Proteomic, Biochemical, and Morphological Analyses of the Effect of Silver Nanoparticles Mixed with Organic and Inorganic Chemicals on Wheat Growth
Abstract
:1. Introduction
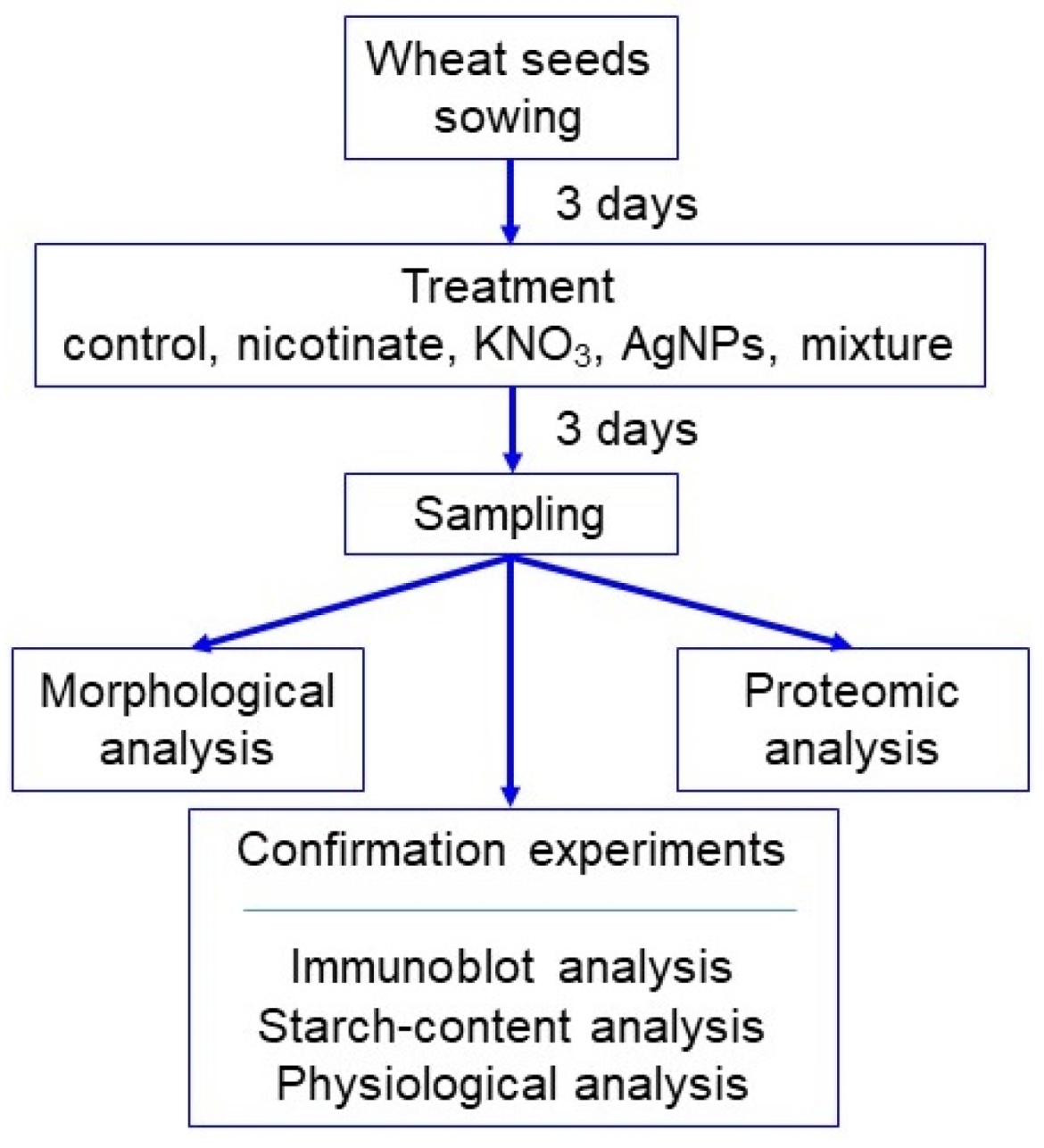
2. Materials and Methods
2.1. Plant Material and Treatment
2.2. Protein Extraction
2.3. Protein Enrichment, Reduction, Alkylation, and Digestion
2.4. Protein Identification using Nano LC-MS/MS and MS Data Analysis
2.5. Differential Analysis of Proteins Using MS Data
2.6. Immunoblot Analysis
2.7. Starch-Content Assay and Starch Staining
2.8. Application of Lipoxygenase Inhibitor
2.9. Statistical Analysis
3. Results
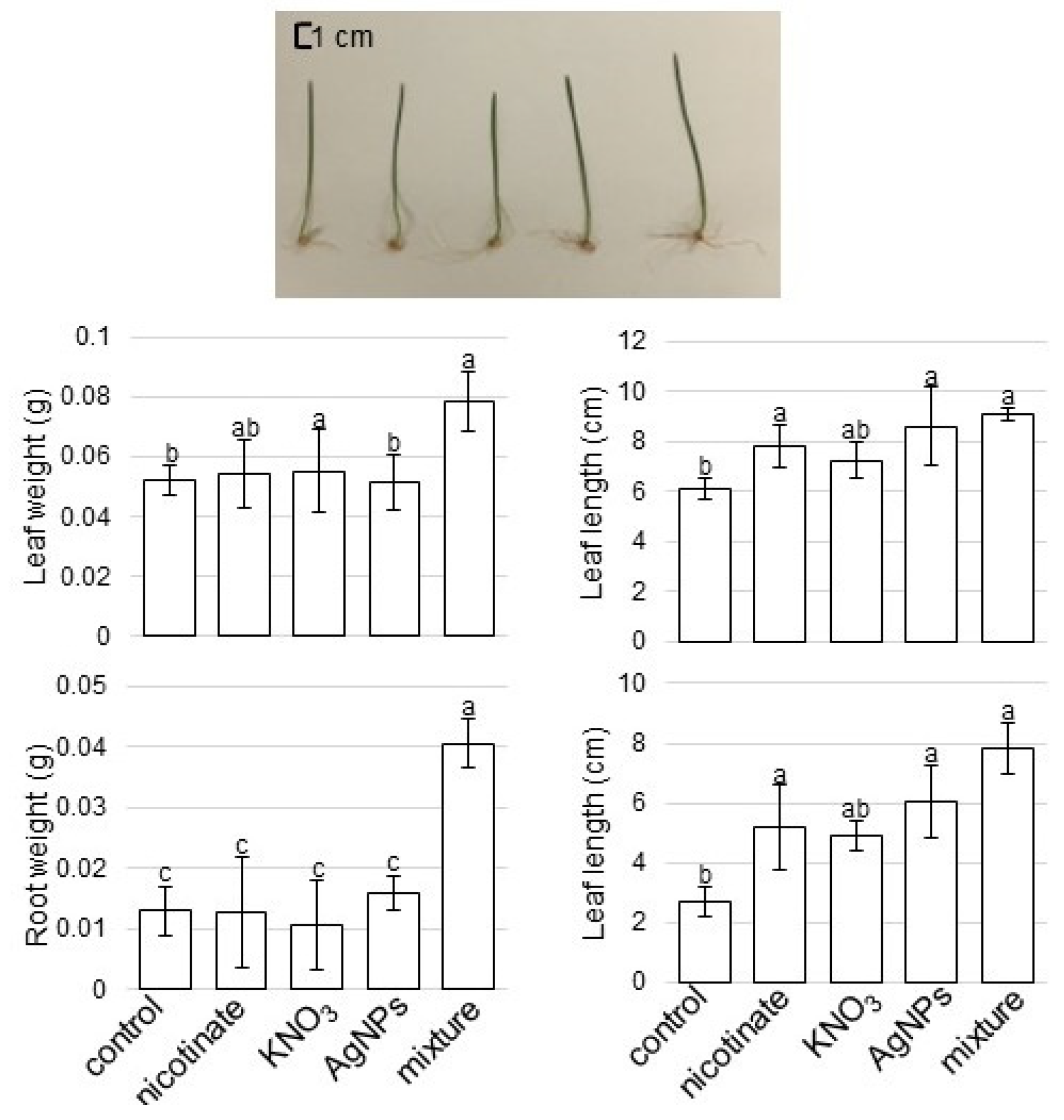
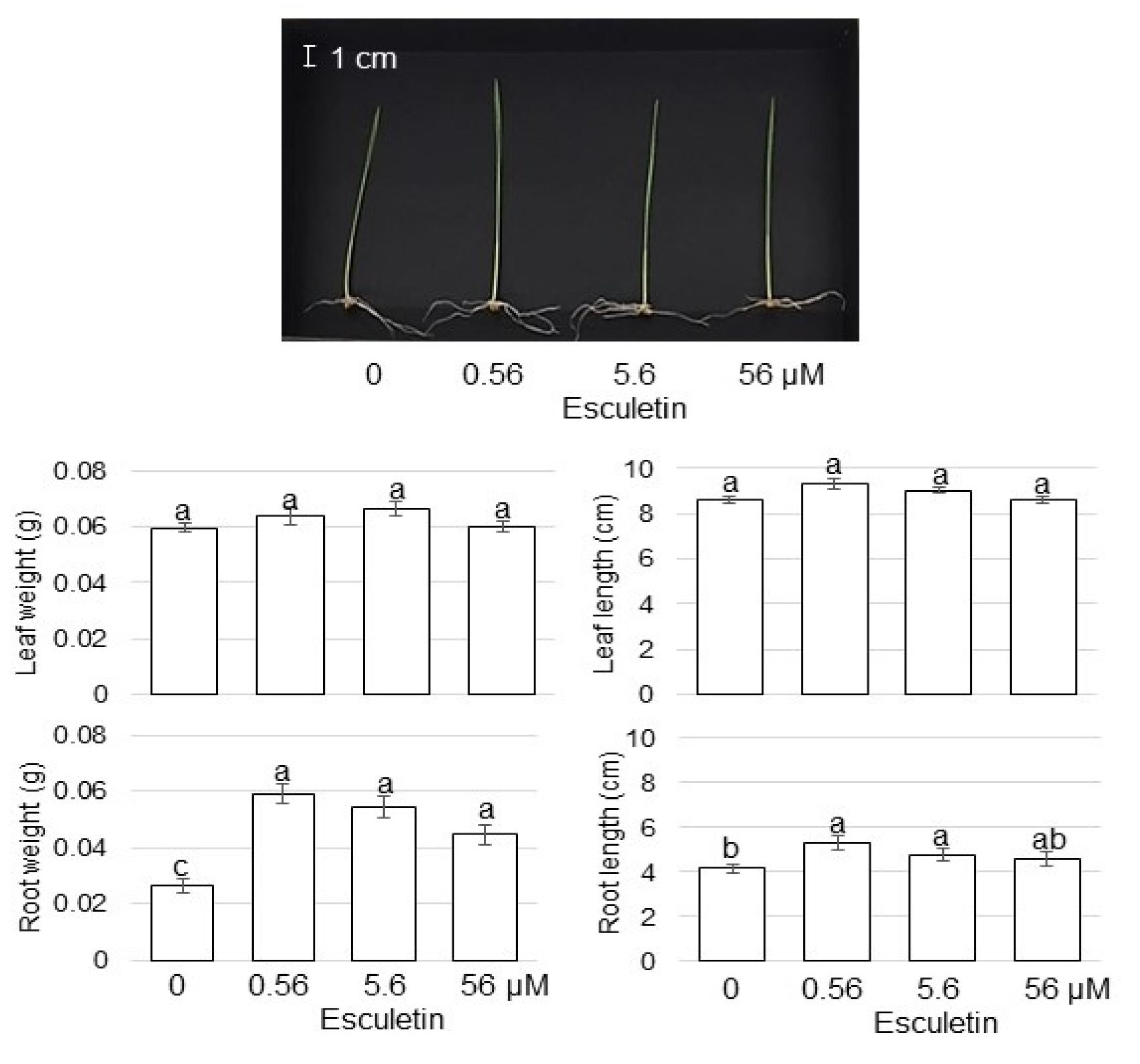
3.1. Growth Response of Wheat to AgNPs Mixed with Organic and Inorganic Chemicals
3.2. Protein Responses of Wheat Roots to AgNPs Mixed with Organic and Inorganic Chemicals
3.3. Amount of ROS Scavenging Enzymes in Wheat Treated with AgNPs Mixed with Organic and Inorganic Chemicals
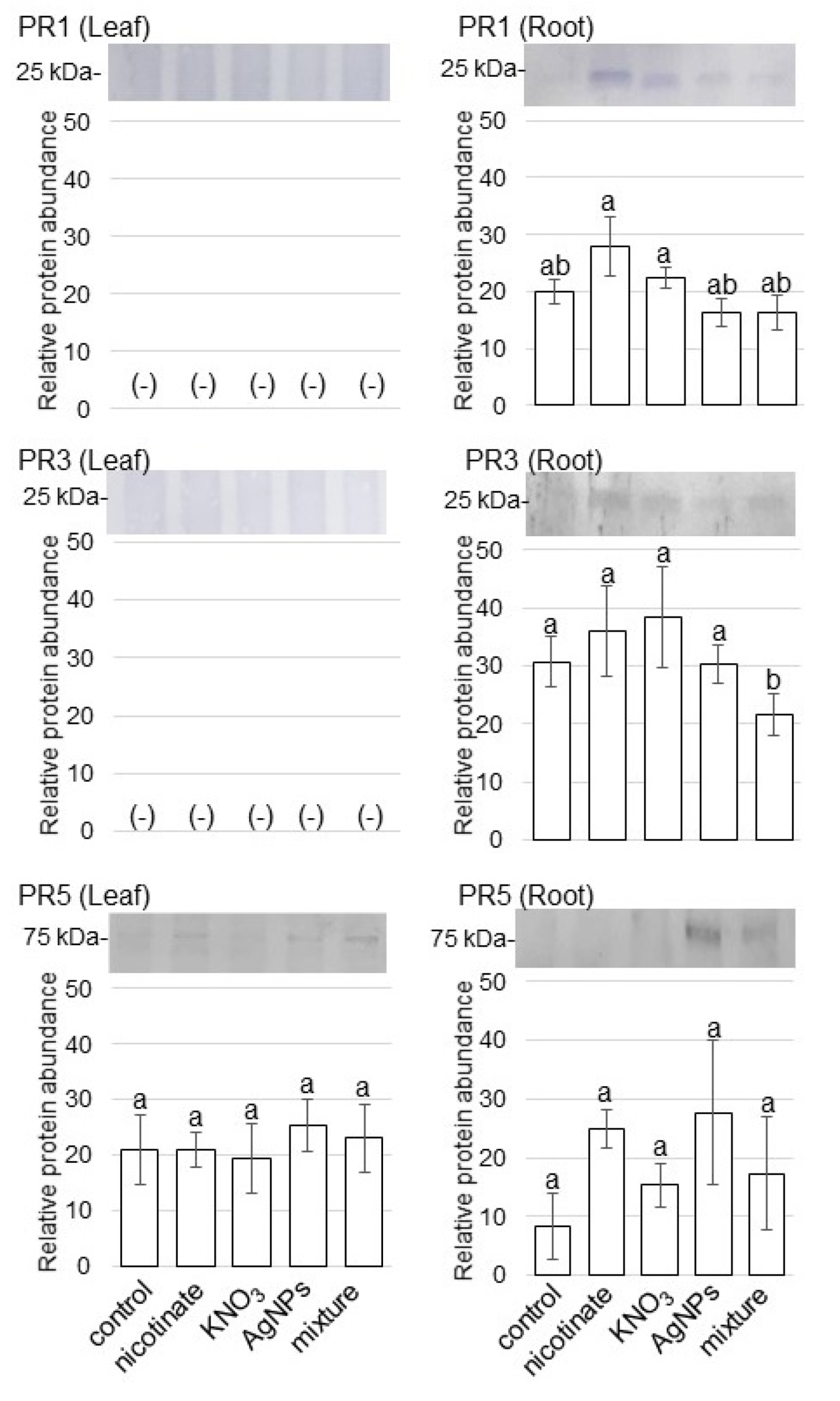
3.4. Accumulation of PR Proteins in Wheat Treated with AgNPs Mixed with Organic and Inorganic Chemicals
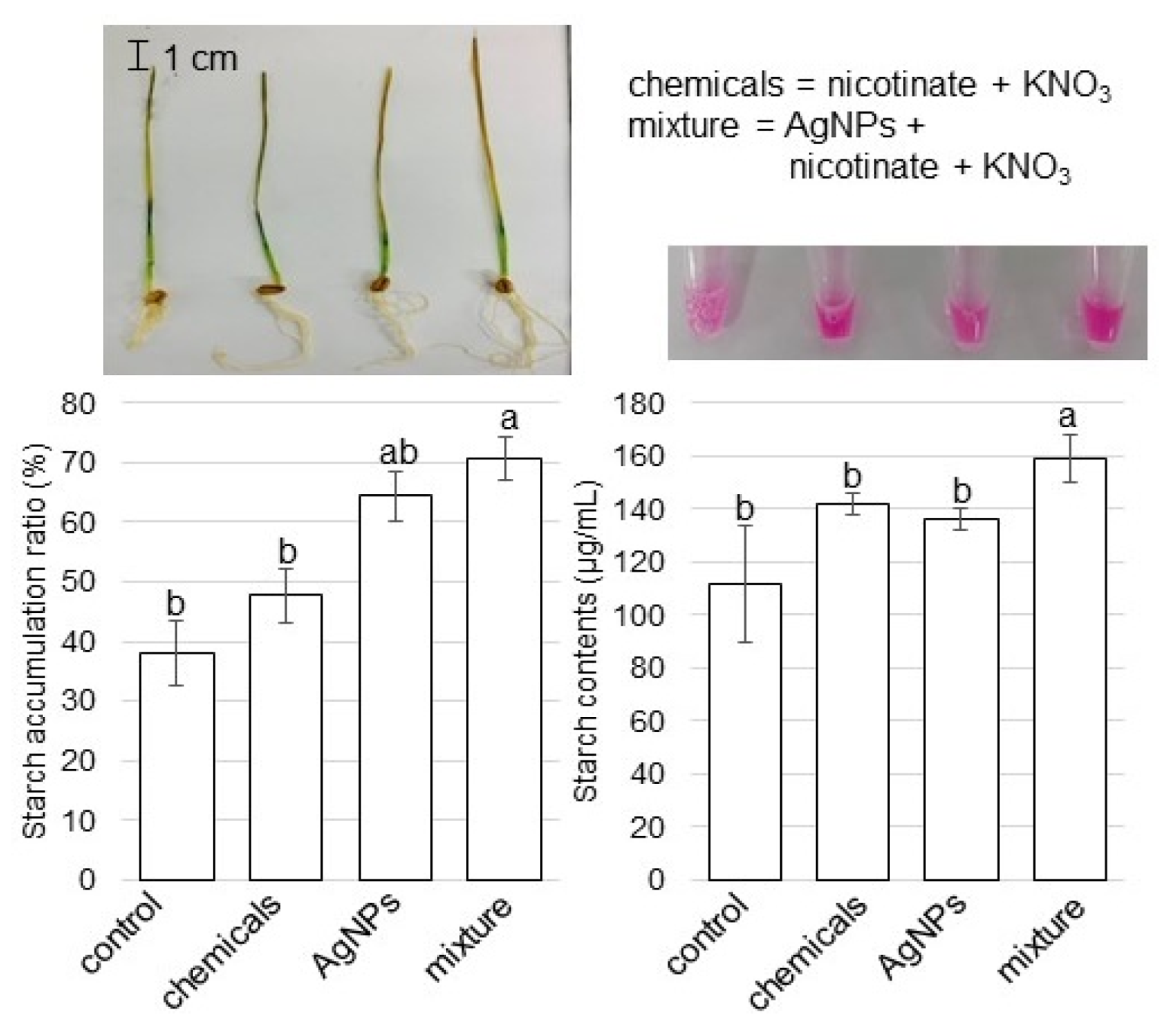
3.5. Starch Accumulation and Contents in Wheat Leaves Treated with AgNPs Mixed with Organic and Inorganic Chemicals
3.6. The Effect of Lipoxygenase Inhibitor on Growth of Wheat Treated with AgNPs Mixed with Organic and Inorganic Chemicals
4. Discussion
4.1. AgNPs Mixed with Organic and Inorganic Chemicals Enhance the Root Growth of Wheat
4.2. ROS Scavenging Pathway Is Suppressed in Wheat Treated with AgNPs Mixed with Organic and Inorganic Chemicals
4.3. PR Protein Is Reduced in Wheat Treated by AgNPs Mixed with Organic and Inorganic Chemicals
4.4. AgNPs Mixed with Organic and Inorganic Chemicals Enhance the Accumulation of Starch in Wheat Leaves
