additionally, type I interferon levels were increased in KSRP−/− mice in response to viral infection as a result of decreased mRNA decay. Resulting from this increased expression of IFN-α and IFN-β, the KSRP−/− mice were more resistant to vesicular stomatitis virus and herpes simplex virus I than WT mice.
In another study, it was demonstrated that Helicobacter pylori infection in mice downregulated KSRP expression and upregulated expression of inflammatory-related genes such as C-X-C motif ligand 2 and TLR2. Increasing mRNA decay of pro-inflammatory factors through KSRP overexpression in H. pylori mice facilitated H. pylori proliferation and colonization and induced aggravated gastric inflammation and mucosal damage, implying that downregulation of KSRP is necessary for an effective innate immune response against H. pylori [101].
Interestingly, modulation of gene expression by KSRP plays an important role not only in Gram-negative pathogens. Lipoteichoic acid (LTA) from the Gram-positive Staphylococcus aureus (aLTA) also constitutes a potent immunostimulation agent. KSRP was downregulated by aLTA in a monocytic cell line (THP-1) at protein level [102]. However, there were no differences of KSRP expression at gene level. Zeng and colleagues hypothesized that aLTA, similar to LPS, may regulate expression of inflammatory genes at the transcriptional level via TLR2-mediated activation of NF-κB. At the post-transcriptional level, aLTA might downregulated the destabilizing factor KSRP. Through these two regulatory mechanisms, aLTA treatment could increase and stabilize the mRNAs, and consequently elevate cytokine production.
Investigations regarding Salmonella enteritidis infection in Caco-2 cells revealed also decreased KSRP expression, similar to H. pylori infection [103]. In addition, overexpression of KSRP in Caco-2 cells resulted in reduced levels of inflammatory factors. Interestingly, Nie and coworkers further demonstrated that the decreased expression of KSRP protein following S. enteritidis infection was diminished when blocking the NF-κB signaling pathway, revealing that changes in the expression of KSRP were regulated by this pathway.
However, infection does not result, in general, in subsequent downregulation of KSRP mRNA as shown for H69 cells infected with Cryptosporidium parvum [46]. Instead, infection by C. parvum activated TLR4/NF-κB signaling and increased miR-27b-3p expression, causing a translational suppression of KSRP in infected host epithelial cells. In turn, downregulation of KSRP stabilized iNOS mRNA and promoted production of nitric oxide, exerting antimicrobial activity, by epithelial cells.
Interestingly, negative regulation of pro-inflammatory factors also plays an important role in the prevention of hepatic fibrosis. Pro-inflammatory factors such as cytokines activate hepatic stellate cells and thereby contribute to the development of hepatic fibrosis [104]. Wang and coworkers showed that soluble egg antigen stimulation and Schistosoma japonicum infection increased KSRP mRNA and protein levels and downregulated miR-27b-3p expression in vitro and in vivo [105]. In accordance, both knockdown of miR-27b-3p and overexpression of KSRP attenuated S. japonicum-induced hepatic fibrosis in vivo, due to increased mRNA decay of proinflammatory factors mediated by KSRP.
Taken together, KSRP seems to be an important negative regulator of inflammatory immune responses in infection. While downregulation of KSRP is important for the generation of a robust immune response, against a pathogen, it is equally important to limit immune responses/production of proinflammatory factors through KSRP-mediated mRNA decay to keep the immune system homeostasis in balance.
4.4. KSRP as a Regulator of Auto-Inflammatory Diseases
As outlined above (see Section 4.1), we observed that, somewhat surprisingly, induction of CAIA, a well-established RA model in C57BL/6 mice, was attenuated in terms of disease onset and severity in KSRP−/− mice, as compared with WT mice [69]. In addition to the reduced number of PMN in KSRP−/− mice, the attenuated course of CAIA in KSRP−/− mice may be explained also by the intrinsic property of KSRP−/− CD4+ T cells to express preferentially Th2-related cytokines such as IL-4, IL-5, IL-10, and IL-13, which have been described to be important for the resolution of RA-associated inflammation in diseased joints [69,70,106].
We analyzed the role of KSRP also in a murine systemic lupus erythematosus (SLE) model. To this end, we made use of MRL-Faslpr mice, which spontaneously develop a SLE-like syndrome [107], and bred those with KSRP−/− mice. The derived MRL-FaslprKSRP−/− mice presented with more severe symptoms of glomerulonephritis, indicative of important protective effects of KSRP in the regulation of immune homeostasis in the kidney [71]. Moreover, we detected that the knockout of KSRP might have different effects on disease progression depending on the organ manifestation. In contrast to glomerulonephritis, lymphadenopathy, a prominent disease symptom in MRL-Faslpr mice, was attenuated in FaslprKSRP/mice, indicating a disease driving force of KSRP. The initial screen of the new mouse strain identified cell types (CD4+ IFN-γ+ T cells, FoxP3+ T cells) and targets (IL-1R, CD11a) of interest for further studies.
Concerning chronic inflammatory disease, Xia and coworkers provided evidence for a new mechanism by which liver epithelial cells maintain homeostasis during inflammation [108]. Previous studies indicated that increased levels of C-X3-C motif chemokine ligand 1 (CX3CL1) in the liver are associated with severe inflammatory liver disease [109,110]. In this study, CX3CL1 mRNA stability was demonstrated as directly regulated by KSRP through its interaction with ARE within the CX3CL1 mRNA 3′-UTR [108]. Thus, upregulation of KSRP destabilized CX3CL1 mRNA in liver epithelial cells. In addition, miR-27b-3p was identified as a negative regulator of immune reaction in response to IFN-γ stimulation: IFN-γ stimulation decreased miR-27b-3p expression and increased KSRP protein contents without changing its mRNA level in vitro and in vivo. Consequently, miR-27b-3p regulated the stabilization of CX3CL1 mRNA by attenuating KSRP mRNA translation efficiency. In agreement, downregulation of miR-27b-3p following IFN-γ stimulation resulted in KSRP induction, providing negative feedback regulation of chemokine expression in liver epithelial cells in response to inflammation.
With regard to autosomal recessive diseases such as cystic fibrosis (CF), caused by massive pro-inflammatory phenotype in the lung [111], Bhattacharyya and colleagues reported that deregulation of miR-155 might be involved in CF pathophysiology [112]. They detected high levels of miR-155 in cultured CF IB3-1 and primary lung epithelial cells and demonstrated the antagonistic role of the RNA-BP KSRP und TTP in the regulation of miR-155 biogenesis in CF cells. Suppression of KSRP led to inhibition of miR-155 maturation, whereas overexpression of TTP suppressed the processing of miR-155 through the induction of miR-1 in CF lung epithelial cells [112].
To sum up, the studies implicate that not only is KSRP an important regulator of immune reactions, KSRP itself is also regulated by miRNAs. Interaction with other RBPs also becomes obvious, raising the question of whether another destabilizing RBP can take over the function of KSRP in the case of KSRP deficiency. To answer this upcoming question, further studies are necessary.
4.5. KSRP Affects Tumorigenesis
Besides its role in shaping immune responses in case of autoimmunity and infection, KSRP also plays a role in tumor pathogenesis. Several reports have demonstrated an association of KSRP with different types of lung cancers. In small cell lung cancer (SCLC) increased KSRP protein levels were detected in tumor tissue, and this correlated with advanced tumor stage [113]. Knockdown of KSRP inhibited SCLC cell proliferation but had no effect on cell migration or invasion. KSRP contributed indirectly to tumor progression by promoting miR-26a maturation that led to inhibition of the tumor suppressor phosphatase and tensin homolog. Bikkavilli et al. described enhanced expression of KSRP in non (N)SCLC tissue, which correlated with poor overall survival [114]. The oncogenic properties of KSRP were attributed to KSRP-mediated downregulation of the tumor suppressor Sprouty RTK signaling antagonist 4. In addition, Yan et al. described KSRP as a metastasis-associated molecule in NSCLC [115]. In that study, interaction of KSRP with heterogeneous nuclear ribonucleoprotein C was observed, which may promote tumor metastasis by activating the IFN-α-Janus kinases—signal transducer and activator of the transcription protein 1 signaling pathway. In contrast to the results of the aforementioned studies, interestingly, KSRP was also shown to act in an anti-tumorigenic manner in NSCLC. In this regard, Chien et al. detected reduced KSRP protein expression in tumor tissue and a strong correlation between KSRP expression and overall survival [116]. The authors proposed that KSRP was necessary to promote miR-23a maturation, thus leading to destabilization of early growth response 3 mRNA, resulting in inhibition of NSCLC cell mobility.
