Long-Distance Repression by Human Silencers: Chromatin Interactions and Phase Separation in Silencers
Abstract
1. Introduction
| Silencer Position | Target Gene | Proximal or Distal | Reference |
|---|---|---|---|
| Promoter | synapsin I (SYN1) | proximal | [16] |
| Promoter | interferon gamma (IFNG) | proximal | [17] |
| Promoter | platelet-derived growth factor subunit A (PDGFA) | proximal | [18] |
| Intron | human CD4 molecule (CD4) | proximal | [19,20] |
| Exon | chimerin 1 (CHN1) | proximal | [21] |
| Intron | collagen type IV alpha 2 chain (COL4A2) | proximal | [22] |
| Promoter | thyroid-stimulating hormone subunit beta (TSHB) | proximal | [23] |
| Promoter | serpin family B member 2 (SERPINB2) | proximal | [24] |
| Promoter | glutathione S-transferase pi 1 (GSTP1) | proximal | [25] |
| Intron | apolipoprotein A2 (APOA2) | proximal | [26] |
| Intron and UTR | methyl CpG-binding protein 2 (MECP2) | proximal | [27] |
| 15 putative silencers | unknown (silencing activity characterized using functional assays) | distal | [28] |
| Intergenic | cyclinD1 (CCND1) | distal | [29] |
| Intron | Rho GTPase-activating protein 6 (ARHGAP6) | proximal | [30] |
| 5 H3K27me3-DNase hypersensitive sites | unknown (silencing activity characterized using functional assays) | distal | [31] |
| Methylation-rich region | human fibroblast growth factor 18 (FGF18) | distal | [32] |
| Methylation-rich region | human insulin like growth factor 2 (IGF2) | distal | [32] |
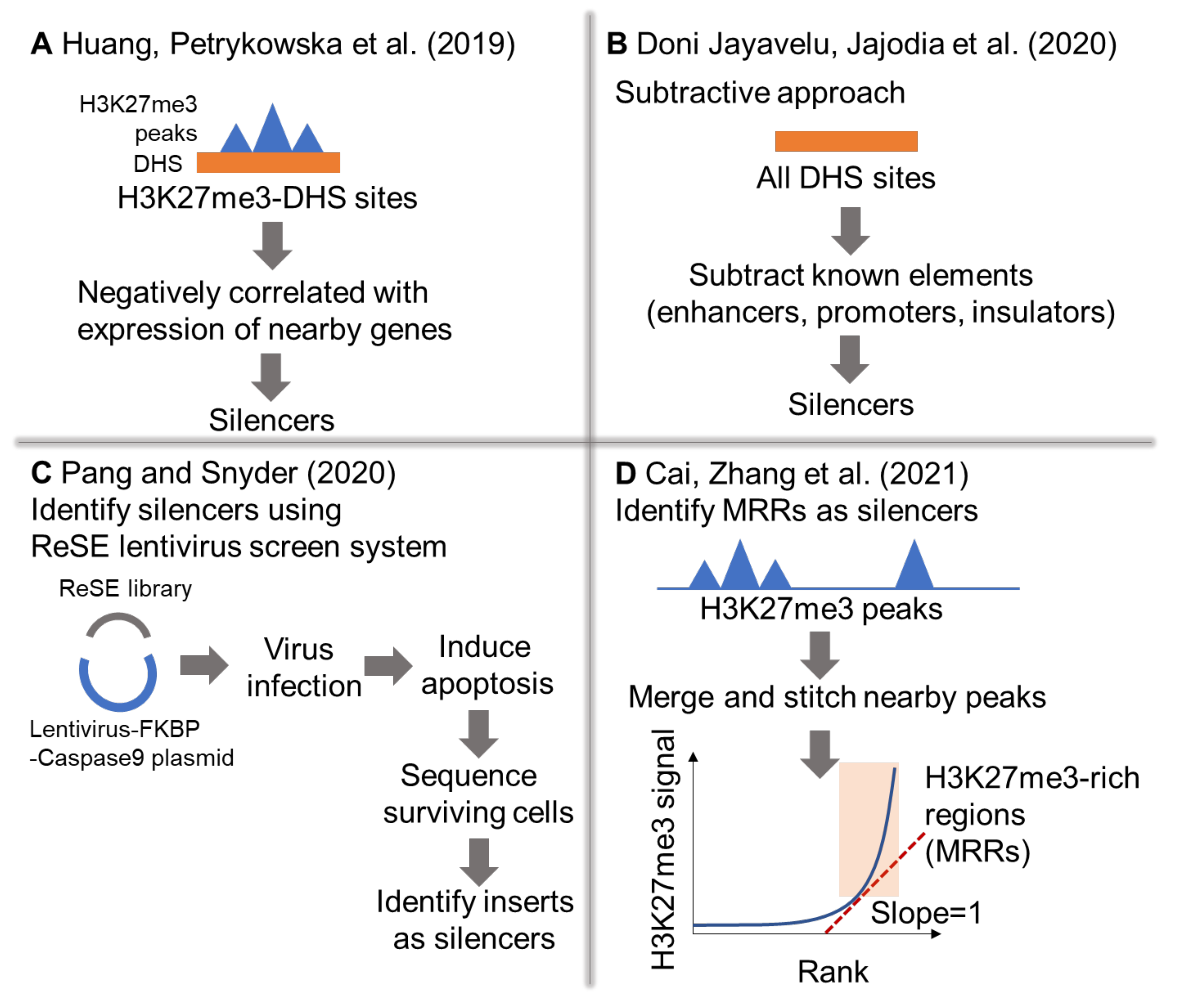
2. Genome-Wide Identification of Silencers
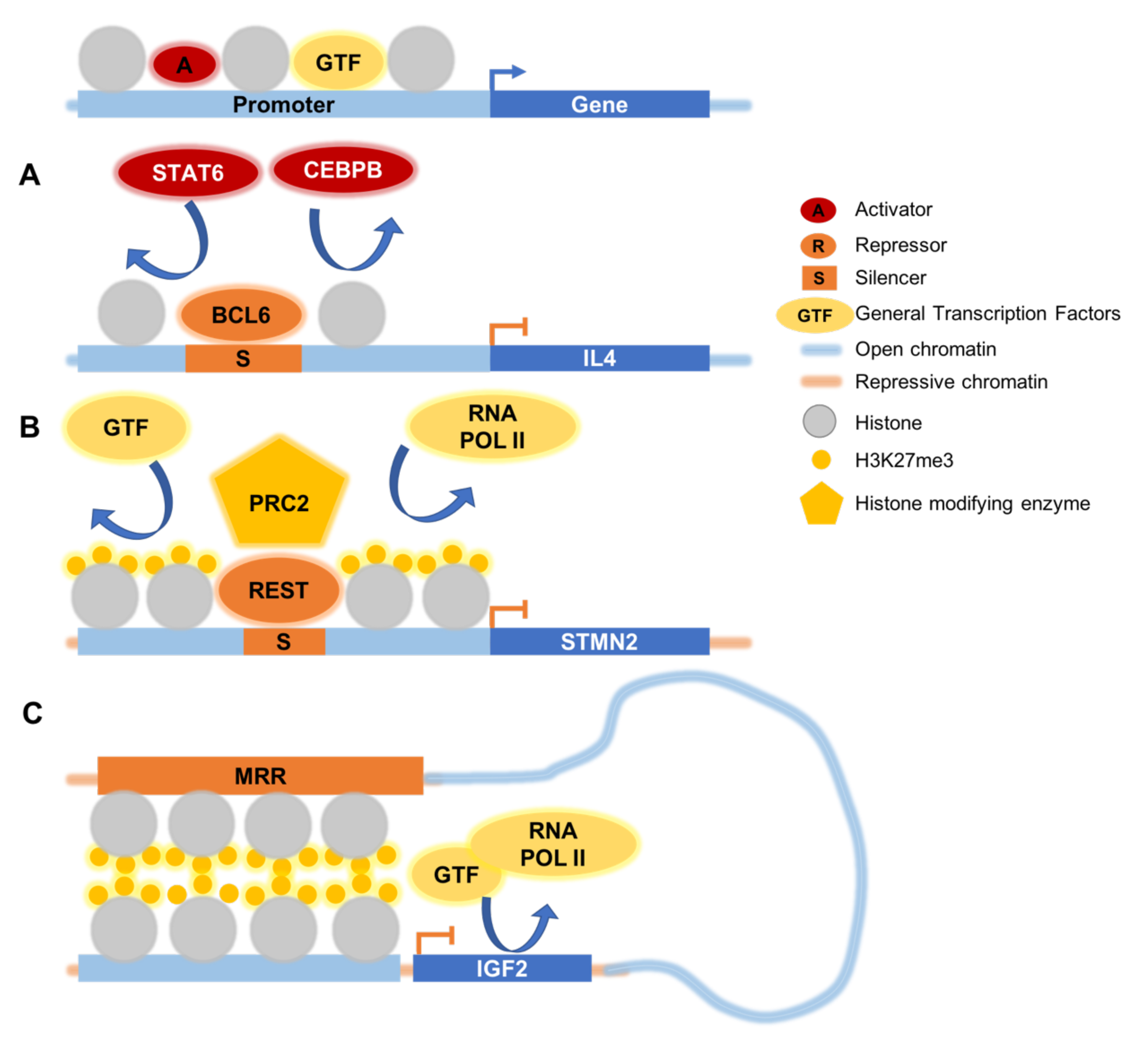
3. Silencers Interfere with Binding of Activators and Transcriptional Machinery
4. Histone Methylation at Silencers
5. Histone Deacetylation at Silencers
6. Distal Silencers Loop to Promoters to Inhibit Gene Expression
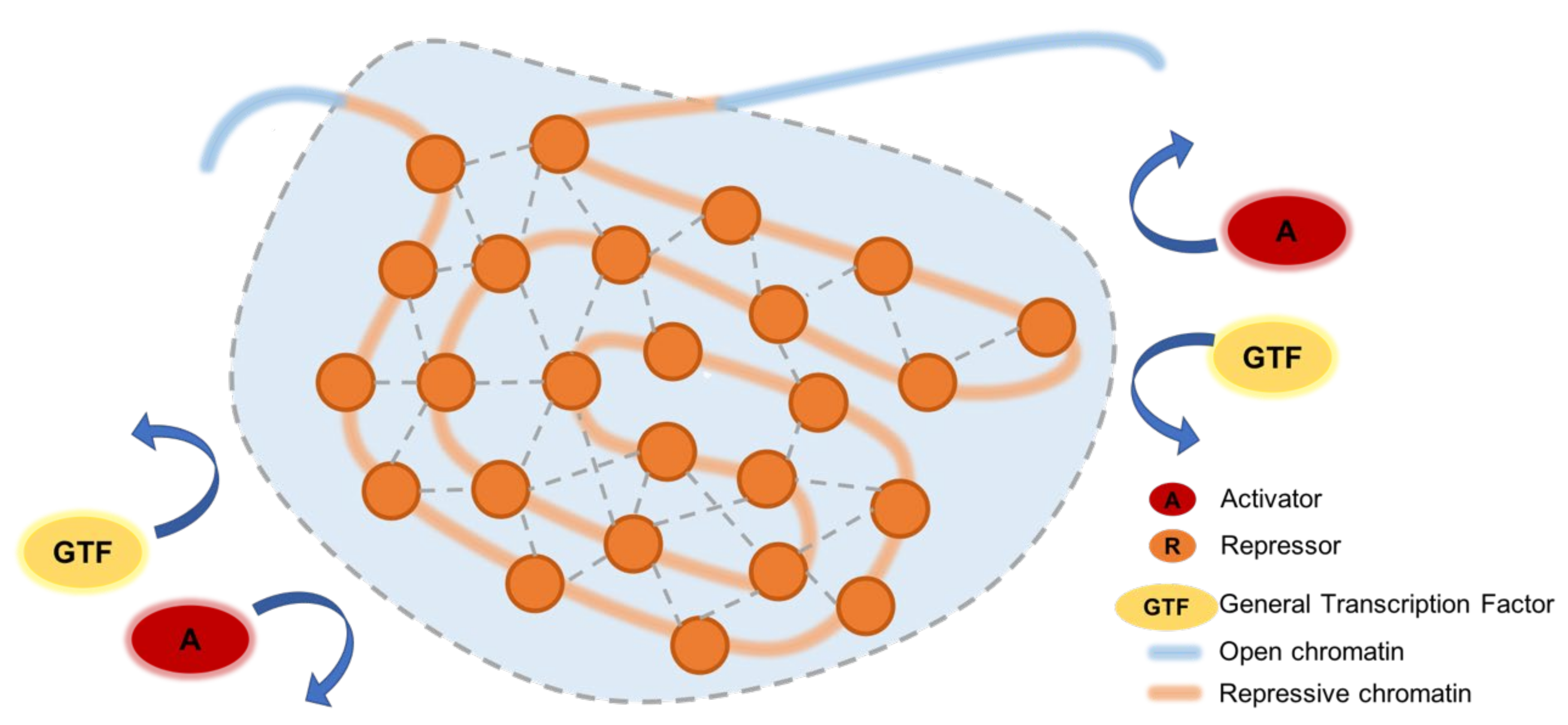
7. Phase Separation in Silencing
8. Potential Role of Non-Coding RNA (ncRNA) in Silencing
9. Silencers in Health and Disease
10. Silencer and Repressor Dysregulation in Cancer
11. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Goode, D.K.; Obier, N.; Vijayabaskar, M.S.; Lie-A-Ling, M.; Lilly, A.J.; Hannah, R.; Lichtinger, M.; Batta, K.; Florkowska, M.; Patel, R.; et al. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation. Dev. Cell 2016, 36, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Heintzman, N.D.; Hon, G.C.; Hawkins, R.D.; Kheradpour, P.; Stark, A.; Harp, L.F.; Ye, Z.; Lee, L.K.; Stuart, R.K.; Ching, C.W.; et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009, 459, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E.M. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Roeder, R.G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996, 21, 327–335. [Google Scholar] [CrossRef]
- Kadonaga, J.T. Perspectives on the RNA polymerase II core promoter. WIREs Dev. Biol. 2012, 1, 40–51. [Google Scholar] [CrossRef]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef]
- Buecker, C.; Wysocka, J. Enhancers as information integration hubs in development: Lessons from genomics. Trends Genet. 2012, 28, 276–284. [Google Scholar] [CrossRef]
- Smallwood, A.; Ren, B. Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol. 2013, 25, 387–394. [Google Scholar] [CrossRef]
- Schoenfelder, S.; Fraser, P. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 20, 437–455. [Google Scholar] [CrossRef]