4.5. AgNPs Mixed with Organic and Inorganic Chemicals Enhance Wheat-Root Growth through Lipoxygenase Regulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Curtis, T.; Halford, N.G. Food Security: The Challenge of Increasing Wheat Yield and the Importance of Not Compromising Food Safety. Ann. Appl. Biol. 2014, 164, 354–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, B.S.; Appels, R.; Botha-Oberholster, A.; Buell, C.R.; Bennetzen, J.L.; Chalhoub, B.; Chumley, F.; Dvorák, J.; Iwanaga, M.; Keller, B.; et al. A workshop report on wheat genome sequencing: International genome research on wheat consortium. Genetics 2004, 168, 1087–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochoa-Meza, L.C.; Quintana-Obregón, E.A.; Vargas-Arispuro, I.; Falcón-Rodríguez, A.B.; Aispuro-Hernández, E.; Virgen-Ortiz, J.J.; Martínez-Téllez, M.Á. Oligosaccharins as Elicitors of Defense Responses in Wheat. Polymers 2021, 13, 3105. [Google Scholar] [CrossRef]
- Duveiller, E.; Singh, R.P.; Nicol, J.M. The challenges of maintaining wheat productivity: Pests, diseases, and potential epidemics. Euphytica 2007, 157, 417–430. [Google Scholar] [CrossRef]
- Das, A.; Dutta, P. Antifungal activity of biogenically synthesized silver and gold nanoparticles against sheath blight of rice. J. Nanosci. Nanotechnol. 2021, 21, 3547–3555. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Bhatt, D.; Zaidi, M.G.; Saradhi, P.P.; Khanna, P.K.; Arora, S. Silver naoparticle-mediated enhancement in growth and antioxidant status of Brassica juncea. Appl. Biochem. Biotechnol. 2012, 167, 2225–2233. [Google Scholar] [CrossRef]
- Sriram, T.; Pandidurai, V. In vitro growth analysis of Zea mays L. using Ag NPs. Int. J. Pharma Bio. Sci. 2017, 8, 30–37. [Google Scholar]
