Diverse Begomoviruses Evolutionarily Hijack Plant Terpenoid-Based Defense to Promote Whitefly Performance
Abstract
1. Introduction
2. Results
2.1. The Olfactory Attraction of the Whitefly Vector Is Conserved in All Betasatellite Subgroups
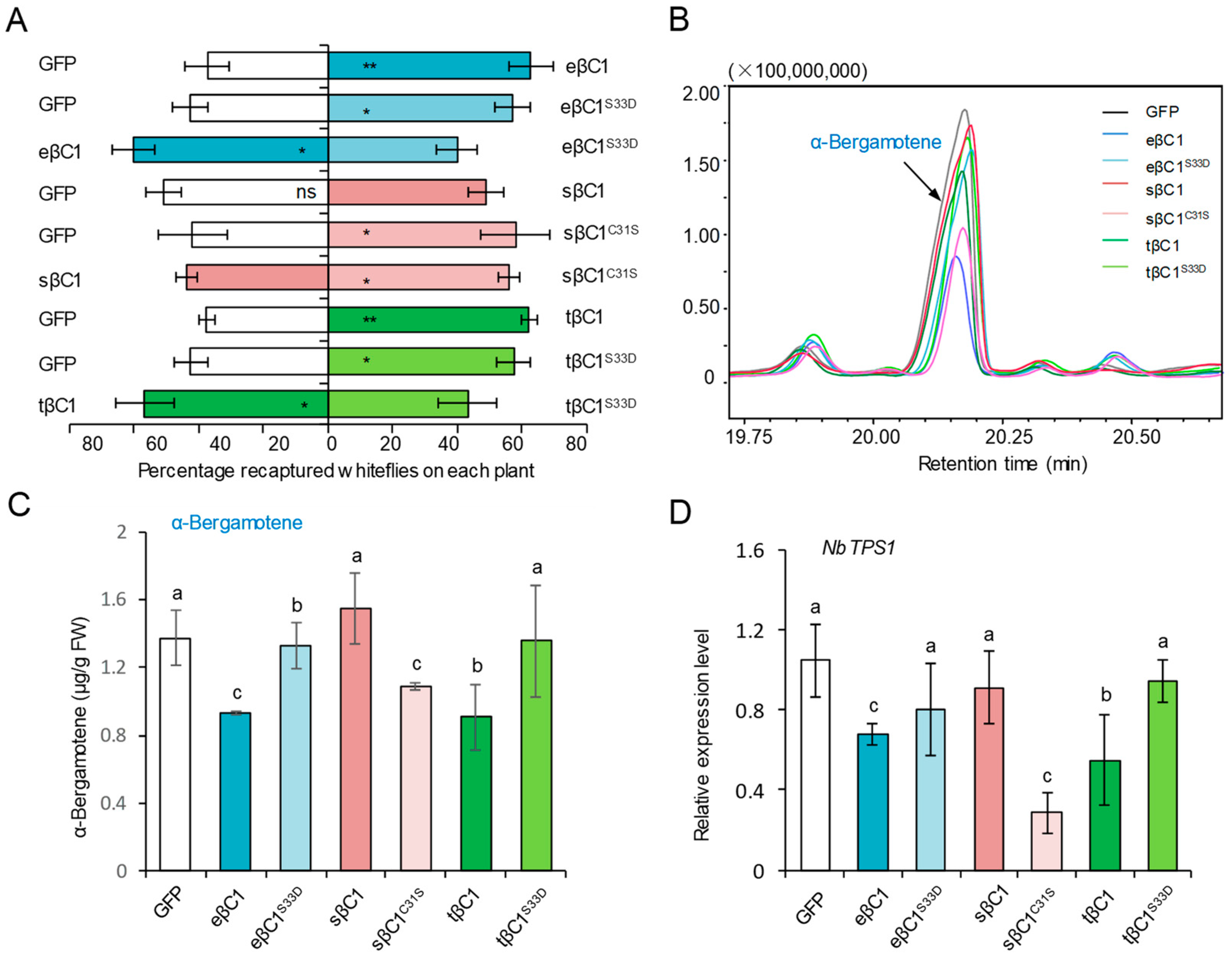
2.2. The Evolutionarily Conserved Serine-33 Is Essential for βC1-Mediated Whitefly Attraction
2.3. The Conserved Serine-33 in βC1 Proteins Is Responsible for Suppressing MYC2 Dimerization
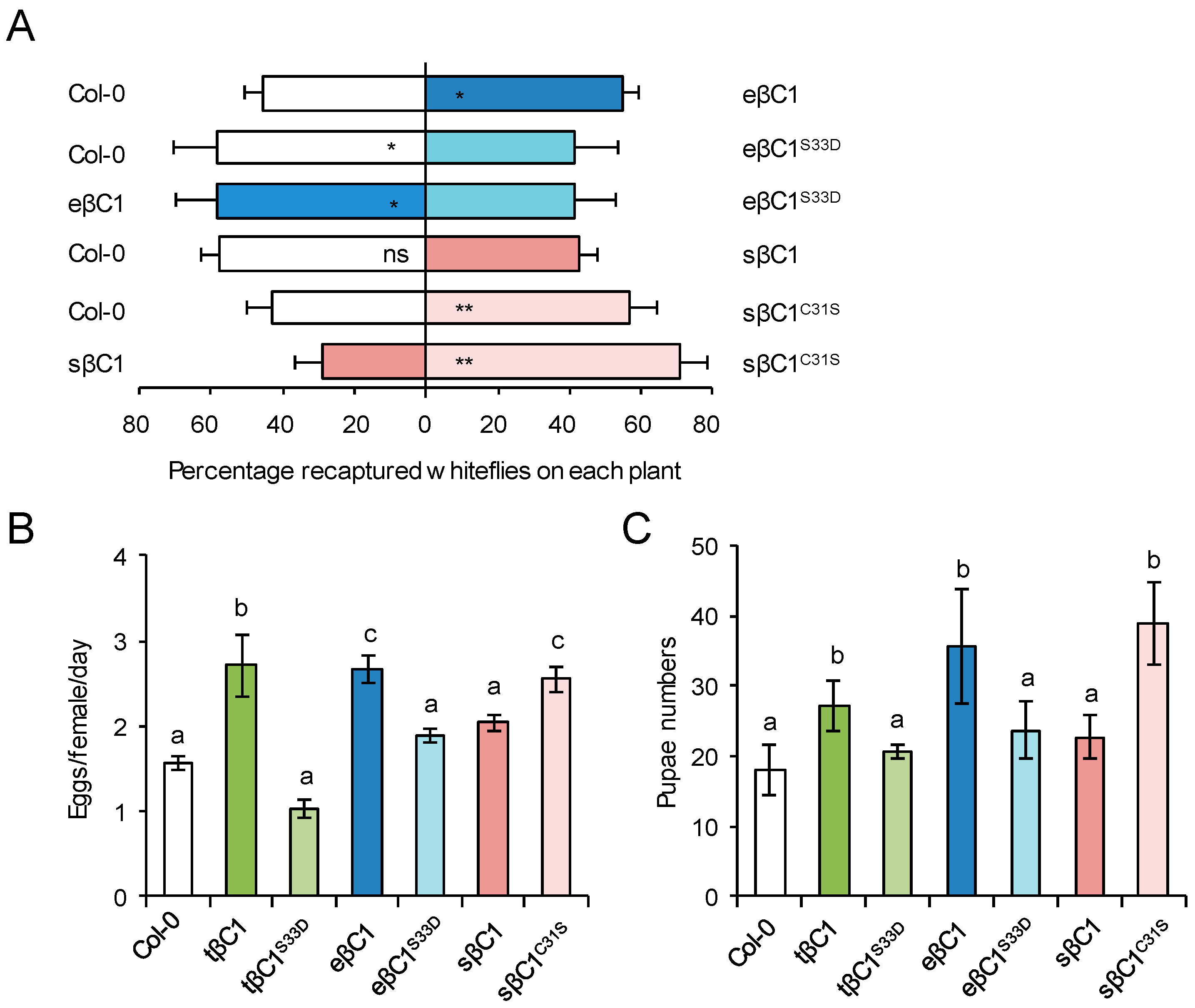
2.4. The Conserved Serine-33 in βC1 Proteins Contributes to Its Compacity to Affect Whitefly Performance
3. Discussion
4. Materials and Methods
4.1. Plant, Virus, and Whitefly Materials and Growth
4.2. Virus Inoculation
4.3. Phylogenetic Analysis
4.4. Insect Preference Experiments
4.5. Volatile Enalysis
4.6. Quantitative PCR
4.7. Plasmid Construction and Agroinfiltration
4.8. Preparation of Recombinant Proteins
4.9. In Vitro Pull-Down Assay
4.10. Bimolecular Fluorescence Complementation (BiFC) Assay
4.11. Luciferase Assays
4.12. Whitefly Bioassays
4.13. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eigenbrode, S.D.; Bosque-Pérez, N.A.; Davis, T.S. Insect-Borne Plant Pathogens and Their Vectors: Ecology, Evolution, and Complex Interactions. Annu. Rev. Entomol. 2018, 63, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, L.; Zhang, X.; Wu, X.; Fang, R. Plant Defense Networks against Insect-Borne Pathogens. Trends Plant Sci. 2020, 26, 272–287. [Google Scholar] [CrossRef] [PubMed]
- Martina, B.E.; Barzon, L.; Pijlman, G.P.; De La Fuente, J.; Rizzoli, A.; Wammes, L.J.; Takken, W.; Van Rij, R.P.; Papa, A. Human to human transmission of arthropod-borne pathogens. Curr. Opin. Virol. 2017, 22, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Yao, X.; Cai, C.; Li, R.; Du, J.; Sun, Y.; Wang, M.; Zou, Z.; Wang, Q.; Kliebenstein, D.J.; et al. Viruses mobilize plant immunity to deter nonvector insect herbivores. Sci. Adv. 2019, 5, eaav9801. [Google Scholar] [CrossRef]
