ANXA11 rs1049550 Associates with Löfgren’s Syndrome and Chronic Sarcoidosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Genotyping
2.3. Meta-Analysis
2.4. Statistical Analysis
3. Results
3.1. Sarcoidosis Patients and Subgroups
3.2. Association of ANXA11 rs1049550 with Sarcoidosis
3.3. Meta-Analysis
3.4. Sarcoidosis Phenotype Studies
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karakaya, B.; Kaiser, Y.; Grunewald, J.; van Moorsel, C. Löfgren’s Syndrome: Diagnosis, Management, and Disease Pathogenesis. Semin. Respir. Crit. Care Med. 2017, 38, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Drent, M.; Crouser, E.D.; Grunewald, J. Challenges of Sarcoidosis and Its Management. N. Engl. J. Med. 2021, 385, 1018–1032. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Franke, A.; Fischer, A.; Jacobs, G.; Nothnagel, M.; Gaede, K.I.; Schürmann, M.; Müller-Quernheim, J.; Krawczak, M.; Rosenstiel, P.; et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat. Genet. 2008, 40, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pabst, S.; Kubisch, C.; Grohé, C.; Wollnik, B. First independent replication study confirms the strong genetic association of ANXA11 with sarcoidosis. Thorax 2010, 65, 939–940. [Google Scholar] [CrossRef]
- Mrazek, F.; Stahelova, A.; Kriegova, E.; Fillerova, R.; Zurkova, M.; Kolek, V.; Petrek, M. Functional variant ANXA11 R230C: True marker of protection and candidate disease modifier in sarcoidosis. Genes Immun. 2011, 12, 490–494. [Google Scholar] [CrossRef][Green Version]
- Levin, A.M.; Iannuzzi, M.C.; Montgomery, C.G.; Trudeau, S.; Datta, I.; McKeigue, P.; Fischer, A.; Nebel, A.; Rybicki, B.A. Association of ANXA11 genetic variation with sarcoidosis in African Americans and European Americans. Genes Immun. 2013, 14, 13–18. [Google Scholar] [CrossRef][Green Version]
- Morais, A.; Lima, B.; Peixoto, M.; Melo, N.; Alves, H.; Marques, J.A.; Delgado, L. Annexin A11 gene polymorphism (R230C variant) and sarcoidosis in a Portuguese population. Tissue Antigens 2013, 82, 186–191. [Google Scholar] [CrossRef]