In colorectal cancer (CRC), enhanced expression of KSRP was found in tumor tissue and this was associated with a worse overall survival [117]. KSRP seemed to drive epithelial cell proliferation in primary and metastatic cells through control of cell cycle progression and promoted, e.g., angiogenesis by enhancing vascular endothelial growth factor secretion. In addition, enhanced KSRP expression in CRC cells was associated with resistance to 5-fluoruracil treatment. Mechanistically, KSRP was demonstrated to down-regulate mRNA levels of the tumor suppressor ERBB receptor feedback inhibitor 1, by enhancing the maturation of miR-501-5p [118].
KAI1 COOH-terminal interacting tetraspanin (KITENIN) contributed to tumor progression and poor clinical outcomes in various cancers including colorectal cancer, most probably by enhancing neoangiogenesis [119]. KSRP contributed to metastasis in CRC by stabilizing the functional KITENIN complex [120]. DKC1125 (disintegrator of KITENIN complex #1125) was reported to suppress KITENIN activity by direct binding to KSRP. This interaction led to destabilization of the KITENIN complex by recruiting the receptor for activated C kinase 1 and miRNA-124, thereby suppressing metastasis in CRC. Of note, KSRP could also have a protective function in CRC by destabilizing Homeobox protein C10 (HOXC10) mRNA [121]. In CRC and in CRC-initiating cells, high expression of circular HOX (cis-HOX) RNA has been detected. cis-HOX blocked KSRP-mediated HOXC10 mRNA destabilization, thus leading to activation of the tumor-promoting Wnt/b-catenin signaling pathway.
In glioblastoma multiforme (GBM) cells, KSRP was demonstrated to inhibit migration of GBM cells, and, therefore, re-sensitized them to chemotherapy [122]. High KSRP expression was detected in GBM patients who survived long after surgery, indicating a link between KSRP and a better overall survival. Moreover, one study linked KSRP to inducing apoptosis in glioma cells in a caspase-dependent manner [123].
The Follistatin-related protein 1 (FSTL1) primary mRNA transcript also encoded for miR-198, and the switch between expression of the FSTL1 protein and miR-198 is an important regulator of tumor metastasis and wound healing [124]. KSRP processed FSTL1 mRNA to generate miR-198 by binding to the FSTL1 3′-UTR [125]. In keratinocytes, TGF-β induced the expression of miR-181a, which bound to the 3′-UTR of KSRP mRNA and thereby promoted its decay. This resulted in impaired miR-198 but enhanced FSTL1 expression. In the case of temozolomide resistance in glioma, TGF-β increased FSTL1 protein expression and decreased miR198 expression without affecting miR-181a or KSRP expression [126]. In these cases, TGF-β enhanced the expression levels of lncRNAs H19 and HOXD cluster antisense RNA 2, which competitively bind to KSRP and prevent KSRP from participating in FSTL1/miR-198 switching. This also resulted in increased expression of O-6-methylguanine DNA methyltransferase, which correlated with bad prognosis and resistance to chemotherapy.
Additionally, in squamous cell carcinoma, KSRP mediated FSTL1/miR-198 processing constitutes an important factor for metastasis [125]. Downregulation of KSRP in malignant epithelial cells inhibited miR-198 processing and thus contributed to FSTL1 expression.
In cervical cancer, interaction of KSRP with lncRNA LINC01305 promoted tumor growth [127]. Inactivation of the breast cancer susceptibility gene 1 (BRCA1) plays a significant role in breast and ovarian cancers and qRT-PCR analyses indicated that KSRP was over-expressed in BRCA1 mutated tumors [128]. In different breast cancer cell lines, mutations of the tumor suppressor gene TP53 led to changes in proteasome gene expression and enhanced proteasomal activity. In this context, anti-oncogenic properties were attributed to KSRP [129]. The p53-mediated proteasomal dysfunction resulted in increased KSRP degradation, and this impaired expression of the tumor-suppressive miRNAs let-7a and miR-30c, whose maturation is dependent on KSRP as outlined above. In breast cancer cells, IL-1 β-mediated replacement of KSRP by the mRNA stabilizing factor HuR at the IL-8 3′-UTR has been described, which in light of the tumor-promoting role of IL-8 may contribute to cancer progression [130]. We have previously demonstrated concurrent binding of KSRP and HuR to the same target 3′-UTR [17].
In hepatocellular carcinoma (HCC) over-expression of FBPs was identified. As outlined above, KSRP, also known as FBP2, has been shown to promote c-myc transcription [5], and c-myc was demonstrated to contribute to HCC progression [131]. The peptidyl-prolyl isomerase Pin1 is over-expressed in several cancer tissues, and may promote tumorigenesis by regulating mRNA decay in cooperation with the ARE-binding proteins AUF1 and KSRP [132].
In esophageal squamous cell carcinoma (ESCC), increased KSRP expression levels were associated with worse overall survival [133]. KSRP promoted growth, migration, and invasion of ESCC cells by enhancing the maturation of cancer-associated miRNAs, such as miR-21, miR-130b, and miR-301. Accordingly, this reduced the expression of the according miRNA target mRNAs, such as bone morphogenetic protein 6, programmed cell death protein 4, and Metalloproteinase inhibitor 3, and promoted epithelial to mesenchymal transition. Additionally, in osteosarcoma cells, KSRP was significantly upregulated, and contributed to enhanced cell proliferation and migration [134]. Likewise, KSRP was associated with enhanced proliferation of melanoma cells [135]. Here, KSRP-mediated destabilization of killin mRNA, a p53-regulated DNA replication inhibitor. Further, KSRP promoted invasiveness and metastasis of pancreatic cancer cells by interaction with the small nucleolar RNAs SNORA18 and SNORA22, and thereby enhanced the number of cell protrusions [136].
Another investigation demonstrated, in mice, that lncRNA Neat1 interacted with KSRP to promote metastasis in soft tissue sarcomas [137]. It was assumed that the Neat1/KSRP complex functioned as an RNA splicing regulator to mediate tumor cell colonization of the lung. Additionally, KSRP may also be involved in papillary thyroid carcinoma (PTC) by interacting with lncRNA AB074169 (lncAB) [138]. In normal cells, lncAB blocked mRNA decay activity of KSRP, whereas in PTC tumor cells lncAB DNA was hypermethylated, resulting in enhanced mRNA degradation of p21 by KSRP, which, in turn, promoted cell proliferation.
Overall, KSRP seems to promote cell proliferation and metastasis, but results strongly depend on tumors investigated, and opposite effects have also been described. In the context of tumorigenesis, a number of different KSRP-mediated mechanisms and KSRP targets has been described. In addition to direct regulation of mRNA stability, KSRP-mediated miRNA maturation and interaction of KSRP with lncRNA on protein level have been observed in different tumor models.