- Wu, X.; Xu, S.; Zhao, P.; Zhang, X.; Yao, X.; Sun, Y.; Fang, R.; Ye, J. The Orthotospovirus nonstructural protein NSs suppresses plant MYC-regulated jasmonate signaling leading to enhanced vector attraction and performance. PLoS Pathog. 2019, 15, e1007897. [Google Scholar] [CrossRef]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. USA 2010, 107, 3600–3605. [Google Scholar] [CrossRef]
- Fang, Y.; Jiao, X.; Xie, W.; Wang, S.; Wu, Q.; Shi, X.; Chen, G.; Su, Q.; Yang, X.; Pan, H.; et al. Tomato yellow leaf curl virus alters the host preferences of its vector Bemisia tabaci. Sci. Rep. 2013, 3, srep02876. [Google Scholar] [CrossRef]
- Su, Q.; Mescher, M.C.; Wang, S.; Chen, G.; Xie, W.; Wu, Q.; Wang, W.; Zhang, Y. Tomato yellow leaf curl virus differentially influences plant defence responses to a vector and a non-vector herbivore. Plant Cell Environ. 2016, 39, 597–607. [Google Scholar] [CrossRef]
- Li, P.; Liu, C.; Deng, W.-H.; Yao, D.-M.; Pan, L.-L.; Li, Y.-Q.; Liu, Y.-Q.; Liang, Y.; Zhou, X.-P.; Wang, X.-W. Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 2019, 15, e1007607. [Google Scholar] [CrossRef]
- Stafford, C.A.; Walker, G.P.; Ullman, D.E. Infection with a plant virus modifies vector feeding behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 9350–9355. [Google Scholar] [CrossRef]
- Luan, J.-B.; Yao, D.; Zhang, T.; Walling, L.L.; Yang, M.; Wang, Y.-J.; Liu, S.-S. Suppression of terpenoid synthesis in plants by a virus promotes its mutualism with vectors. Ecol. Lett. 2012, 16, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Luan, J.-B.; Qi, J.-F.; Huang, C.-J.; Li, M.; Zhou, X.-P.; Liu, S.-S. Begomovirus-whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Mol. Ecol. 2012, 21, 1294–1304. [Google Scholar] [CrossRef]
- Li, R.; Weldegergis, B.T.; Li, J.; Jung, C.; Qu, J.; Sun, Y.; Qian, H.; Tee, C.; Van Loon, J.J.; Dicke, M.; et al. Virulence Factors of Geminivirus Interact with MYC2 to Subvert Plant Resistance and Promote Vector Performance. Plant Cell 2014, 26, 4991–5008. [Google Scholar] [CrossRef]