- Feng, X.; Zang, S.; Yang, Y.; Zhao, S.; Li, Y.; Gao, X.; Zhang, L. Annexin A11 (ANXA11) gene polymorphisms are associated with sarcoidosis in a Han Chinese population: A case-control study. BMJ Open 2014, 4, 1–6. [Google Scholar] [CrossRef]
- Sikorova, K.; Kishore, A.; Rapti, A.; Adam, K.; Kocourkova, L.; Zizkova, V.; Charikiopoulou, M.; Kalianos, A.; Bouros, E.; Bouros, D.; et al. Association of TGF-β3 and ANXA11 with pulmonary sarcoidosis in Greek population. Expert Rev. Respir. Med. 2020, 14, 1065–1069. [Google Scholar] [CrossRef]
- Mirsaeidi, M.; Vu, A.; Zhang, W.; Arbieva, Z.; Zhang, C.; Abbasi, T.; Hakim, A.; Schraufnagel, D.; Sweiss, N.; Baughman, R.; et al. Annexin A11 is associated with pulmonary fibrosis in African American patients with sarcoidosis. Sarcoidosis Vasc. Diffus. Lung Dis. Off. J. WASOG 2016, 33, 418–422. [Google Scholar]
- Calender, A.; Weichhart, T.; Valeyre, D.; Pacheco, Y. Current insights in genetics of sarcoidosis: Functional and clinical impacts. J. Clin. Med. 2020, 9, 2633. [Google Scholar] [CrossRef]
- Van Moorsel, C.H.M.; Petrek, M.; Rivera, N. Unravelling the genetic basis of sarcoidosis. Eur. Respir. Monogr. 2022, 96, 41–56. [Google Scholar]
- Statement on sarcoidosis. Joint Statement of the American Thoracic Society (ATS), the European Respiratory Society (ERS) and the World Association of Sarcoidosis and Other Granulomatous Disorders (WASOG) adopted by the ATS Board of Directors and by the ER. Am. J. Respir. Crit. Care Med. 1999, 160, 736–755. [CrossRef]
- Scadding, J.G. Prognosis of Intrathoracic Sarcoidosis in England: A review of 136 cases after 5 years’ observation. Br. Med. J. 1961, 1165–1172. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- Martorell-Marugan, J.; Toro-Dominguez, D.; Alarcon-Riquelme, M.E.; Carmona-Saez, P. MetaGenyo: A web tool for meta-analysis of genetic association studies. BMC Bioinform. 2017, 18, 1–6. [Google Scholar] [CrossRef]