- Song, U.; Jun, H.; Waldman, B.; Roh, J.; Kim Yi, J.; Lee, E.J. Functional analysis of nanoparticle toxicity: A comparative study of the effects of TiO2 and Ag on tomatoes (Lycopersicon escolentum). Ecotoxicol. Environ. Saf. 2013, 93, 60–67. [Google Scholar] [CrossRef]
- Syu, Y.Y.; Hung, J.H.; Chen, J.C.; Chuang, H.W. Impact of size and shape of silver NPs on Arabidopsis plant growth and gene expression. Plant Physiol. Biochem. 2014, 83, 57–64. [Google Scholar] [CrossRef]
- Bello-Bello, J.J.; Chavez-Santoscoy, R.A.; Lecona-Guzman, C.A.; Bogdanchikova, N.; Salinase-Ruiz, J.; Gornez-Merino, F.C.; Pestryakov, A. Harmetic response of Ag NPs on invitro multiplication of sugarcane using a temperorary immersion system. Dose Response 2017, 15, 1559325817744945. [Google Scholar] [CrossRef] [Green Version]
- Sajjad, Y.; Jaskani, M.; Ashraf, M.Y.; Ahamd, R. Response of morphological and physiological growth attributes to foliar application of plant growth regulators in gladiolous ‘white prosperity’. Pak. J. Agric. Sci. 2014, 51, 123–129. [Google Scholar]
- Berglund, T.; Wallström, A.; Nguyen, T.V.; Laurell, C.; Ohlsson, A.B. Nicotinamide; antioxidative and DNA hypomethylation e_ects in plant cells. Plant Physiol. Biochem. 2017, 118, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Lara, T.S.; Lira, J.M.S.; Rodrigues, A.C.; Rakocevic, M.; Alvarenga, A.A. Potassium nitrate priming affects the activity of nitrate reductase and antioxidant enzymes in tomato germination. J. Agric. Sci. 2014, 6, 72. [Google Scholar] [CrossRef]
- Anosheh, H.P.; Sadeghi, H.; Emam, Y. Chemical priming with urea and KNO3 enhances maize hybrids (Zeamays L.) seed viability under abiotic stress. J. Crop. Sci. Biotechnol. 2011, 14, 289–295. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, T.; Mustafa, G.; Nishiuchi, T.; Komatsu, S. Comparative analysis of the effect of inorganic and organic chemicals with silver nanoparticles on soybean under flooding stress. Int. J. Mol. Sci. 2020, 21, 1300. [Google Scholar] [CrossRef] [Green Version]