5. Conclusions
KSRP constitutes an important regulator of both innate and adaptive immune cells in addition to tumorigenesis. To date, KSRP may be considered as an important break of immune activation by inhibiting the production of proinflammatory cytokines by the initiation of mRNA decay and the promotion of miRNA maturation. This break is released in response to infection and also due to post-transcriptional modulation of KSRP activity. Aside from the role of KSRP to regulate the overall extent of immune activation, it also serves to finetune the character of an (adaptive) immune response, as reflected, e.g., by the intrinsic Th2 bias of CD4+ T cells in case of KSRP deficiency. However, further in-depth analyses are required to gain full insight in KSRP targets, especially with regard to the issue of whether this RBP acts only on the level of effector cytokines, or orchestrates the shape of immune responses also, by affecting the expression of key transcription factors. Despite the emerging evidence of the regulatory importance of KSRP, its function has been addressed in few immune cell types to date. Therefore, further studies need to address the functional role of KSRP in other innate (e.g., NK cells, innate lymphoid cells) and adaptive (B cells) immune cell types under basal conditions and in response to activation in suitable disease models. With regard to the latter, it will be important to employ mice with conditional KSRP deficiency in order to address the cell type-specific role of this RBP. Finally, it will also be important to shed light on the exact role of KSRP in tumorigenesis; depending on the tumor type, KSRP may either block or promote tumor induction and progression.
Altogether, deeper understanding into the role of KSRP in the immune system and for tumor induction and progression is a necessary prerequisite for the development of drugs which, when applied by suitable nano-carriers, may allow control of KSRP in a cell type-specific manner for therapeutic purposes. According to our data, in particular the ability of KSRP to shift T helper cell polarization towards a distinct direction (enhancing KSRP activity favors Th1 response, inhibition of KSRP activity promotes Th2 response), may be an interesting tool to restore immune homeostasis in different chronic inflammatory diseases.
Author Contributions
Writing—original draft preparation, K.-A.P., V.B., H.K., M.B. and A.P.; creating figures and tables, K.-A.P., V.B., M.B., R.K. and A.P.; writing—review and editing, K.-A.P., V.B., M.B., A.P. and H.K.; funding acquisition, M.B., A.P. and H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by DFG, grant number BR 3880/4-1, PA 1933/7-1, PA 1933/2-3 and KL1020/10-1 (to HK).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| aa | Amino acid |
| ALAE | Axon-enriched long intergenic noncoding RNA regulating axon elongation |
| AMD | ARE-mediated mRNA decay |
| APC | Antigen presenting cell |
| ARE | AU-rich element |
| ATM | Ataxia telangiectasia mutated |
| AUF1 | AU-binding factor 1 |
| BRCA1 | Breast cancer susceptibility gene 1 |
| CAIA | Collagen antibody induced-arthritis |
| CD | Cluster of differentiation |
| CF | Cystic fibrosis |
| Cis-HOX | circular HOX |
| CRC | Colorectal cancer |
| CTL | Cytotoxic T lymphocyte |
| CX3CL1 | C-X3-C motif chemokine ligand 1 |
| CXCL1 | C-X-C motif chemokine Ligand 1 |
| DC | Dendritic cell |
| ESCC | Esophageal squamous cell carcinoma |
| FBP | Far upstream element binding protein |
| FSTL1 | Follistatin-related protein 1 |
| FUSE | Far upstream element |
| GBM | Glioblastoma multiforme |
| HCC | Hepatocellular carcinoma |
| HuR | Human antigen R |
| HOXC10 | Homeobox protein C10 |
| IFN | Interferon |
| IL | Interleukin |
| iNOS | Inducible nitric oxide synthase |
| KH | K homology |
| KITENIN | Kai 1 COOH-terminal interacting tetraspanin |
| KSRP | KH-type splicing regulatory protein |
| lncRNA | Long non-coding RNA |
| LPS | lipopolysaccharide |
| LTA | Lipoteichoic acid |
| MAPK | Mitogen actiaved protein kinase |
| miRNA | MicroRNA |
| NF-κB | Nuclear factor k-light-chain-enhancer of activated B cells |
| NSCLC | Non-small cell lung cancer |
| PKB | Protein kinase B |
| PMN | Polymorphonuclear neutrophilic leukocytes |
| PTC | Papillary thyroid carcinoma |
| RA | Rheumatoid arthritis |
| RBP | RNA-binding protein |
| RIG-I | Retinoic acid-inducible gene I |
| RISC | RNA-induced silencing complex |
| ROS | Reactive oxygen species |
| RUNX1 | Runt-related transcription factor 1 |
| SCLC | Small cell lung cancer |
| SLE | Systemic lupus erythematosus |
| TGF-β | Transforming growth factor β |
| TTP | Tristetraprolin |
| TLR | Toll-like receptor |
| TNF-α | Tumor necrosis factor-α |
| Th | T helper cell |
| USE | Upstream sequence element |
| UTR | Untranslated region |
| WT | Wild type |
References
- García-Mauriño, S.M.; Rivero-Rodríguez, F.; Velázquez-Cruz, A.; Hernández-Vellisca, M.; Díaz-Quintana, A.; De la Rosa, M.A.; Díaz-Moreno, I. Rna binding protein regulation and cross-talk in the control of au-rich mrna fate. Front. Mol. Biosci. 2017, 4, 71. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.J.D.; Man, J.H.S.; Schatz, J.H.; Marsden, P.A. Translational remodeling by rna-binding proteins and noncoding rnas. Wiley Interdiscip. Rev. RNA 2021, 12, e1647. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.F.; Lin, W.J.; Lin, C.C.; Luber, C.A.; Godbout, R.; Mann, M.; Chen, C.Y. Dead box protein ddx1 regulates cytoplasmic localization of ksrp. PLoS ONE 2013, 8, e73752. [Google Scholar]
- Ring, H.Z.; Vameghi-Meyers, V.; Nikolic, J.M.; Min, H.; Black, D.L.; Francke, U. Mapping of the khsrp gene to a region of conserved synteny on human chromosome 19p13.3 and mouse chromosome 17. Genomics 1999, 56, 350–352. [Google Scholar] [CrossRef]
- Davis-Smyth, T.; Duncan, R.C.; Zheng, T.; Michelotti, G.; Levens, D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 1996, 271, 31679–31687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Chen, Q.M. Far upstream element binding protein 1: A commander of transcription, translation and beyond. Oncogene 2013, 32, 2907–2916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Chung, Y.J.; Parrilla Castellar, E.R.; Zheng, Y.; Chung, H.J.; Bandle, R.; Liu, J.; Tessarollo, L.; Batchelor, E.; Aplan, P.D.; et al. Far upstream element binding protein plays a crucial role in embryonic development, hematopoiesis, and stabilizing myc expression levels. Am. J. Pathol. 2016, 186, 701–715. [Google Scholar] [CrossRef] [Green Version]