- Akincilar, S.C.; Khattar, E.; Boon, P.L.; Unal, B.; Fullwood, M.J.; Tergaonkar, V. Long-Range Chromatin Interactions Drive Mutant TERT Promoter Activation. Cancer Discov. 2016, 6, 1276–1291. [Google Scholar] [CrossRef] [PubMed]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931. [Google Scholar] [CrossRef] [PubMed]
- Ogbourne, S.; Antalis, T.M. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998, 331 Pt 1, 1–14. [Google Scholar] [CrossRef]
- Maston, G.A.; Evans, S.K.; Green, M.R. Transcriptional Regulatory Elements in the Human Genome. Annu. Rev. Genom. Hum. Genet. 2006, 7, 29–59. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Suzuki, T.; Mori, N.; Greengard, P. Identification of a functional silencer element involved in neuron-specific expression of the synapsin I gene. Proc. Natl. Acad. Sci. USA 1993, 90, 1460–1464. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Ghosh, P.; Cippitelli, M.; Subleski, J.; Hardy, K.J.; Ortaldo, J.R.; Young, H.A. Characterization of a silencer regulatory element in the human interferon-gamma promoter. J. Biol. Chem. 1994, 269, 25728–25734. [Google Scholar] [CrossRef]
- Kaetzel, D.M., Jr.; Maul, R.S.; Liu, B.; Bonthron, D.; Fenstermaker, R.A.; Coyne, D.W. Platelet-derived growth factor A-chain gene transcription is mediated by positive and negative regulatory regions in the promoter. Biochem. J. 1994, 301 Pt 2, 321–327. [Google Scholar] [CrossRef]
- Donda, A.; Schulz, M.; Bürki, K.; De Libero, G.; Uematsu, Y. Identification and characterization of a human CD4 silencer. Eur. J. Immunol. 1996, 26, 493–500. [Google Scholar] [CrossRef]
- Sawada, S.; Scarborough, J.D.; Killeen, N.; Littman, D.R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 1994, 77, 917–929. [Google Scholar] [CrossRef]
- Dong, J.M.; Smith, P.; Hall, C.; Lim, L. Promoter region of the transcriptional unit for human alpha 1-chimaerin, a neuron-specific GTPase-activating protein for p21rac. Eur. J. Biochem. 1995, 227, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Haniel, A.; Welge-Lussen, U.; Kuhn, K.; Poschl, E. Identification and characterization of a novel transcriptional silencer in the human collagen type IV gene COL4A2. J. Biol. Chem. 1995, 270, 11209–11215. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Lesoon-Wood, L.A.; Weintraub, B.D.; Chung, J.H. A soluble transcription factor, Oct-1, is also found in the insoluble nuclear matrix and possesses silencing activity in its alanine-rich domain. Mol. Cell Biol. 1996, 16, 4366–4377. [Google Scholar] [CrossRef] [PubMed]
- Antalis, T.M.; Costelloe, E.; Muddiman, J.; Ogbourne, S.; Donnan, K. Regulation of the plasminogen activator inhibitor type-2 gene in monocytes: Localization of an upstream transcriptional silencer. Blood 1996, 88, 3686–3697. [Google Scholar] [CrossRef] [PubMed]
- Moffat, G.J.; McLaren, A.W.; Wolf, C.R. Functional characterization of the transcription silencer element located within the human Pi class glutathione S-transferase promoter. J. Biol. Chem. 1996, 271, 20740–20747. [Google Scholar] [CrossRef]
- Bossu, J.P.; Chartier, F.L.; Fruchart, J.C.; Auwerx, J.; Staels, B.; Laine, B. Two regulatory elements of similar structure and placed in tandem account for the repressive activity of the first intron of the human apolipoprotein A-II gene. Biochem. J. 1996, 318 Pt 2, 547–553. [Google Scholar] [CrossRef]