- Jhanzab, H.M.; Razzaq, A.; Bibi, Y.; Yasmeen, F.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Komatsu, S. Proteomic analysis of the effect of inorganic and organic chemicals on silver nanoparticles in wheat. Int. J. Mol. Sci. 2019, 20, 825. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Komatsu, S.; Han, C.; Nanjo, Y.; Altaf-Un-Nahar, M.; Wang, K.; He, D.; Yang, P. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteome Res. 2013, 12, 4769–4784. [Google Scholar] [CrossRef]
- Zhang, Y.; Wen, Z.; Washburn, M.P.; Florens, L. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal. Chem. 2009, 81, 6317–6326. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Siniteyn, P.; Carlson, A.; Hein, Y.; Gieger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of proteomics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Nagel, A.; Thimm, O.; Redestig, H.; Blaesing, O.E.; Rofas, N.P.; Selbig, J.; Hannemann, J.; Piques, M.C.; Steinhauser, D.; et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes and comparison with known responses. Plant Physiol. 2005, 138, 1195–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yamamoto, A.; Nakamura, T.; Nouri, M.Z.; Nanjo, Y.; Nishizawa, K.; Furukawa, K. Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J. Proteome Res. 2011, 10, 3993–4004. [Google Scholar] [CrossRef]
- Yin, X.; Komatsu, S. Comprehensive analysis of response and tolerant mechanisms in early-stage soybean at initial-flooding stress. J. Proteom. 2017, 169, 225–232. [Google Scholar] [CrossRef]
- Konishi, H.; Ishiguro, K.; Komatsu, S. A proteomics approach towards understanding blast fungus infection of rice grown under different levels of nitrogen fertilization. Proteomics 2001, 1, 1162–1171. [Google Scholar] [CrossRef]