- Gherzi, R.; Chen, C.Y.; Trabucchi, M.; Ramos, A.; Briata, P. The role of ksrp in mrna decay and microrna precursor maturation. Wiley Interdiscip. Rev. RNA 2010, 1, 230–239. [Google Scholar] [CrossRef]
- García-Mayoral, M.F.; Hollingworth, D.; Masino, L.; Díaz-Moreno, I.; Kelly, G.; Gherzi, R.; Chou, C.F.; Chen, C.Y.; Ramos, A. The structure of the c-terminal kh domains of ksrp reveals a noncanonical motif important for mrna degradation. Structure 2007, 15, 485–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briata, P.; Chen, C.Y.; Giovarelli, M.; Pasero, M.; Trabucchi, M.; Ramos, A.; Gherzi, R. Ksrp, many functions for a single protein. Front. Biosci. 2011, 16, 1787–1796. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Maeda, K. Control of rna stability in immunity. Annu. Rev. Immunol. 2021, 39, 481–509. [Google Scholar] [CrossRef]
- King, P.H.; Chen, C.Y. Role of ksrp in control of type i interferon and cytokine expression. J. Interf. Cytokine Res. 2014, 34, 267–274. [Google Scholar] [CrossRef] [Green Version]
- Min, H.; Turck, C.W.; Nikolic, J.M.; Black, D.L. A new regulatory protein, ksrp, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997, 11, 1023–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briata, P.; Chen, C.Y.; Ramos, A.; Gherzi, R. Functional and molecular insights into ksrp function in mrna decay. Biochim. Biophys. Acta 2013, 1829, 689–694. [Google Scholar] [CrossRef]
- Gherzi, R.; Chen, C.Y.; Ramos, A.; Briata, P. Ksrp controls pleiotropic cellular functions. Semin. Cell Dev. Biol. 2014, 34, 2–8. [Google Scholar] [CrossRef]
- Schmidtke, L.; Schrick, K.; Saurin, S.; Kafer, R.; Gather, F.; Weinmann-Menke, J.; Kleinert, H.; Pautz, A. The kh-type splicing regulatory protein (ksrp) regulates type iii interferon expression post-transcriptionally. Biochem. J. 2019, 476, 333–352. [Google Scholar] [CrossRef]
- Linker, K.; Pautz, A.; Fechir, M.; Hubrich, T.; Greeve, J.; Kleinert, H. Involvement of ksrp in the post-transcriptional regulation of human inos expression-complex interplay of ksrp with ttp and hur. Nucleic Acids Res. 2005, 33, 4813–4827. [Google Scholar] [CrossRef] [Green Version]
- Milone, J.; Wilusz, J.; Bellofatto, V. Characterization of deadenylation in trypanosome extracts and its inhibition by poly(a)-binding protein pab1p. RNA 2004, 10, 448–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwede, A.; Manful, T.; Jha, B.A.; Helbig, C.; Bercovich, N.; Stewart, M.; Clayton, C. The role of deadenylation in the degradation of unstable mrnas in trypanosomes. Nucleic Acids Res. 2009, 37, 5511–5528. [Google Scholar] [CrossRef] [Green Version]
- Li, C.H.; Irmer, H.; Gudjonsdottir-Planck, D.; Freese, S.; Salm, H.; Haile, S.; Estévez, A.M.; Clayton, C. Roles of a trypanosoma brucei 5′->3′ exoribonuclease homolog in mrna degradation. RNA 2006, 12, 2171–2186. [Google Scholar] [CrossRef] [Green Version]
- Tourrière, H.; Chebli, K.; Tazi, J. Mrna degradation machines in eukaryotic cells. Biochimie 2002, 84, 821–837. [Google Scholar] [CrossRef]
- Gherzi, R.; Lee, K.Y.; Briata, P.; Wegmuller, D.; Moroni, C.; Karin, M.; Chen, C.Y. A kh domain rna binding protein, ksrp, promotes are-directed mrna turnover by recruiting the degradation machinery. Mol. Cell 2004, 14, 571–583. [Google Scholar] [CrossRef]
- Lellek, H.; Kirsten, R.; Diehl, I.; Apostel, F.; Buck, F.; Greeve, J. Purification and molecular cloning of a novel essential component of the apolipoprotein b mrna editing enzyme-complex. J. Biol. Chem. 2000, 275, 19848–19856. [Google Scholar] [CrossRef] [Green Version]
- Winzen, R.; Thakur, B.K.; Dittrich-Breiholz, O.; Shah, M.; Redich, N.; Dhamija, S.; Kracht, M.; Holtmann, H. Functional analysis of ksrp interaction with the au-rich element of interleukin-8 and identification of inflammatory mrna targets. Mol. Cell. Biol. 2007, 27, 8388–8400. [Google Scholar] [CrossRef] [Green Version]
- Dhamija, S.; Kuehne, N.; Winzen, R.; Doerrie, A.; Dittrich-Breiholz, O.; Thakur, B.K.; Kracht, M.; Holtmann, H. Interleukin-1 activates synthesis of interleukin-6 by interfering with a kh-type splicing regulatory protein (ksrp)-dependent translational silencing mechanism. J. Biol. Chem. 2011, 286, 33279–33288. [Google Scholar] [CrossRef] [Green Version]
- Hammond, S.M. An overview of micrornas. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Yi, R.; Cullen, B.R. Micrornas and small interfering rnas can inhibit mrna expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef] [Green Version]
- Trabucchi, M.; Briata, P.; Garcia-Mayoral, M.; Haase, A.D.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. The rna-binding protein ksrp promotes the biogenesis of a subset of micrornas. Nature 2009, 459, 1010–1014. [Google Scholar] [CrossRef] [Green Version]
- Gulei, D.; Raduly, L.; Broseghini, E.; Ferracin, M.; Berindan-Neagoe, I. The extensive role of mir-155 in malignant and non-malignant diseases. Mol. Asp. Med. 2019, 70, 33–56. [Google Scholar] [CrossRef]
- Yazarlou, F.; Kadkhoda, S.; Ghafouri-Fard, S. Emerging role of let-7 family in the pathogenesis of hematological malignancies. Biomed. Pharmacother. 2021, 144, 112334. [Google Scholar] [CrossRef] [PubMed]
- Nicastro, G.; García-Mayoral, M.F.; Hollingworth, D.; Kelly, G.; Martin, S.R.; Briata, P.; Gherzi, R.; Ramos, A. Noncanonical g recognition mediates ksrp regulation of let-7 biogenesis. Nat. Struct. Mol. Biol. 2012, 19, 1282–1286. [Google Scholar] [CrossRef] [Green Version]
- Deng, B.; Tang, X.; Wang, Y. Role of microrna-129 in cancer and non-cancerous diseases (review). Exp. Ther. Med. 2021, 22, 918. [Google Scholar] [CrossRef]
- Lu, D.; Nakagawa, R.; Lazzaro, S.; Staudacher, P.; Abreu-Goodger, C.; Henley, T.; Boiani, S.; Leyland, R.; Galloway, A.; Andrews, S.; et al. The mir-155-pu.1 axis acts on pax5 to enable efficient terminal b cell differentiation. J. Exp. Med. 2014, 211, 2183–2198. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Gao, D.; Shao, Z.; Zheng, Q.; Yu, Q. Mir-155 indicates the fate of cd4(+) t cells. Immunol. Lett. 2020, 224, 40–49. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Niazi, V.; Taheri, M. Role of mirnas and lncrnas in hematopoietic stem cell differentiation. Non-Coding RNA Res. 2021, 6, 8–14. [Google Scholar] [CrossRef]