- Liu, J.; Francke, U. Identification of cis-regulatory elements for MECP2 expression. Hum. Mol. Genet. 2006, 15, 1769–1782. [Google Scholar] [CrossRef][Green Version]
- Petrykowska, H.M.; Vockley, C.M.; Elnitski, L. Detection and characterization of silencers and enhancer-blockers in the greater CFTR locus. Genome Res. 2008, 18, 1238–1246. [Google Scholar] [CrossRef]
- French, J.D.; Ghoussaini, M.; Edwards, S.L.; Meyer, K.B.; Michailidou, K.; Ahmed, S.; Khan, S.; Maranian, M.J.; O’Reilly, M.; Hillman, K.M.; et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. 2013, 92, 489–503. [Google Scholar] [CrossRef]
- Qi, H.; Liu, M.; Emery, D.W.; Stamatoyannopoulos, G. Functional validation of a constitutive autonomous silencer element. PLoS ONE 2015, 10, e0124588. [Google Scholar] [CrossRef]
- Huang, D.; Petrykowska, H.M.; Miller, B.F.; Elnitski, L.; Ovcharenko, I. Identification of human silencers by correlating cross-tissue epigenetic profiles and gene expression. Genome Res. 2019, 29, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, Y.; Loh, Y.P.; Tng, J.Q.; Lim, M.C.; Cao, Z.; Raju, A.; Lieberman Aiden, E.; Li, S.; Manikandan, L.; et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat. Commun. 2021, 12, 719. [Google Scholar] [CrossRef] [PubMed]
- Doni Jayavelu, N.; Jajodia, A.; Mishra, A.; Hawkins, R.D. Candidate silencer elements for the human and mouse genomes. Nat. Commun. 2020, 11, 1061. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Snyder, M.P. Systematic identification of silencers in human cells. Nat. Genet. 2020, 52, 254–263. [Google Scholar] [CrossRef]
- Ngan, C.Y.; Wong, C.H.; Tjong, H.; Wang, W.; Goldfeder, R.L.; Choi, C.; He, H.; Gong, L.; Lin, J.; Urban, B.; et al. Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat. Genet. 2020, 52, 264–272. [Google Scholar] [CrossRef]
- Harris, M.B.; Mostecki, J.; Rothman, P.B. Repression of an interleukin-4-responsive promoter requires cooperative BCL-6 function. J. Biol. Chem 2005, 280, 13114–13121. [Google Scholar] [CrossRef]
- Mori, N.; Schoenherr, C.; Vandenbergh, D.J.; Anderson, D.J. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron 1992, 9, 45–54. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Kim, J.-M.; Kim, K.; Punj, V.; Liang, G.; Ulmer, T.S.; Lu, W.; An, W. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Sci. Rep. 2015, 5, 16714. [Google Scholar] [CrossRef]
- Healton Sean, E.; Pinto Hugo, D.; Mishra Laxmi, N.; Hamilton Gregory, A.; Wheat Justin, C.; Swist-Rosowska, K.; Shukeir, N.; Dou, Y.; Steidl, U.; Jenuwein, T.; et al. H1 linker histones silence repetitive elements by promoting both histone H3K9 methylation and chromatin compaction. Proc. Natl. Acad. Sci. USA 2020, 117, 14251–14258. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, S.; Wang, G.G. Polycomb Gene Silencing Mechanisms: PRC2 Chromatin Targeting, H3K27me3 ‘Readout’, and Phase Separation-Based Compaction. Trends Genet. TIG 2021, 37, 547–565. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Akopyan, G.; Garban, H.; Bonavida, B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene 2006, 25, 1125–1142. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, L.; Atchison, M.L. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004, 18, 2596–2601. [Google Scholar] [CrossRef]
- Basu, A.; Wilkinson, F.H.; Colavita, K.; Fennelly, C.; Atchison, M.L. YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment. Nucleic Acids Res. 2014, 42, 2208–2223. [Google Scholar] [CrossRef]