- Tran, Q.H.; Nguyen, V.Q.; Le, A.T. Silver nanoparticles: Synthesis, properties, toxicology, applications and perspectives. Adv. Nat. Sci. 2013, 4, 033001. [Google Scholar] [CrossRef] [Green Version]
- Teper, P.; Oleszko-Torbus, N.; Bochenek, M.; Hajduk, B.; Kubacki, J.; Jałowiecki, Ł.; Płaza, G.; Kowalczuk, A.; Mendrek, B. Hybrid nanolayers of star polymers and silver nanoparticles with antibacterial activity. Colloids Surf. B Biointerfaces 2022, 213, 112404. [Google Scholar] [CrossRef]
- Vannini, C.; Domingo, G.; Onelli, E.; Prinsi, B.; Marsoni, M.; Espen, L.; Bracale, M. Morphological and proteomic responses of Eruca sativa exposed to silver nanoparticles or silver nitrate. PLoS ONE 2013, 8, e68752. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, K.; Singh, V.P.; Prasad, S.M.; Chauhan, D.K.; Tripathi, D.K.; Sharma, S. Silicon and plant growth promoting rhizobacteria differentially regulate AgNP-induced toxicity in Brassica juncea: Implication of nitric oxide. J. Hazard Mater. 2020, 390, 121806. [Google Scholar] [CrossRef]
- Yin, L.; Colman, B.P.; McGill, B.M.; Wright, J.P.; Bernhardt, E.S. Effects of silver nanoparticle exposure on germination and early growth of eleven wetland plants. PLoS ONE 2012, 7, e47674. [Google Scholar] [CrossRef] [Green Version]
- Salama, H.M.H. Effects of silver nanoparticles in some crop plants, common bean (Phaseolus vulgaris L.) and corn (Zea mays L.). Int. Res. J. Biotechnol. 2012, 3, 190–197. [Google Scholar]
- Mustafa, G.; Sakata, K.; Komatsu, S. Proteomic analysis of soybean root exposed to varying sizes of silver nanoparticles under flooding stress. J. Proteom. 2016, 148, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, G.; Hasan, M.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Komatsu, S. A comparative proteomic analysis of engineered and bio synthesized silver nanoparticles on soybean seedlings. J. Proteom. 2020, 224, 103833. [Google Scholar] [CrossRef] [PubMed]