- Hofmann, S.; Franke, A.; Fischer, A.; Jacobs, G.; Nothnagel, M.; Gaede, K.I.; Schürmann, M.; Müller-quernheim, J.; Krawczak, M.; Rosenstiel, P.; et al. Corrigendum: Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat. Genet. 2009, 41, 504. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Zhou, H.; Diao, M.; Zhang, M. The Association between ANXA11 Gene Polymorphisms and Sarcoidosis: A Meta-Analysis and systematic review. Sarcoidosis Vasc. Diffus. Lung Dis. Off. J. WASOG 2016, 33, 102–111. [Google Scholar]
- Grunewald, J.; Brynedal, B.; Darlington, P.; Nisell, M.; Cederlund, K.; Hillert, J.; Eklund, A. Different HLA-DRB1 allele distributions in distinct clinical subgroups of sarcoidosis patients. Respir. Res. 2010, 11, 25. [Google Scholar] [CrossRef]
- Karakaya, B.; van Moorsel, C.H.M.; Veltkamp, M.; Roodenburg-Benschop, C.; Kazemier, K.M.; van der Helm-van Mil, A.H.M.; Huizinga, T.W.J.; Grutters, J.C.; Rijkers, G.T. A Polymorphism in C-C Chemokine Receptor 5 (CCR5) Associates with Löfgren’s Syndrome and Alters Receptor Expression as well as Functional Response. Cells 2021, 10. [Google Scholar] [CrossRef]
- Spagnolo, P.; Sato, H.; Grunewald, J.; Brynedal, B.; Hillert, J.; Mañá, J.; Wells, A.U.; Eklund, A.; Welsh, K.I.; Du Bois, R.M. A common haplotype of the C-C chemokine receptor 2 gene and HLA-DRB1*0301 are independent genetic risk factors for Löfgren’s syndrome. J. Intern. Med. 2008, 264, 433–441. [Google Scholar] [CrossRef]
- van Moorsel, C.H.M.; Christiani, D.C. Genetic susceptibility to sarcoidosis, a chronic inflammatory disorder. Am. J. Respir. Crit. Care Med. 2012, 186, 816–818. [Google Scholar] [CrossRef]
- Tomas, A.; Moss, S.E. Calcium- and cell cycle-dependent association of annexin 11 with the nuclear envelope. J. Biol. Chem. 2003, 278, 20210–20216. [Google Scholar] [CrossRef]
- Moss, S.E.; Morgan, R.O. The annexins. Genome Biol. 2004, 5, 219. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, C.; Liu, S.; Qi, H.; Yin, Y.; Liang, R.; Sun, M.-Z.; Greenaway, F.T. Annexin A11 in disease. Clin. Chim. Acta 2014, 431, 164–168. [Google Scholar] [CrossRef]