- Zhang, Y.; Dufau, M.L. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem 2002, 277, 33431–33438. [Google Scholar] [CrossRef]
- Zhang, Y.; Dufau, M.L. Repression of the luteinizing hormone receptor gene promoter by cross talk among EAR3/COUP-TFI, Sp1/Sp3, and TFIIB. Mol. Cell Biol. 2003, 23, 6958–6972. [Google Scholar] [CrossRef]
- Won, J.; Yim, J.; Kim, T.K. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem. 2002, 277, 38230–38238. [Google Scholar] [CrossRef]
- Enya, K.; Hayashi, H.; Takii, T.; Ohoka, N.; Kanata, S.; Okamoto, T.; Onozaki, K. The interaction with Sp1 and reduction in the activity of histone deacetylase 1 are critical for the constitutive gene expression of IL-1α in human melanoma cells. J. Leukoc. Biol. 2008, 83, 190–199. [Google Scholar] [CrossRef]
- Lagger, G.; Doetzlhofer, A.; Schuettengruber, B.; Haidweger, E.; Simboeck, E.; Tischler, J.; Chiocca, S.; Suske, G.; Rotheneder, H.; Wintersberger, E.; et al. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol. Cell Biol. 2003, 23, 2669–2679. [Google Scholar] [CrossRef]
- Biddlestone, J.; Batie, M.; Bandarra, D.; Munoz, I.; Rocha, S. SINHCAF/FAM60A and SIN3A specifically repress HIF-2α expression. Biochem. J. 2018, 475, 2073–2090. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, I.; Osato, M.; Egawa, T.; Sunshine, M.J.; Bae, S.-C.; Komori, T.; Ito, Y.; Littman, D.R. Differential Requirements for Runx Proteins in CD4 Repression and Epigenetic Silencing during T Lymphocyte Development. Cell 2002, 111, 621–633. [Google Scholar] [CrossRef]
- Lutterbach, B.; Westendorf, J.J.; Linggi, B.; Isaac, S.; Seto, E.; Hiebert, S.W. A Mechanism of Repression by Acute Myeloid Leukemia-1, the Target of Multiple Chromosomal Translocations in Acute Leukemia. J. Biol. Chem. 2000, 275, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Siu, G.; Wurster, A.L.; Duncan, D.D.; Soliman, T.M.; Hedrick, S.M. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 1994, 13, 3570–3579. [Google Scholar] [CrossRef] [PubMed]
- Shakya, A.; Kang, J.; Chumley, J.; Williams, M.A.; Tantin, D. Oct1 Is a Switchable, Bipotential Stabilizer of Repressed and Inducible Transcriptional States. J. Biol. Chem. 2011, 286, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.; Weaver, J.A.; Sterling, K.M.; Bresnick, E. Nuclear transcription factor Oct-1 binds to the 5’-upstream region of CYP1A1 and negatively regulates its expression. Int. J. Biochem. Cell Biol. 1996, 28, 217–227. [Google Scholar] [CrossRef]
- Delhase, M.; Castrillo, J.-L.; de la Hoya, M.; Rajas, F.; Hooghe-Peters, E.L. AP-1 and Oct-1 Transcription Factors Down-regulate the Expression of the Human PIT1/GHF1 Gene. J. Biol. Chem. 1996, 271, 32349–32358. [Google Scholar] [CrossRef]
- Bentrari, F.; Chantôme, A.; Knights, A.; Jeannin, J.-F.; Pance, A. Oct-2 forms a complex with Oct-1 on the iNOS promoter and represses transcription by interfering with recruitment of RNA PolII by Oct-1. Nucleic Acids Res. 2015, 43, 9757–9765. [Google Scholar] [CrossRef]
- Deng, W.; Lee, J.; Wang, H.; Miller, J.; Reik, A.; Gregory, P.D.; Dean, A.; Blobel, G.A. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 2012, 149, 1233–1244. [Google Scholar] [CrossRef]
- Tolhuis, B.; Palstra, R.J.; Splinter, E.; Grosveld, F.; de Laat, W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 2002, 10, 1453–1465. [Google Scholar] [CrossRef]