- Hossain, Z.; Mustafa, G.; Sakata, K.; Komatsu, S. Insights into the proteomic response of soybean towards Al₂O₃, ZnO, and Ag nanoparticles stress. J. Hazard. Mater. 2016, 304, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.S.; Li, M.; Chang, F.Y.; Li, W.; Yin, L.Y. Physiological analysis of silver nanoparticles and AgNO3 toxicity to Spirodela polyrrhiza. Environ. Toxicol. Chem. 2012, 31, 1880–1886. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.S.; Yin, L.Y.; Ren, N.N.; Zhao, S.T.; Li, Z.; Zhi, Y.; Shao, H.; Li, W.; Gontero, B. Silver nanoparticles induced reactive oxygen species via photosynthetic energy transport imbalance in an aquatic plant. Nanotoxicology 2017, 11, 157–167. [Google Scholar] [CrossRef]
- Abbas, Q.; Yousaf, B.; Ullah, H.; Ali, M.U.; Zia-Ur-Rehman, M.; Rizwan, M.; Rinklebe, J. Biochar-induced immobilization and transformation of silver-nanoparticles affect growth, intracellular-radicles generation and nutrients assimilation by reducing oxidative stress in maize. J. Hazard. Mater. 2020, 390, 121976. [Google Scholar] [CrossRef]
- Kannaujia, R.; Srivastava, C.M.; Prasad, V.; Singh, B.N.; Pandey, V. Phyllanthus emblica fruit extract stabilized biogenic silver nanoparticles as a growth promoter of wheat varieties by reducing ROS toxicity. Plant Physiol. Biochem. 2019, 142, 460–471. [Google Scholar] [CrossRef]
- Nishad, R.; Ahmed, T.; Rahman, V.J.; Kareem, A. Modulation of plant defense system in response to microbial interactions. Front. Plant Sci. 2020, 11, 1298. [Google Scholar] [CrossRef]