- Luan, J.-B.; Wang, X.-W.; Colvin, J.; Liu, S.-S. Plant-mediated whitefly–begomovirus interactions: Research progress and future prospects. Bull. Entomol. Res. 2014, 104, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cao, X.; Zhong, W.; Zhou, S.; Li, Z.; An, H.; Liu, X.; Wu, R.; Bohora, S.; Wu, Y.; et al. A viral protein orchestrates rice ethylene signaling to coordinate viral infection and insect vector-mediated transmission. Mol. Plant 2022, 15, 689–705. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, X.; Chen, D.; Ye, J. First Report of Sida leaf curl virus and Associated Betasatellite from Tobacco. Plant Dis. 2022, 106, 1078. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Huang, G.; Shi, T.; Wang, G.; Fang, R.; Ye, J. Surveillance and distribution of the emergent Sri Lankan cassava mosaic virus in China. Phytopathol. Res. 2020, 2, 18. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Lett, J.-M.; Martin, D.P.; Roumagnac, P.; Varsani, A.; Zerbini, F.M.; Navas-Castillo, J. ICTV Virus Taxonomy Profile: Geminiviridae 2021. J. Gen. Virol. 2021, 102, 001696. [Google Scholar] [CrossRef]
- Gnanasekaran, P.; KishoreKumar, R.; Bhattacharyya, D.; Kumar, R.V.; Chakraborty, S. Multifaceted role of geminivirus associated betasatellite in pathogenesis. Mol. Plant Pathol. 2019, 20, 1019–1033. [Google Scholar] [CrossRef]
- King, A.M.Q.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch. Virol. 2018, 162, 2505–2538. [Google Scholar] [CrossRef]
- Jia, Q.; Liu, N.; Xie, K.; Dai, Y.; Han, S.; Zhao, X.; Qian, L.; Wang, Y.; Zhao, J.; Gorovits, R.; et al. CLCuMuB βC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana. PLoS Pathog. 2016, 12, e1005668. [Google Scholar] [CrossRef] [PubMed]
- Ismayil, A.; Yang, M.; Haxim, Y.; Wang, Y.; Li, J.; Han, L.; Wang, Y.; Zheng, X.; Wei, X.; Nagalakshmi, U.; et al. Cotton leaf curl Multan virus βC1 Protein Induces Autophagy by Disrupting the Interaction of Autophagy-Related Protein 3 with Glyceraldehyde-3-Phosphate Dehydrogenases. Plant Cell 2020, 32, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. Hijack to escape: A geminivirus seizes a host imprinted E3 ligase to escape epigenetic repression. Sci. China Life Sci. 2020, 64, 323–325. [Google Scholar] [CrossRef]