- Elton, T.S.; Selemon, H.; Elton, S.M.; Parinandi, N.L. Regulation of the mir155 host gene in physiological and pathological processes. Gene 2013, 532, 1–12. [Google Scholar] [CrossRef]
- Spoerl, D.; Duroux-Richard, I.; Louis-Plence, P.; Jorgensen, C. The role of mir-155 in regulatory t cells and rheumatoid arthritis. Clin. Immunol. 2013, 148, 56–65. [Google Scholar] [CrossRef]
- Banerjee, A.; Schambach, F.; DeJong, C.S.; Hammond, S.M.; Reiner, S.L. Micro-rna-155 inhibits ifn-gamma signaling in cd4+ t cells. Eur. J. Immunol. 2010, 40, 225–231. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Yi, P.; Si, Y.; Tan, P.; He, J.; Yu, S.; Ren, Y.; Ma, Y.; Zhang, J.; et al. Ksrp specifies monocytic and granulocytic differentiation through regulating mir-129 biogenesis and runx1 expression. Nat. Commun. 2017, 8, 1428. [Google Scholar] [CrossRef]
- Liu, G.; Du, X.; Xiao, L.; Zeng, Q.; Liu, Q. Activation of fgd5-as1 promotes progression of cervical cancer through regulating bst2 to inhibit macrophage m1 polarization. J. Immunol. Res. 2021, 2021, 5857214. [Google Scholar] [CrossRef]
- Zhao, H.; Shao, X.; Liu, H.; Liu, Q.; Lu, J.; Li, W. The circrna_102911/mir-129-5p/sox6 axis is involved with t lymphocyte immune function in elderly patients with laparoscopic left hepatectomy for hepatolithiasis. Exp. Ther. Med. 2021, 21, 150. [Google Scholar] [CrossRef]
- Jiang, S.; Yan, W.; Wang, S.E.; Baltimore, D. Dual mechanisms of posttranscriptional regulation of tet2 by let-7 microrna in macrophages. Proc. Natl. Acad. Sci. USA 2019, 116, 12416–12421. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Dutta, B.; Tse, S.W.; Gupta, N.; Tan, C.F.; Low, J.K.; Yeoh, K.W.; Kon, O.L.; Tam, J.P.; Sze, S.K. Hypoxia-induced tumor exosomes promote m2-like macrophage polarization of infiltrating myeloid cells and microrna-mediated metabolic shift. Oncogene 2019, 38, 5158–5173. [Google Scholar] [CrossRef]
- Amirouche, A.; Tadesse, H.; Miura, P.; Bélanger, G.; Lunde, J.A.; Côté, J.; Jasmin, B.J. Converging pathways involving microrna-206 and the rna-binding protein ksrp control post-transcriptionally utrophin a expression in skeletal muscle. Nucleic Acids Res. 2014, 42, 3982–3997. [Google Scholar] [CrossRef]
- Puppo, M.; Bucci, G.; Rossi, M.; Giovarelli, M.; Bordo, D.; Moshiri, A.; Gorlero, F.; Gherzi, R.; Briata, P. Mirna-mediated khsrp silencing rewires distinct post-transcriptional programs during tgf-β-induced epithelial-to-mesenchymal transition. Cell Rep. 2016, 16, 967–978. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Gong, A.Y.; Eischeid, A.N.; Chen, X.M. Mir-27b targets ksrp to coordinate tlr4-mediated epithelial defense against cryptosporidium parvum infection. PLoS Pathog. 2012, 8, e1002702. [Google Scholar] [CrossRef] [Green Version]
- Pullmann, R., Jr.; Kim, H.H.; Abdelmohsen, K.; Lal, A.; Martindale, J.L.; Yang, X.; Gorospe, M. Analysis of turnover and translation regulatory rna-binding protein expression through binding to cognate mrnas. Mol. Cell. Biol. 2007, 27, 6265–6278. [Google Scholar] [CrossRef] [Green Version]
- Tadesse, H.; Deschênes-Furry, J.; Boisvenue, S.; Côté, J. Kh-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum. Mol. Genet. 2008, 17, 506–524. [Google Scholar] [CrossRef] [Green Version]
- Briata, P.; Bordo, D.; Puppo, M.; Gorlero, F.; Rossi, M.; Perrone-Bizzozero, N.; Gherzi, R. Diverse roles of the nucleic acid-binding protein khsrp in cell differentiation and disease. Wiley Interdiscip. Rev. RNA 2016, 7, 227–240. [Google Scholar] [CrossRef] [Green Version]
- Briata, P.; Forcales, S.V.; Ponassi, M.; Corte, G.; Chen, C.Y.; Karin, M.; Puri, P.L.; Gherzi, R. P38-dependent phosphorylation of the mrna decay-promoting factor ksrp controls the stability of select myogenic transcripts. Mol. Cell 2005, 20, 891–903. [Google Scholar] [CrossRef]
- Díaz-Moreno, I.; Hollingworth, D.; Frenkiel, T.A.; Kelly, G.; Martin, S.; Howell, S.; García-Mayoral, M.; Gherzi, R.; Briata, P.; Ramos, A. Phosphorylation-mediated unfolding of a kh domain regulates ksrp localization via 14-3-3 binding. Nat. Struct. Mol. Biol. 2009, 16, 238–246. [Google Scholar] [CrossRef] [Green Version]
- Danckwardt, S.; Gantzert, A.S.; Macher-Goeppinger, S.; Probst, H.C.; Gentzel, M.; Wilm, M.; Gröne, H.J.; Schirmacher, P.; Hentze, M.W.; Kulozik, A.E. P38 mapk controls prothrombin expression by regulated rna 3′ end processing. Mol. Cell 2011, 41, 298–310. [Google Scholar] [CrossRef]
- Bollmann, F.; Art, J.; Henke, J.; Schrick, K.; Besche, V.; Bros, M.; Li, H.; Siuda, D.; Handler, N.; Bauer, F.; et al. Resveratrol post-transcriptionally regulates pro-inflammatory gene expression via regulation of ksrp rna binding activity. Nucleic Acids Res. 2014, 42, 12555–12569. [Google Scholar] [CrossRef]
- Moshiri, A.; Puppo, M.; Rossi, M.; Gherzi, R.; Briata, P. Resveratrol limits epithelial to mesenchymal transition through modulation of khsrp/hnrnpa1-dependent alternative splicing in mammary gland cells. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 291–298. [Google Scholar] [CrossRef]
- Kung, Y.A.; Hung, C.T.; Chien, K.Y.; Shih, S.R. Control of the negative ires trans-acting factor khsrp by ubiquitination. Nucleic Acids Res. 2017, 45, 271–287. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, C.P.; MacGurn, J.A. Coupling conjugation and deconjugation activities to achieve cellular ubiquitin dynamics. Trends Biochem. Sci. 2020, 45, 427–439. [Google Scholar] [CrossRef]
- Lin, J.Y.; Li, M.L.; Shih, S.R. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009, 37, 47–59. [Google Scholar] [CrossRef]
- Yuan, H.; Deng, R.; Zhao, X.; Chen, R.; Hou, G.; Zhang, H.; Wang, Y.; Xu, M.; Jiang, B.; Yu, J. Sumo1 modification of khsrp regulates tumorigenesis by preventing the tl-g-rich mirna biogenesis. Mol. Cancer 2017, 16, 157. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.M.; Yeh, E.T.H. Sumo: From bench to bedside. Physiol. Rev. 2020, 100, 1599–1619. [Google Scholar] [CrossRef]
- Wang, C.; Xu, W.; Chao, Y.; Liang, M.; Zhang, F.; Huang, K. E3 ligase fbxw2 is a new therapeutic target in obesity and atherosclerosis. Adv. Sci. 2020, 7, 2001800. [Google Scholar] [CrossRef]
- Ard, R.; Allshire, R.C.; Marquardt, S. Emerging properties and functional consequences of noncoding transcription. Genetics 2017, 207, 357–367. [Google Scholar]