- González-Bosch, C. Priming plant resistance by activation of redox-sensitive genes. Free Radic. Biol. Med. 2018, 122, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Le, K.D.; Kim, J.; Nguyen, H.T.; Yu, N.H.; Park, A.R.; Lee, C.W.; Kim, J.C. Streptomyces sp. JCK-6131 protects plants against bacterial and fungal diseases via two mechanisms. Front. Plant Sci. 2021, 12, 726266. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Petruzzelli, L.; Kunz, C.; Waldvogel, R.; Meins, F., Jr.; Leubner-Metzger, G. Distinct ethylene- and tissue-specific regulation of beta-1,3-glucanases and chitinases during pea seed germination. Planta 1999, 209, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kirubakaran, S.I.; Sakthivel, N. Heterologous expression of new antifungal chitinase from wheat. Protein Expr. Purif. 2007, 56, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Numan, M.; Bukhari, S.A.; Rehman, M.U.; Mustafa, G.; Sadia, B. Phylogenetic analyses, protein modeling and active site prediction of two pathogenesis related (PR2 and PR3) genes from bread wheat. PLoS ONE 2021, 16, e0257392. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Westereng, B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sørlie, M.; Eijsink, V.G. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Pandey, A.; Singh, S.; Pillai, S. Biotechnological potential of Beauveria bassiana as a source of novel biocatalysts and metabolites. Crit. Rev. Biotechnol. 2020, 40, 1019–1034. [Google Scholar] [CrossRef]
- Abdel-Aziz, H.M.M.; Rizwan, M. Chemically synthesized silver nanoparticles induced physio-chemical and chloroplast ultrastructural changes in broad bean seedlings. Chemosphere 2019, 235, 1066–1072. [Google Scholar] [CrossRef]
- Mahakham, W.; Sarmah, A.K.; Maensiri, S.; Theerakulpisut, P. Nanopriming technology for enhancing germination and starch metabolism of aged rice seeds using phytosynthesized silver nanoparticles. Sci. Rep. 2017, 7, 8263. [Google Scholar] [CrossRef]