- Gui, X.; Liu, C.; Qi, Y.; Zhou, X. Geminiviruses employ host DNA glycosylases to subvert DNA methylation-mediated defense. Nat. Commun. 2022, 13, 575. [Google Scholar] [CrossRef]
- Wu, X.; Ye, J. Manipulation of jasmonate signaling by plant viruses and their insect vectors. Viruses 2020, 12, 148. [Google Scholar] [CrossRef]
- Yang, X.; Guo, W.; Li, F.; Sunter, G.; Zhou, X. Geminivirus-Associated Betasatellites: Exploiting Chinks in the Antiviral Arsenal of Plants. Trends Plant Sci. 2019, 24, 519–529. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.W.; Lemey, P.; Gray, A.J.A.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.-M.; Varsani, A.; et al. The Spread of Tomato Yellow Leaf Curl Virus from the Middle East to the World. PLoS Pathog. 2010, 6, e1001164. [Google Scholar] [CrossRef] [PubMed]
- Masood, M.; Amin, I.; Hassan, I.; Mansoor, S.; Brown, J.K.; Briddon, R.W. Diversity and Distribution of Cryptic Species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex in Pakistan. J. Econ. Entomol. 2017, 110, 2295–2300. [Google Scholar] [CrossRef]
- Pan, L.L.; Cui, X.Y.; Chen, Q.F.; Wang, X.W.; Liu, S.S. Cotton Leaf Curl Disease: Which Whitefly Is the Vector? Phytopathology 2018, 108, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Bedford, I.D.; Briddon, R.W.; Markham, P.G.; Wong, S.M.; Stanley, J. A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA 2000, 97, 6890–6895. [Google Scholar] [CrossRef]
- Saunders, K.; Bedford, I.D.; Yahara, T.; Stanley, J. Aetiology: The earliest recorded plant virus disease. Nature 2003, 422, 831. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. Advances in Understanding Begomovirus Satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yang, X.; Bisaro, D.M.; Zhou, X. The βC1 Protein of Geminivirus–Betasatellite Complexes: A Target and Repressor of Host Defenses. Mol. Plant 2018, 11, 1424–1426. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.L.; Wong, W.S. Production of santalenes and bergamotene in Nicotiana tabacum plants. PLoS ONE 2019, 14, e0203249. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Liu, Z.; Song, F.; Xie, Q.; Hanley-Bowdoin, L.; Zhou, X. Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiol. 2011, 157, 1394–1406. [Google Scholar] [CrossRef]