- Cao, F.; Fang, Y.; Tan, H.K.; Goh, Y.; Choy, J.Y.H.; Koh, B.T.H.; Hao Tan, J.; Bertin, N.; Ramadass, A.; Hunter, E.; et al. Super-Enhancers and Broad H3K4me3 Domains Form Complex Gene Regulatory Circuits Involving Chromatin Interactions. Sci. Rep. 2017, 7, 2186. [Google Scholar] [CrossRef]
- Ogiyama, Y.; Schuettengruber, B.; Papadopoulos, G.L.; Chang, J.-M.; Cavalli, G. Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Mol. Cell 2018, 71, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Stathopoulos, A.; Levine, M. Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev. Biol. 2005, 280, 482–493. [Google Scholar] [CrossRef] [PubMed]
- Gisselbrecht, S.S.; Palagi, A.; Kurland, J.V.; Rogers, J.M.; Ozadam, H.; Zhan, Y.; Dekker, J.; Bulyk, M.L. Transcriptional Silencers in Drosophila Serve a Dual Role as Transcriptional Enhancers in Alternate Cellular Contexts. Mol. Cell 2020, 77, 324–337.e328. [Google Scholar] [CrossRef] [PubMed]
- Comet, I.; Savitskaya, E.; Schuettengruber, B.; Nègre, N.; Lavrov, S.; Parshikov, A.; Juge, F.; Gracheva, E.; Georgiev, P.; Cavalli, G. PRE-Mediated Bypass of Two Su(Hw) Insulators Targets PcG Proteins to a Downstream Promoter. Dev. Cell 2006, 11, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Eagen, K.P.; Aiden, E.L.; Kornberg, R.D. Polycomb-mediated chromatin loops revealed by a subkilobase-resolution chromatin interaction map. Proc. Natl. Acad. Sci. USA 2017, 114, 8764–8769. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588. [Google Scholar] [CrossRef]
- Leicher, R.; Ge, E.J.; Lin, X.; Reynolds, M.J.; Xie, W.; Walz, T.; Zhang, B.; Muir, T.W.; Liu, S. Single-molecule and in silico dissection of the interaction between Polycomb repressive complex 2 and chromatin. Proc. Natl. Acad. Sci. USA 2020, 117, 30465. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Cavalli, G. Polycomb domain formation depends on short and long distance regulatory cues. PLoS ONE 2013, 8, e56531. [Google Scholar] [CrossRef]
- Kundu, S.; Ji, F.; Sunwoo, H.; Jain, G.; Lee, J.T.; Sadreyev, R.I.; Dekker, J.; Kingston, R.E. Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol. Cell 2017, 65, 432–446.e435. [Google Scholar] [CrossRef]
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e1816. [Google Scholar] [CrossRef] [PubMed]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Yu, D.; Hansen, A.S.; Ganguly, S.; Liu, R.; Heckert, A.; Darzacq, X.; Zhou, Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558, 318–323. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Bhat, P.; Honson, D.; Guttman, M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 2021, 22, 653–670. [Google Scholar] [CrossRef]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef]
- Bannister, A.J.; Zegerman, P.; Partridge, J.F.; Miska, E.A.; Thomas, J.O.; Allshire, R.C.; Kouzarides, T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001, 410, 120–124. [Google Scholar] [CrossRef]
- Lachner, M.; O’Carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Coffey, E.L.; Dall’Agnese, A.; Hannett, N.M.; Tang, X.; Henninger, J.E.; Platt, J.M.; Oksuz, O.; Zamudio, A.V.; Afeyan, L.K.; et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 2020, 586, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Ji, L.; Zhang, Y.; Lv, P.; Cao, X.; Wang, Q.; Yan, Z.; Dong, S.; Du, D.; Zhang, F.; et al. The Nuclear Matrix Protein SAFB Cooperates with Major Satellite RNAs to Stabilize Heterochromatin Architecture Partially through Phase Separation. Mol. Cell 2020, 77, 368–383.e367. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, Y.; Zheng, X.; Liu, C.; Dong, S.; Li, R.; Zhang, G.; Wei, Y.; Qu, H.; Li, Y.; et al. Histone Modifications Regulate Chromatin Compartmentalization by Contributing to a Phase Separation Mechanism. Mol. Cell 2019, 76, 646–659.e646. [Google Scholar] [CrossRef] [PubMed]