- Giovarelli, M.; Bucci, G.; Ramos, A.; Bordo, D.; Wilusz, C.J.; Chen, C.Y.; Puppo, M.; Briata, P.; Gherzi, R. H19 long noncoding rna controls the mrna decay promoting function of ksrp. Proc. Natl. Acad. Sci. USA 2014, 111, E5023–E5028. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.; Bucci, G.; Rizzotto, D.; Bordo, D.; Marzi, M.J.; Puppo, M.; Flinois, A.; Spadaro, D.; Citi, S.; Emionite, L.; et al. Lncrna epr controls epithelial proliferation by coordinating cdkn1a transcription and mrna decay response to tgf-β. Nat. Commun. 2019, 10, 1969. [Google Scholar] [CrossRef]
- Wei, M.; Huang, J.; Li, G.W.; Jiang, B.; Cheng, H.; Liu, X.; Jiang, X.; Zhang, X.; Yang, L.; Bao, L.; et al. Axon-enriched lincrna alae is required for axon elongation via regulation of local mrna translation. Cell Rep. 2021, 35, 109053. [Google Scholar] [CrossRef]
- Stögerer, T.; Stäger, S. Innate immune sensing by cells of the adaptive immune system. Front. Immunol. 2020, 11, 1081. [Google Scholar] [CrossRef]
- Fleshner, M.; Crane, C.R. Exosomes, damps and mirna: Features of stress physiology and immune homeostasis. Trends Immunol. 2017, 38, 768–776. [Google Scholar] [CrossRef]
- Gaudino, S.J.; Kumar, P. Cross-talk between antigen presenting cells and t cells impacts intestinal homeostasis, bacterial infections, and tumorigenesis. Front. Immunol. 2019, 10, 360. [Google Scholar] [CrossRef] [Green Version]
- Farber, D.L. Form and function for t cells in health and disease. Nat. Rev. Immunol. 2020, 20, 83–84. [Google Scholar] [CrossRef]
- Kafer, R.; Schrick, K.; Schmidtke, L.; Montermann, E.; Hobernik, D.; Bros, M.; Chen, C.Y.; Kleinert, H.; Pautz, A. Inactivation of the ksrp gene modifies collagen antibody induced arthritis. Mol. Immunol. 2017, 87, 207–216. [Google Scholar] [CrossRef]
- Kafer, R.; Schmidtke, L.; Schrick, K.; Montermann, E.; Bros, M.; Kleinert, H.; Pautz, A. The rna-binding protein ksrp modulates cytokine expression of cd4(+) t cells. J. Immunol. Res. 2019, 2019, 4726532. [Google Scholar] [CrossRef] [Green Version]
- Schmidtke, L.; Meineck, M.; Saurin, S.; Otten, S.; Gather, F.; Schrick, K.; Käfer, R.; Roth, W.; Kleinert, H.; Weinmann-Menke, J.; et al. Knockout of the kh-type splicing regulatory protein drives glomerulonephritis in mrl-fas(lpr) mice. Cells 2021, 10, 3167. [Google Scholar] [CrossRef]
- Lin, W.J.; Zheng, X.; Lin, C.C.; Tsao, J.; Zhu, X.; Cody, J.J.; Coleman, J.M.; Gherzi, R.; Luo, M.; Townes, T.M.; et al. Posttranscriptional control of type i interferon genes by ksrp in the innate immune response against viral infection. Mol. Cell. Biol. 2011, 31, 3196–3207. [Google Scholar] [CrossRef] [Green Version]
- Soonthornvacharin, S.; Rodriguez-Frandsen, A.; Zhou, Y.; Galvez, F.; Huffmaster, N.J.; Tripathi, S.; Balasubramaniam, V.R.; Inoue, A.; de Castro, E.; Moulton, H.; et al. Systems-based analysis of rig-i-dependent signalling identifies khsrp as an inhibitor of rig-i receptor activation. Nat. Microbiol. 2017, 2, 17022. [Google Scholar] [CrossRef] [PubMed]
- Sehnert, B.; Cavcic, A.; Böhm, B.; Kalden, J.R.; Nandakumar, K.S.; Holmdahl, R.; Burkhardt, H. Antileukoproteinase: Modulation of neutrophil function and therapeutic effects on anti-type ii collagen antibody-induced arthritis. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2004, 50, 2347–2359. [Google Scholar] [CrossRef] [PubMed]
- Austyn, J.M. Dendritic cells in the immune system-history, lineages, tissues, tolerance, and immunity. Microbiol. Spectr. 2016, 4, 4–6. [Google Scholar] [CrossRef]
- Silva, L.M.; Muñoz-Caro, T.; Burgos, R.A.; Hidalgo, M.A.; Taubert, A.; Hermosilla, C. Far beyond phagocytosis: Phagocyte-derived extracellular traps act efficiently against protozoan parasites in vitro and in vivo. Mediat. Inflamm. 2016, 2016, 5898074. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.Y.; Hsieh, S.C.; Liu, C.W.; Lu, C.S.; Wu, C.H.; Liao, H.T.; Chen, M.H.; Li, K.J.; Shen, C.Y.; Kuo, Y.M.; et al. Cross-talk among polymorphonuclear neutrophils, immune, and non-immune cells via released cytokines, granule proteins, microvesicles, and neutrophil extracellular trap formation: A novel concept of biology and pathobiology for neutrophils. Int. J. Mol. Sci. 2021, 22, 3119. [Google Scholar] [CrossRef]
- Cassatella, M.A.; Östberg, N.K.; Tamassia, N.; Soehnlein, O. Biological roles of neutrophil-derived granule proteins and cytokines. Trends Immunol. 2019, 40, 648–664. [Google Scholar] [CrossRef] [PubMed]
- Kraus, R.F.; Gruber, M.A. Neutrophils-from bone marrow to first-line defense of the innate immune system. Front. Immunol. 2021, 12, 767175. [Google Scholar] [CrossRef]
- Turner, J.S.; Benet, Z.L.; Grigorova, I.L. Signals 1, 2 and b cell fate or: Where, when and for how long? Immunol Rev 2020, 296, 9–23. [Google Scholar] [CrossRef]
- Zhu, X.; Zhu, J. Cd4 t helper cell subsets and related human immunological disorders. Int. J. Mol. Sci. 2020, 21, 8011. [Google Scholar] [CrossRef]
- Bedoui, S.; Heath, W.R.; Mueller, S.N. Cd4(+) t-cell help amplifies innate signals for primary cd8(+) t-cell immunity. Immunol. Rev. 2016, 272, 52–64. [Google Scholar] [CrossRef]
- Eisenbarth, S.C.; Baumjohann, D.; Craft, J.; Fazilleau, N.; Ma, C.S.; Tangye, S.G.; Vinuesa, C.G.; Linterman, M.A. Cd4(+) t cells that help b cells—A proposal for uniform nomenclature. Trends Immunol. 2021, 42, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Halle, S.; Halle, O.; Förster, R. Mechanisms and dynamics of t cell-mediated cytotoxicity in vivo. Trends Immunol. 2017, 38, 432–443. [Google Scholar] [CrossRef]
- Tindemans, I.; Serafini, N.; Di Santo, J.P.; Hendriks, R.W. Gata-3 function in innate and adaptive immunity. Immunity 2014, 41, 191–206. [Google Scholar] [CrossRef] [Green Version]
- Tudor, C.; Marchese, F.P.; Hitti, E.; Aubareda, A.; Rawlinson, L.; Gaestel, M.; Blackshear, P.J.; Clark, A.R.; Saklatvala, J.; Dean, J.L. The p38 mapk pathway inhibits tristetraprolin-directed decay of interleukin-10 and pro-inflammatory mediator mrnas in murine macrophages. FEBS Lett. 2009, 583, 1933–1938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Wagner, B.J.; Ehrenman, K.; Schaefer, A.W.; DeMaria, C.T.; Crater, D.; DeHaven, K.; Long, L.; Brewer, G. Purification, characterization, and cdna cloning of an au-rich element rna-binding protein, auf1. Mol. Cell Biol. 1993, 13, 7652–7665. [Google Scholar]