- Ghoussaini, M.; Mountjoy, E.; Carmona, M.; Peat, G.; Schmidt, E.M.; Hercules, A.; Fumis, L.; Miranda, A.; Carvalho-Silva, D.; Buniello, A.; et al. Open Targets Genetics: Systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021, 49, D1311–D1320. [Google Scholar] [CrossRef]
- Javierre, B.M.; Sewitz, S.; Cairns, J.; Wingett, S.W.; Várnai, C.; Thiecke, M.J.; Freire-Pritchett, P.; Spivakov, M.; Fraser, P.; Burren, O.S.; et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 2016, 167, 1369–1384. [Google Scholar] [CrossRef]
- Qazi, K.R.; Paredes, P.T.; Dahlberg, B.; Grunewald, J.; Eklund, A.; Gabrielsson, S. Proinflammatory exosomes in bronchoalveolar lavage fluid of patients with sarcoidosis. Thorax 2010, 65, 1016–1024. [Google Scholar] [CrossRef]
- Mirsaeidi, M.; Gidfar, S.; Vu, A.; Schraufnagel, D. Annexins family: Insights into their functions and potential role in pathogenesis of sarcoidosis. J. Transl. Med. 2016, 14, 1–9. [Google Scholar] [CrossRef]
| Controls | Sarcoidosis All | Löfgren’s Syndrome | Chronic Sarcoidosis | ||
|---|---|---|---|---|---|
| N | 363 | 262 | 149 | 113 | |
| Age yrs | 40 | 38 | 36 | 42 | |
| Female n (%) | 185 (50.1) | 130 (49.6) | 94 (63.1) | 36 (31.8) | |
| Scadding Stage | n = 228 | n = 115 | n = 113 | ||
| n (%) | 0 | 5 (2.2) | 5 (4.3) | ||
| I | 101 (44.3) | 101 (87.8) | |||
| II | 38 (16.7) | 9 (7.8) | 29 (25.7) | ||
| III | 13 (5.7) | 13 (11.5) | |||
| IV | 71 (31.1) | 71 (62.8) |
| Controls | Sarcoidosis All | Löfgren’s Syndrome | Chronic Sarcoidosis | ||
|---|---|---|---|---|---|
| N | 363 | 262 | 149 | 113 | |
| Genotype | |||||
| n (%) | CC | 130 (35.8) | 125 (47.7) | 66 (44.3) | 59 (52.2) |
| CT | 167 (46.0) | 118 (45.0) | 69 (46.3) | 49 (43.4) | |
| TT | 66 (18.2) | 19 (7.3) | 14 (9.4) | 5 (4.4) | |
| Allele | |||||
| n (%) | C | 427 (58.8) | 368 (70.2) | 201 (67.4) | 167 (73.9) |
| T | 299 (41.2) | 156 (29.8) | 97 (32.6) | 59 (26.1) | |
| OR * | 0.61 | 0.69 | 0.51 | ||
| 95% CI | 0.48–0.77 | 0.52–0.92 | 0.36–0.70 | ||
| p | 3 × 10−5 | 0.01 | 4 × 10−5 |
| Additive | Dominant (CC vs. CT+TT) | Recessive (CC+CT vs. TT) | |||||
|---|---|---|---|---|---|---|---|
| n | OR, 95% CI * | p Value | OR, 95% CI * | p Value | OR, 95% CI * | p Value | |