- Keenen, M.M.; Brown, D.; Brennan, L.D.; Renger, R.; Khoo, H.; Carlson, C.R.; Huang, B.; Grill, S.W.; Narlikar, G.J.; Redding, S. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. eLife 2021, 10, e64563. [Google Scholar] [CrossRef]
- Zhen, C.Y.; Tatavosian, R.; Huynh, T.N.; Duc, H.N.; Das, R.; Kokotovic, M.; Grimm, J.B.; Lavis, L.D.; Lee, J.; Mejia, F.J.; et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. eLife 2016, 5, e17667. [Google Scholar] [CrossRef]
- Wani, A.H.; Boettiger, A.N.; Schorderet, P.; Ergun, A.; Münger, C.; Sadreyev, R.I.; Zhuang, X.; Kingston, R.E.; Francis, N.J. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat. Commun. 2016, 7, 10291. [Google Scholar] [CrossRef]
- Grau, D.J.; Chapman, B.A.; Garlick, J.D.; Borowsky, M.; Francis, N.J.; Kingston, R.E. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011, 25, 2210–2221. [Google Scholar] [CrossRef]
- Plys, A.J.; Davis, C.P.; Kim, J.; Rizki, G.; Keenen, M.M.; Marr, S.K.; Kingston, R.E. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019, 33, 799–813. [Google Scholar] [CrossRef]
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019, 294, 1451–1463. [Google Scholar] [CrossRef]
- Long, Y.; Hwang, T.; Gooding, A.R.; Goodrich, K.J.; Rinn, J.L.; Cech, T.R. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet. 2020, 52, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, A.A.; Sultanov, R.I.; Magnitov, M.D.; Galitsyna, A.A.; Dashinimaev, E.B.; Lieberman Aiden, E.; Razin, S.V. RedChIP identifies noncoding RNAs associated with genomic sites occupied by Polycomb and CTCF proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2116222119. [Google Scholar] [CrossRef]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Barutcu, A.R.; Blencowe, B.J.; Rinn, J.L. Differential contribution of steady-state RNA and active transcription in chromatin organization. EMBO Rep. 2019, 20, e48068. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011, 12, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Keohane, A.M.; Lavender, J.S.; O’Neill, L.P.; Turner, B.M. Histone acetylation and X inactivation. Dev. Genet. 1998, 22, 65–73. [Google Scholar] [CrossRef]
- Plath, K.; Fang, J.; Mlynarczyk-Evans, S.K.; Cao, R.; Worringer, K.A.; Wang, H.; de la Cruz, C.C.; Otte, A.P.; Panning, B.; Zhang, Y. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003, 300, 131–135. [Google Scholar] [CrossRef]
- Naughton, C.; Sproul, D.; Hamilton, C.; Gilbert, N. Analysis of active and inactive X chromosome architecture reveals the independent organization of 30 nm and large-scale chromatin structures. Mol. Cell 2010, 40, 397–409. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Kallunki, P.; Edelman, G.M.; Jones, F.S. The neural restrictive silencer element can act as both a repressor and enhancer of L1 cell adhesion molecule gene expression during postnatal development. Proc. Natl. Acad. Sci. USA 1998, 95, 3233–3238. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-Y.; Zukin, R.S. REST, a master transcriptional regulator in neurodegenerative disease. Curr. Opin. Neurobiol. 2018, 48, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef]