- Kwon, O.S.; Choi, J.S.; Islam, M.N.; Kim, Y.S.; Kim, H.P. Inhibition of 5-lipoxygenase and skin inflammation by the aerial parts of Artemisia capillaris and its constituents. Arch. Pharm. Res. 2011, 34, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Feussner, I. The Oxylipin pathways: Biochemistry and function. Annu. Rev. Plant Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Siedow, J. Plant lipoxygenase: Structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 145–188. [Google Scholar] [CrossRef]
- Singh, P.; Arif, Y.; Miszczuk, E.; Bajguz, A.; Hayat, S. Specific Roles of Lipoxygenases in Development and Responses to Stress in Plants. Plants 2022, 11, 979. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Côté, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. jPOSTTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komatsu, S.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K. Proteomic, Biochemical, and Morphological Analyses of the Effect of Silver Nanoparticles Mixed with Organic and Inorganic Chemicals on Wheat Growth. Cells 2022, 11, 1579. https://doi.org/10.3390/cells11091579
Komatsu S, Yamaguchi H, Hitachi K, Tsuchida K. Proteomic, Biochemical, and Morphological Analyses of the Effect of Silver Nanoparticles Mixed with Organic and Inorganic Chemicals on Wheat Growth. Cells. 2022; 11(9):1579. https://doi.org/10.3390/cells11091579
Chicago/Turabian StyleKomatsu, Setsuko, Hisateru Yamaguchi, Keisuke Hitachi, and Kunihiro Tsuchida. 2022. "Proteomic, Biochemical, and Morphological Analyses of the Effect of Silver Nanoparticles Mixed with Organic and Inorganic Chemicals on Wheat Growth" Cells 11, no. 9: 1579. https://doi.org/10.3390/cells11091579
APA StyleKomatsu, S., Yamaguchi, H., Hitachi, K., & Tsuchida, K. (2022). Proteomic, Biochemical, and Morphological Analyses of the Effect of Silver Nanoparticles Mixed with Organic and Inorganic Chemicals on Wheat Growth. Cells, 11(9), 1579. https://doi.org/10.3390/cells11091579