| Controls | 363 | ||||||
| Sarcoidosis | 262 | 0.61, 0.48–0.77 | <0.0001 | 0.61, 0.44–0.85 | 0.0029 | 0.35, 0.21–0.60 | <0.0001 |
| Löfgren’s syndrome | 149 | 0.69, 0.52–0.92 | 0.01 | 0.70, 0.48–1.03 | 0.074 | 0.47, 0.25–0.86 | 0.0095 |
| Chronic Sarcoidosis | 113 | 0.51, 0.36–0.71 | <0.0001 | 0.51, 0.33–0.78 | 0.002 | 0.21, 0.08–0.53 | 0.0001 |
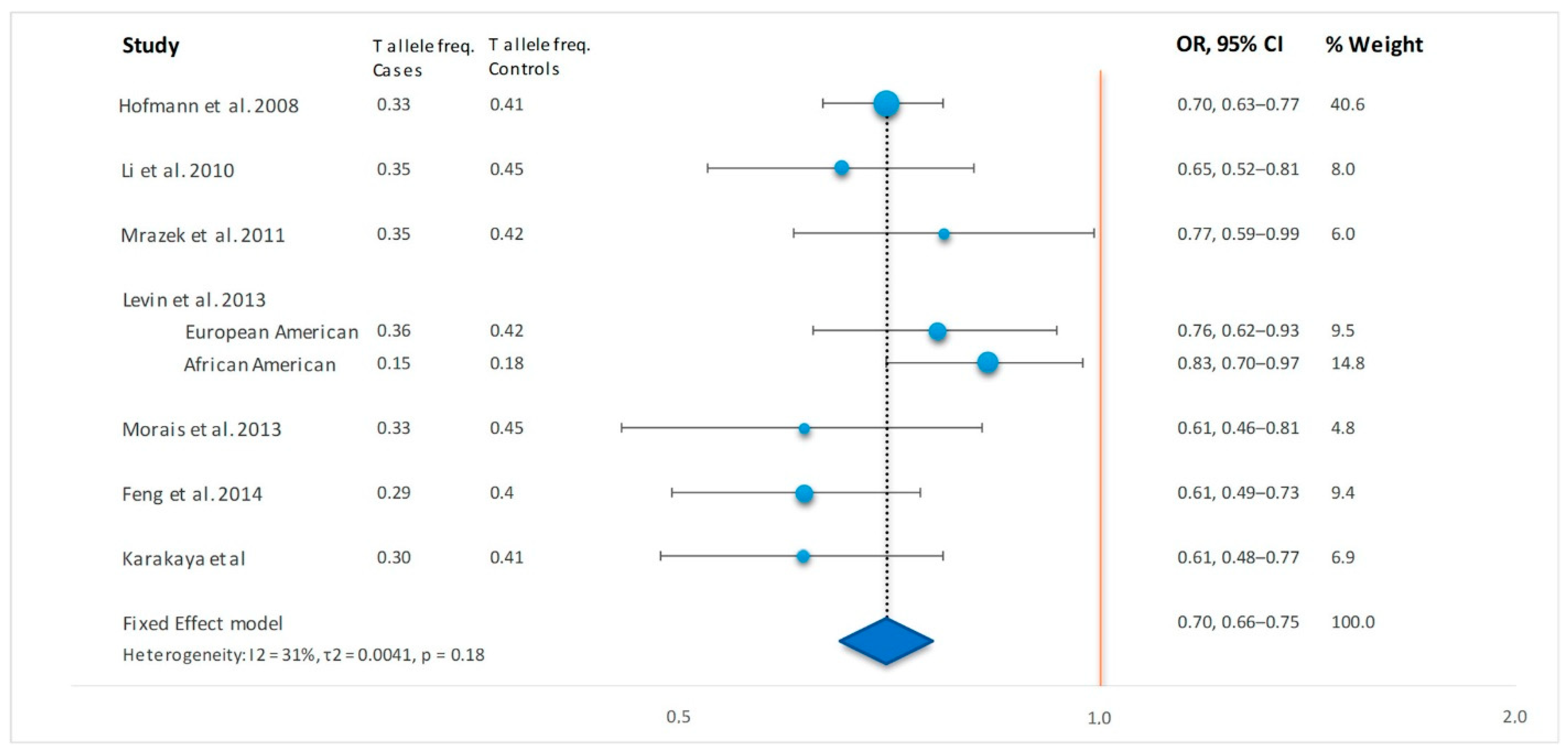
| Author | Year | Country | Ethnicity | Case/Control, N | Case T Allele Freq | Control T Allele Freq | OR, 95% CI, p Value * | Population T Allele Frequency gnomAD |
|---|---|---|---|---|---|---|---|---|
| Hofmann S et al. [3] | 2008 | Germany | Caucasian | 1636/1811 | 0.33 | 0.41 | 0.70, 0.63–0.77, 1 × 10−12 | European: 0.42 |
| Li et al. [4] | 2010 | Germany | Caucasian | 349/313 | 0.35 | 0.45 | 0.65, 0.52–0.81, 1 × 10−4 | European: 0.42 |
| Mrazek et al. [5] | 2011 | Czech | Caucasian | 245/254 | 0.35 | 0.42 | 0.77, 0.59–0.99, 0.04 | European: 0.42 |
| Levin et al. [6] # | 2013 | USA | European American | 446/350 | 0.36 | 0.42 | 0.76, 0.62–0.93, 0.008 | European: 0.42 |
| African American | 1232/893 | 0.15 | 0.18 | 0.83, 0.70–0.97, 0.02 | African/African American: 0.20 | |||
| Morais et al. [7] | 2013 | Portugal | Caucasian | 208/197 | 0.33 | 0.45 | 0.61, 0.46–0.81, 6 × 10−4 | European: 0.42 |
| Feng et al. [8] | 2014 | China | Chinese-Han | 412/418 | 0.29 | 0.40 | 0.60, 0.49–0.73, 8 × 10−7 | East Asian: 0.66 § |
| Sikorova et al. [9] | 2020 | Greece | Caucasian | 103/100 | Not available | Not available | 0.59, 0.39–0.89, 0.01 † | European: 0.42 |
| Karakaya et al. | Netherlands | Caucasian | 262/363 | 0.30 | 0.41 | 0.61, 0.48–0.77, 3 × 10−5 | European: 0.42 |
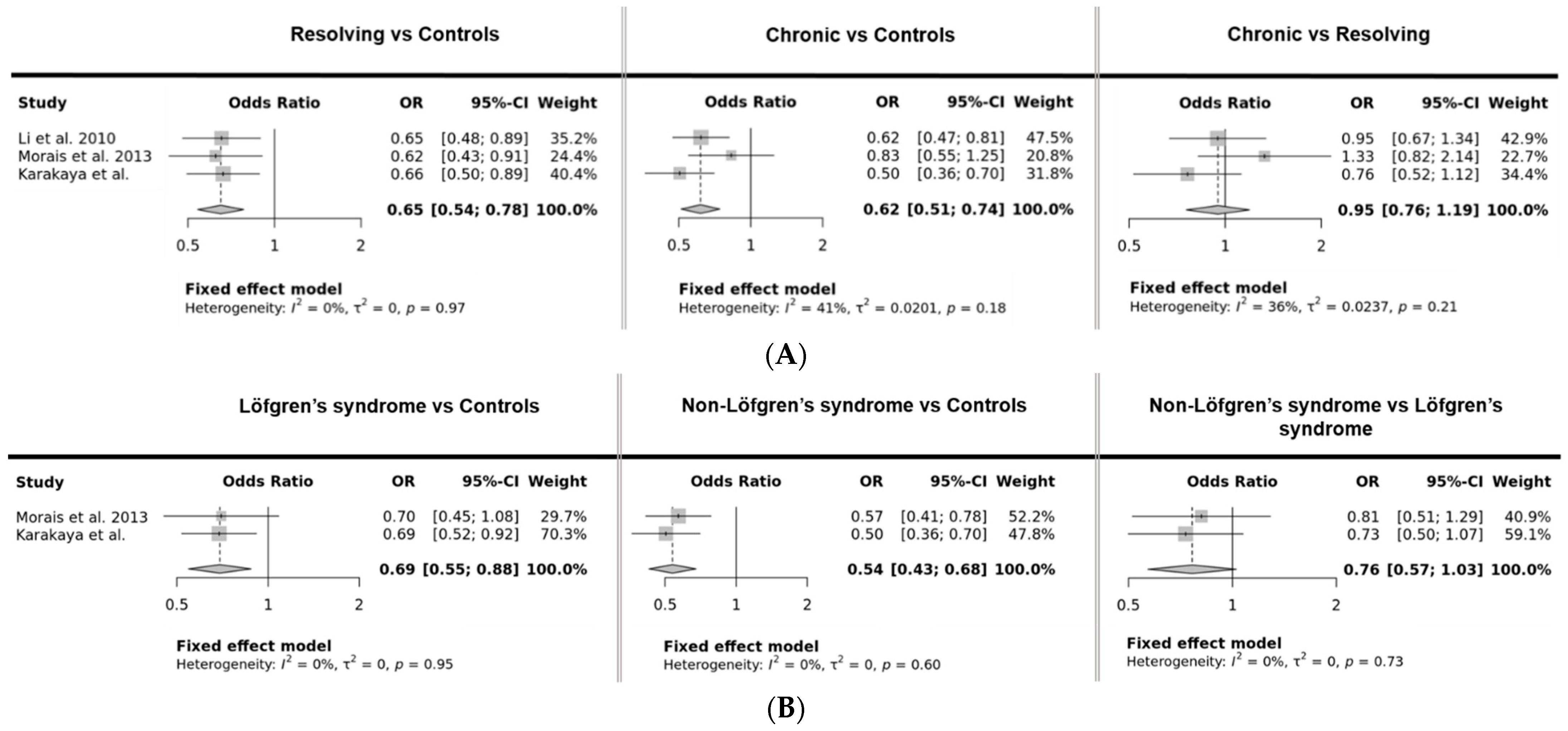
| Study | T Allele Frequency | OR, 95% CI, p Value * | ||||
|---|---|---|---|---|---|---|
| Controls | Resolving | Chronic | Resolving vs. Controls | Chronic vs. Controls | Chronic vs. Resolving | |
| Li et al, 2010 [4] | 0.45 (n = 313) | 0.35 (n = 117) | 0.34 (n = 176) | 0.65, 0.48–0.89, 0.007 | 0.62, 0.47–0.81, 5 × 10−4 | 0.95, 0.67–1.34, 0.76 |