- Bödör, C.; O’Riain, C.; Wrench, D.; Matthews, J.; Iyengar, S.; Tayyib, H.; Calaminici, M.; Clear, A.; Iqbal, S.; Quentmeier, H.; et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia 2011, 25, 726–729. [Google Scholar] [CrossRef]
- McCabe, M.T.; Graves, A.P.; Ganji, G.; Diaz, E.; Halsey, W.S.; Jiang, Y.; Smitheman, K.N.; Ott, H.M.; Pappalardi, M.B.; Allen, K.E.; et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl. Acad. Sci. USA 2012, 109, 2989–2994. [Google Scholar] [CrossRef]
- Majer, C.R.; Jin, L.; Scott, M.P.; Knutson, S.K.; Kuntz, K.W.; Keilhack, H.; Smith, J.J.; Moyer, M.P.; Richon, V.M.; Copeland, R.A.; et al. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 2012, 586, 3448–3451. [Google Scholar] [CrossRef]
- Li, H.; Cai, Q.; Wu, H.; Vathipadiekal, V.; Dobbin, Z.C.; Li, T.; Hua, X.; Landen, C.N.; Birrer, M.J.; Sánchez-Beato, M.; et al. SUZ12 promotes human epithelial ovarian cancer by suppressing apoptosis via silencing HRK. Mol. Cancer Res. 2012, 10, 1462–1472. [Google Scholar] [CrossRef]
- Liu, C.; Shi, X.; Wang, L.; Wu, Y.; Jin, F.; Bai, C.; Song, Y. SUZ12 is involved in progression of non-small cell lung cancer by promoting cell proliferation and metastasis. Tumour Biol. 2014, 35, 6073–6082. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-C.; Li, C.-L.; Cui, J.; Jiao, M.; Wu, T.; Jing, L.I.; Nan, K.-J. BMI-1, a promising therapeutic target for human cancer. Oncol. Lett. 2015, 10, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, M.; Li, Y.; Chang, I.; Yuan, Q.; Ekimyan-Salvo, M.; Deng, P.; Yu, B.; Yu, Y.; Dong, J.; et al. Targeting BMI1+ Cancer Stem Cells Overcomes Chemoresistance and Inhibits Metastases in Squamous Cell Carcinoma. Cell Stem Cell 2017, 20, 621–634.e626. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; van Galen, P.; Pedley, N.M.; Lima-Fernandes, E.; Frelin, C.; Davis, T.; Cao, L.; Baiazitov, R.; Du, W.; Sydorenko, N.; et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 2014, 20, 29–36. [Google Scholar] [CrossRef]
- Westbrook, T.F.; Martin, E.S.; Schlabach, M.R.; Leng, Y.; Liang, A.C.; Feng, B.; Zhao, J.J.; Roberts, T.M.; Mandel, G.; Hannon, G.J.; et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 2005, 121, 837–848. [Google Scholar] [CrossRef]
- Huang, Z.; Bao, S. Ubiquitination and deubiquitination of REST and its roles in cancers. FEBS Lett. 2012, 586, 1602–1605. [Google Scholar] [CrossRef]
- Chang, Y.-T.; Lin, T.-P.; Campbell, M.; Pan, C.-C.; Lee, S.-H.; Lee, H.-C.; Yang, M.-H.; Kung, H.-J.; Chang, P.-C. REST is a crucial regulator for acquiring EMT-like and stemness phenotypes in hormone-refractory prostate cancer. Sci. Rep. 2017, 7, 42795. [Google Scholar] [CrossRef]
- Thandapani, P. Super-enhancers in cancer. Pharmacology 2019, 199, 129–138. [Google Scholar] [CrossRef]
- Andricovich, J.; Perkail, S.; Kai, Y.; Casasanta, N.; Peng, W.; Tzatsos, A. Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors. Cancer Cell 2018, 33, 512–526.e518. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-Enhancers in the Control of Cell Identity and Disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef]