- Casolaro, V.; Fang, X.; Tancowny, B.; Fan, J.; Wu, F.; Srikantan, S.; Asaki, S.Y.; De Fanis, U.; Huang, S.K.; Gorospe, M.; et al. Posttranscriptional regulation of il-13 in t cells: Role of the rna-binding protein hur. J. Allergy Clin. Immunol. 2008, 121, 853–859. [Google Scholar] [CrossRef] [Green Version]
- Umland, S.P.; Razac, S.; Shah, H.; Nahrebne, D.K.; Egan, R.W.; Billah, M.M. Interleukin-5 mrna stability in human t cells is regulated differently than interleukin-2, interleukin-3, interleukin-4, granulocyte/macrophage colony-stimulating factor, and interferon-gamma. Am. J. Respir. Cell Mol. Biol. 1998, 18, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Stassen, M.; Schmitt, E.; Bopp, T. From interleukin-9 to t helper 9 cells. Ann. N. Y. Acad. Sci. 2012, 1247, 56–68. [Google Scholar] [CrossRef]
- Redpath, S.A.; Heieis, G.; Perona-Wright, G. Spatial regulation of il-4 signalling in vivo. Cytokine 2015, 75, 51–56. [Google Scholar] [CrossRef]
- Akdis, C.A.; Arkwright, P.D.; Brüggen, M.C.; Busse, W.; Gadina, M.; Guttman-Yassky, E.; Kabashima, K.; Mitamura, Y.; Vian, L.; Wu, J.; et al. Type 2 immunity in the skin and lungs. Allergy 2020, 75, 1582–1605. [Google Scholar] [CrossRef]
- Sherwood, E.R.; Burelbach, K.R.; McBride, M.A.; Stothers, C.L.; Owen, A.M.; Hernandez, A.; Patil, N.K.; Williams, D.L.; Bohannon, J.K. Innate immune memory and the host response to infection. J. Immunol. 2022, 208, 785–792. [Google Scholar] [CrossRef]
- Vidya, M.K.; Kumar, V.G.; Sejian, V.; Bagath, M.; Krishnan, G.; Bhatta, R. Toll-like receptors: Significance, ligands, signaling pathways, and functions in mammals. Nat. Rev. Immunol. 2018, 37, 20–36. [Google Scholar] [CrossRef]
- Greene, J.T.; Brian, B.F.t.; Senevirathne, S.E.; Freedman, T.S. Regulation of myeloid-cell activation. Curr. Opin. Immunol. 2021, 73, 34–42. [Google Scholar] [CrossRef]
- Chan, L.; Karimi, N.; Morovati, S.; Alizadeh, K.; Kakish, J.E.; Vanderkamp, S.; Fazel, F.; Napoleoni, C.; Alizadeh, K.; Mehrani, Y.; et al. The roles of neutrophils in cytokine storms. Viruses 2021, 13, 2318. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, L.; Pandeya, A.; Cui, J.; Zhang, Y.; Li, Z. Pyroptosis-induced inflammation and tissue damage. J. Mol. Biol. 2022, 434, 167301. [Google Scholar] [CrossRef] [PubMed]
- Minaga, K.; Watanabe, T.; Hara, A.; Yoshikawa, T.; Kamata, K.; Kudo, M. Plasmacytoid dendritic cells as a new therapeutic target for autoimmune pancreatitis and igg4-related disease. Front. Immunol. 2021, 12, 713779. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.W.; Forero, A. Beyond good and evil: Molecular mechanisms of type i and iii ifn functions. J. Immunol. 2022, 208, 247–256. [Google Scholar] [CrossRef]
- Chiang, H.S.; Liu, H.M. The molecular basis of viral inhibition of irf- and stat-dependent immune responses. Front. Immunol. 2018, 9, 3086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Cao, M.; Yi, S.; Cheng, J.; Wang, L.; Tao, Y.; Wu, D.; Peng, J.; Zhang, M.; Qi, P.; et al. Effects of the rna-binding protein, ksrp, on innate immune response against helicobacter pylori infection in mice. Biochem. Biophys. Res. Commun. 2018, 495, 1573–1579. [Google Scholar] [CrossRef]
- Zeng, R.Z.; Kim, H.G.; Kim, N.R.; Lee, H.Y.; Jung, B.J.; Ko, M.Y.; Lee, S.Y.; Chung, D.K. Protein expression changes in human monocytic thp-1 cells treated with lipoteichoic acid from lactobacillus plantarum and staphylococcus aureus. Mol. Cells 2010, 29, 585–594. [Google Scholar] [CrossRef]
- Nie, Y.; Cao, M.; Wu, D.; Li, N.; Peng, J.; Yi, S.; Yang, X.; Zhang, M.; Hu, G.; Zhao, J. Kh-type splicing regulatory protein is regulated by nuclear factor-κb signaling to mediate innate immunity in caco-2 cells infected by salmonella enteritidis. Folia. Microbiol. 2018, 63, 669–676. [Google Scholar] [CrossRef]
- Ignat, S.R.; Dinescu, S.; Hermenean, A.; Costache, M. Cellular interplay as a consequence of inflammatory signals leading to liver fibrosis development. Cells 2020, 9, 461. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Li, M.; Zhao, X.; Wang, H.; Zhu, J.; Wang, C.; Zhou, M.; Dong, H.; Zhou, R. Upregulation of ksrp by mir-27b attenuates schistosomiasis-induced hepatic fibrosis by targeting tgf-β1. FASEB J. 2020, 34, 4120–4133. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Bozec, A.; Ramming, A.; Schett, G. Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nat. Rev. Rheumatol. 2019, 15, 9–17. [Google Scholar] [CrossRef]
- Gordon, R.A.; Tilstra, J.S.; Marinov, A.; Nickerson, K.M.; Bastacky, S.I.; Shlomchik, M.J. Murine lupus is neutrophil elastase-independent in the mrl.Faslpr model. PLoS ONE 2020, 15, e0226396. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Lu, Y.; Li, X.; Mao, T.; Chen, X.M.; Zhou, R. Upregulation of ksrp by mir-27b provides ifn-γ-induced post-transcriptional regulation of cx3cl1 in liver epithelial cells. Sci. Rep. 2015, 5, 17590. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Kim, M.H.; Yi, H.S.; Kim, S.Y.; Kim, H.H.; Kim, J.H.; Yeon, J.E.; Byun, K.S.; Byun, J.S.; Jeong, W.I. Cx(3)cr1 differentiates f4/80(low) monocytes into pro-inflammatory f4/80(high) macrophages in the liver. Sci. Rep. 2018, 8, 15076. [Google Scholar] [CrossRef]
- Sutti, S.; Heymann, F.; Bruzzì, S.; Peusquens, J.; Trautwein, C.; Albano, E.; Tacke, F. Cx(3)cr1 modulates the anti-inflammatory activity of hepatic dendritic cells in response to acute liver injury. Clin. Sci. 2017, 131, 2289–2301. [Google Scholar] [CrossRef]
- Roesch, E.A.; Nichols, D.P.; Chmiel, J.F. Inflammation in cystic fibrosis: An update. Pediatr. Pulmonol. 2018, 53, S30–S50. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, S.; Kumar, P.; Tsuchiya, M.; Bhattacharyya, A.; Biswas, R. Regulation of mir-155 biogenesis in cystic fibrosis lung epithelial cells: Antagonistic role of two mrna-destabilizing proteins, ksrp and ttp. Biochem. Biophys. Res. Commun. 2013, 433, 484–488. [Google Scholar] [CrossRef]
- Tong, L.; Luo, Y.; Wei, T.; Guo, L.; Wang, H.; Zhu, W.; Zhang, J. Kh-type splicing regulatory protein (khsrp) contributes to tumorigenesis by promoting mir-26a maturation in small cell lung cancer. Mol. Cell. Biochem. 2016, 422, 61–74. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Zerayesus, S.A.; Van Scoyk, M.; Wilson, L.; Wu, P.Y.; Baskaran, A.; Tang, K.; Raheem, S.; Samuelson, B.A.; Reddy, N.M.; et al. K-homology splicing regulatory protein (ksrp) promotes post-transcriptional destabilization of spry4 transcripts in non-small cell lung cancer. J. Biol. Chem. 2017, 292, 7423–7434. [Google Scholar] [CrossRef] [Green Version]