| Levin et al. 2013 [6] # | 0.18 (n = 893) | n = 304 | n = 650 | 0.82, 0.64–1.06, 0.13 §,† | 0.79, 0.65–0.95, 0.02 §,† | |
| Morais et al. 2013 [7] | 0.45 (n = 197) | 0.34 (n = 86) | 0.40 (n = 62) | 0.62, 0.43–0.91, 0.01 | 0.83, 0.55–1.25, 0.37 | 1.33, 0.82–2.14, 0.24 |
| Karakaya et al. | 0.41 (n = 363) | 0.32 (n = 142) | 0.26 (n = 113) | 0.66, 0.50–0.89, 0.005 | 0.51, 0.36–0.70, 4 × 10−5 | 0.76, 0.52–1.12, 0.65 |
| Controls | Löfgren’s Syndrome | Non-Löfgren’s Syndrome | Löfgren’s Syndrome vs. Controls | Non-Löfgren’s Syndrome vs. Controls | Non-Löfgren’s Syndrome vs. Löfgren’s Syndrome | |
| Morais, 2013 [7] | 0.45 (n = 197) | 0.36 (n = 55) | 0.32 (n = 145) | 0.70, 0.45–1.08, 0.11 | 0.57, 0.42–0.78, 5 × 10−4 | 0.81, 0.51–1.29, 0.38 |
| Karakaya et al. | 0.41 (n = 363) | 0.33 (n = 149) | 0.26 (n = 113) | 0.69, 0.52–0.92, 0.01 | 0.51, 0.36–0.70, 4 × 10−5 | 0.73, 0.50–1.07, 0.11 |
| Mrazek, 2011 [5] | TT frequency: 0.15 (n = 254) | TT frequency: 0.21 (n = 39) | TT frequency: 0.07 (n = 147) | 0.31, 0.11–0.84, 0.02 † | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karakaya, B.; van der Vis, J.J.; Veltkamp, M.; Biesma, D.H.; Grutters, J.C.; van Moorsel, C.H.M. ANXA11 rs1049550 Associates with Löfgren’s Syndrome and Chronic Sarcoidosis. Cells 2022, 11, 1557. https://doi.org/10.3390/cells11091557
Karakaya B, van der Vis JJ, Veltkamp M, Biesma DH, Grutters JC, van Moorsel CHM. ANXA11 rs1049550 Associates with Löfgren’s Syndrome and Chronic Sarcoidosis. Cells. 2022; 11(9):1557. https://doi.org/10.3390/cells11091557
Chicago/Turabian StyleKarakaya, Bekir, Joanne J. van der Vis, Marcel Veltkamp, Douwe H. Biesma, Jan C. Grutters, and Coline H. M. van Moorsel. 2022. "ANXA11 rs1049550 Associates with Löfgren’s Syndrome and Chronic Sarcoidosis" Cells 11, no. 9: 1557. https://doi.org/10.3390/cells11091557
APA StyleKarakaya, B., van der Vis, J. J., Veltkamp, M., Biesma, D. H., Grutters, J. C., & van Moorsel, C. H. M. (2022). ANXA11 rs1049550 Associates with Löfgren’s Syndrome and Chronic Sarcoidosis. Cells, 11(9), 1557. https://doi.org/10.3390/cells11091557