- Wang, X.; Cairns, M.J.; Yan, J. Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Res. 2019, 47, 11481–11496. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ovcharenko, I. Enhancer–silencer transitions in the human genome. Genome Res. 2022, 32, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; Kaul, R.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, S.D.; Davidson, S.; Shchuka, V.M.; Singh, G.; Malek-Gilani, N.; Langroudi, L.; Martchenko, A.; So, V.; Macpherson, N.N.; Mitchell, J.A. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 2017, 27, 246–258. [Google Scholar] [CrossRef]
- Pott, S.; Lieb, J.D. What are super-enhancers? Nat. Genet. 2015, 47, 8–12. [Google Scholar] [CrossRef]
- Shin, H.Y.; Willi, M.; Yoo, K.H.; Zeng, X.; Wang, C.; Metser, G.; Hennighausen, L. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016, 48, 904–911. [Google Scholar] [CrossRef]
- Hay, D.; Hughes, J.R.; Babbs, C.; Davies, J.O.J.; Graham, B.J.; Hanssen, L.; Kassouf, M.T.; Marieke Oudelaar, A.M.; Sharpe, J.A.; Suciu, M.C.; et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat. Genet. 2016, 48, 895–903. [Google Scholar] [CrossRef]
- Osterwalder, M.; Barozzi, I.; Tissières, V.; Fukuda-Yuzawa, Y.; Mannion, B.J.; Afzal, S.Y.; Lee, E.A.; Zhu, Y.; Plajzer-Frick, I.; Pickle, C.S.; et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 2018, 554, 239. [Google Scholar] [CrossRef]
- Mansour, M.R.; Abraham, B.J.; Anders, L.; Berezovskaya, A.; Gutierrez, A.; Durbin, A.D.; Etchin, J.; Lawton, L.; Sallan, S.E.; Silverman, L.B.; et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014, 346, 1373–1377. [Google Scholar] [CrossRef]
- Kandaswamy, R.; Sava, G.P.; Speedy, H.E.; Beà, S.; Martín-Subero, J.I.; Studd, J.B.; Migliorini, G.; Law, P.J.; Puente, X.S.; Martín-García, D.; et al. Genetic Predisposition to Chronic Lymphocytic Leukemia Is Mediated by a BMF Super-Enhancer Polymorphism. Cell Rep. 2016, 16, 2061–2067. [Google Scholar] [CrossRef]
- Loven, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511, 616–620. [Google Scholar] [CrossRef]
- Chipumuro, E.; Marco, E.; Christensen, C.L.; Kwiatkowski, N.; Zhang, T.; Hatheway, C.M.; Abraham, B.J.; Sharma, B.; Yeung, C.; Altabef, A.; et al. CDK7 Inhibition Suppresses Super-Enhancer-Linked Oncogenic Transcription in MYCN-Driven Cancer. Cell 2014, 159, 1126–1139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, K.; Cai, Y.; Nambu, A.; See, Y.X.; Fu, C.; Raju, A.; Lakshmanan, M.; Osato, M.; Tergaonkar, V.; et al. Super-silencers regulated by chromatin interactions control apoptotic genes. bioRxiv 2022. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; See, Y.X.; Tergaonkar, V.; Fullwood, M.J. Long-Distance Repression by Human Silencers: Chromatin Interactions and Phase Separation in Silencers. Cells 2022, 11, 1560. https://doi.org/10.3390/cells11091560
Zhang Y, See YX, Tergaonkar V, Fullwood MJ. Long-Distance Repression by Human Silencers: Chromatin Interactions and Phase Separation in Silencers. Cells. 2022; 11(9):1560. https://doi.org/10.3390/cells11091560
Chicago/Turabian StyleZhang, Ying, Yi Xiang See, Vinay Tergaonkar, and Melissa Jane Fullwood. 2022. "Long-Distance Repression by Human Silencers: Chromatin Interactions and Phase Separation in Silencers" Cells 11, no. 9: 1560. https://doi.org/10.3390/cells11091560
APA StyleZhang, Y., See, Y. X., Tergaonkar, V., & Fullwood, M. J. (2022). Long-Distance Repression by Human Silencers: Chromatin Interactions and Phase Separation in Silencers. Cells, 11(9), 1560. https://doi.org/10.3390/cells11091560