- Yan, M.; Sun, L.; Li, J.; Yu, H.; Lin, H.; Yu, T.; Zhao, F.; Zhu, M.; Liu, L.; Geng, Q.; et al. Rna-binding protein khsrp promotes tumor growth and metastasis in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2019, 38, 478. [Google Scholar] [CrossRef]
- Chien, M.H.; Lee, W.J.; Yang, Y.C.; Li, Y.L.; Chen, B.R.; Cheng, T.Y.; Yang, P.W.; Wang, M.Y.; Jan, Y.H.; Lin, Y.K.; et al. Ksrp suppresses cell invasion and metastasis through mir-23a-mediated egr3 mrna degradation in non-small cell lung cancer. Biochim. Biophys. Acta Gene. Regul. Mech. 2017, 1860, 1013–1024. [Google Scholar] [CrossRef]
- Caiazza, F.; Oficjalska, K.; Tosetto, M.; Phelan, J.J.; Noonan, S.; Martin, P.; Killick, K.; Breen, L.; O’Neill, F.; Nolan, B.; et al. Kh-type splicing regulatory protein controls colorectal cancer cell growth and modulates the tumor microenvironment. Am. J. Pathol. 2019, 189, 1916–1932. [Google Scholar] [CrossRef] [Green Version]
- Pan, R.; Cai, W.; Sun, J.; Yu, C.; Li, P.; Zheng, M. Inhibition of khsrp sensitizes colorectal cancer to 5-fluoruracil through mir-501-5p-mediated errfi1 mrna degradation. J. Cell. Physiol. 2020, 235, 1576–1587. [Google Scholar] [CrossRef]
- Oh, H.H.; Park, K.J.; Kim, N.; Park, S.Y.; Park, Y.L.; Oak, C.Y.; Myung, D.S.; Cho, S.B.; Lee, W.S.; Kim, K.K.; et al. Impact of kitenin on tumor angiogenesis and lymphangiogenesis in colorectal cancer. Oncol. Rep. 2016, 35, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Bae, J.A.; Bae, W.K.; Kim, S.J.; Ko, Y.S.; Kim, K.Y.; Park, S.Y.; Yu, Y.H.; Kim, E.A.; Chung, I.J.; Kim, H.; et al. A new ksrp-binding compound suppresses distant metastasis of colorectal cancer by targeting the oncogenic kitenin complex. Mol. Cancer 2021, 20, 78. [Google Scholar] [CrossRef]
- Chen, M.; Wang, F.; Xia, H.; Yao, S. Microrna-155: Regulation of immune cells in sepsis. Mediat. Inflamm. 2021, 2021, 8874854. [Google Scholar] [CrossRef]
- Yang, J.; Fan, J.; Li, Y.; Li, F.; Chen, P.; Fan, Y.; Xia, X.; Wong, S.T. Genome-wide rnai screening identifies genes inhibiting the migration of glioblastoma cells. PLoS ONE 2013, 8, e61915. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Wei, B.; Wei, J.; Li, Z.; Tian, Y.; Du, C. Caspase-related apoptosis genes in gliomas by rna-seq and bioinformatics analysis. J. Clin. Neurosci. 2016, 33, 259–263. [Google Scholar] [CrossRef]
- Mattiotti, A.; Prakash, S.; Barnett, P.; van den Hoff, M.J.B. Follistatin-like 1 in development and human diseases. Exp.Cell. Mol. Life Sci. 2018, 75, 2339–2354. [Google Scholar] [CrossRef]
- Sundaram, G.M.; Quah, S.; Guang, L.G.; Sampath, P. Hur enhances fstl1 transcript stability to promote invasion and metastasis of squamous cell carcinoma. Am. J. Cancer Res. 2021, 11, 4981–4993. [Google Scholar]
- Nie, E.; Jin, X.; Miao, F.; Yu, T.; Zhi, T.; Shi, Z.; Wang, Y.; Zhang, J.; Xie, M.; You, Y. Tgf-β1 modulates temozolomide resistance in glioblastoma via altered microrna processing and elevated mgmt. Neuro Oncol. 2021, 23, 435–446. [Google Scholar] [CrossRef]
- Huang, X.; Liu, X.; Du, B.; Liu, X.; Xue, M.; Yan, Q.; Wang, X.; Wang, Q. Lncrna linc01305 promotes cervical cancer progression through khsrp and exosome-mediated transfer. Aging 2021, 13, 19230–19242. [Google Scholar] [CrossRef]
- Santarosa, M.; Del Col, L.; Viel, A.; Bivi, N.; D’Ambrosio, C.; Scaloni, A.; Tell, G.; Maestro, R. Brca1 modulates the expression of hnrnpa2b1 and khsrp. Cell Cycle 2010, 9, 4666–4673. [Google Scholar] [CrossRef] [Green Version]
- Walerych, D.; Lisek, K.; Sommaggio, R.; Piazza, S.; Ciani, Y.; Dalla, E.; Rajkowska, K.; Gaweda-Walerych, K.; Ingallina, E.; Tonelli, C.; et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat. Cell Biol. 2016, 18, 897–909. [Google Scholar] [CrossRef]
- Suswam, E.A.; Nabors, L.B.; Huang, Y.; Yang, X.; King, P.H. Il-1beta induces stabilization of il-8 mrna in malignant breast cancer cells via the 3′ untranslated region: Involvement of divergent rna-binding factors hur, ksrp and tiar. Int. J. Cancer 2005, 113, 911–919. [Google Scholar] [CrossRef]
- Zubaidah, R.M.; Tan, G.S.; Tan, S.B.; Lim, S.G.; Lin, Q.; Chung, M.C. 2-d dige profiling of hepatocellular carcinoma tissues identified isoforms of far upstream binding protein (fubp) as novel candidates in liver carcinogenesis. Proteomics 2008, 8, 5086–5096. [Google Scholar] [CrossRef]
- Krishnan, N.; Titus, M.A.; Thapar, R. The prolyl isomerase pin1 regulates mrna levels of genes with short half-lives by targeting specific rna binding proteins. PLoS ONE 2014, 9, e85427. [Google Scholar] [CrossRef]
- Fujita, Y.; Masuda, K.; Hamada, J.; Shoda, K.; Naruto, T.; Hamada, S.; Miyakami, Y.; Kohmoto, T.; Watanabe, M.; Takahashi, R.; et al. Kh-type splicing regulatory protein is involved in esophageal squamous cell carcinoma progression. Oncotarget 2017, 8, 101130–101145. [Google Scholar] [CrossRef] [Green Version]
- Pruksakorn, D.; Teeyakasem, P.; Klangjorhor, J.; Chaiyawat, P.; Settakorn, J.; Diskul-Na-Ayudthaya, P.; Chokchaichamnankit, D.; Pothacharoen, P.; Srisomsap, C. Overexpression of kh-type splicing regulatory protein regulates proliferation, migration, and implantation ability of osteosarcoma. Int. J. Oncol. 2016, 49, 903–912. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Chou, C.F.; Liu, S.; Crossman, D.; Yusuf, N.; Wu, Y.; Chen, C.Y. Ksrp modulates melanoma growth and efficacy of vemurafenib. Biochim. Biophys. Acta Gene. Regul. Mech. 2019, 1862, 759–770. [Google Scholar] [CrossRef]
- Taniuchi, K.; Ogasawara, M. Khsrp-bound small nucleolar rnas associate with promotion of cell invasiveness and metastasis of pancreatic cancer. Oncotarget 2020, 11, 131–147. [Google Scholar] [CrossRef]
- Huang, J.; Sachdeva, M.; Xu, E.; Robinson, T.J.; Luo, L.; Ma, Y.; Williams, N.T.; Lopez, O.; Cervia, L.D.; Yuan, F.; et al. The long noncoding rna neat1 promotes sarcoma metastasis by regulating rna splicing pathways. Mol. Cancer Res. 2020, 18, 1534–1544. [Google Scholar] [CrossRef]
- Gou, Q.; Gao, L.; Nie, X.; Pu, W.; Zhu, J.; Wang, Y.; Liu, X.; Tan, S.; Zhou, J.K.; Gong, Y.; et al. Long noncoding rna ab074169 inhibits cell proliferation via modulation of khsrp-mediated cdkn1a expression in papillary thyroid carcinoma. Cancer Res. 2018, 78, 4163–4174. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).