- Zhong, X.; Wang, Z.Q.; Xiao, R.; Cao, L.; Wang, Y.; Xie, Y.; Zhou, X. Mimic Phosphorylation of a beta C1 Protein Encoded by TYLCCNB Impairs Its Functions as a Viral Suppressor of RNA Silencing and a Symptom Determinant. J. Virol. 2017, 91, e00300-17. [Google Scholar] [CrossRef]
- Zubair, M.; Zaidi, S.S.-E.; Shakir, S.; Amin, I.; Mansoor, S. An Insight into Cotton Leaf Curl Multan Betasatellite, the Most Important Component of Cotton Leaf Curl Disease Complex. Viruses 2017, 9, 280. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Song, C.; Cao, Y.; Dai, J.; Li, G.; Manzoor, M.A.; Chen, C.; Deng, H. The Multifaceted Roles of MYC2 in Plants: Toward Transcriptional Reprogramming and Stress Tolerance by Jasmonate Signaling. Front. Plant Sci. 2022, 13, 989. [Google Scholar] [CrossRef]
- Wang, D.; Dawadi, B.; Qu, J.; Ye, J. Light-Engineering Technology for Enhancing Plant Disease Resistance. Front Plant Sci. 2022, 12, 805614. [Google Scholar] [CrossRef]
- Li, F.; Qiao, R.; Wang, Z.; Yang, X.; Zhou, X. Occurrence and distribution of geminiviruses in China. Sci. China Life Sci. 2022, 65, 1498–1503. [Google Scholar] [CrossRef] [PubMed]
- Shingote, P.R.; Wasule, D.L.; Parma, V.S.; Holkar, S.K.; Karkute, S.G.; Parlawar, N.D.; Senanayake, D.M.J.B. An Overview of Chili Leaf Curl Disease: Molecular Mechanisms, Impact, Challenges, and Disease Management Strategies in Indian Subcontinent. Front. Microbiol. 2022, 13, 899512. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, X.; Gong, Y.; Wang, D.; Xu, D.; Wang, N.; Sun, Y.; Gao, L.; Liu, S.-S.; Deng, X.W.; et al. Red-light is an environmental effector for mutualism between begomovirus and its vector whitefly. PLoS Pathog. 2021, 17, e1008770. [Google Scholar] [CrossRef]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 2017, 6, e23897. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; He, C.; Wang, L.; Ge, B. Post-translational regulation of antiviral innate signaling. Eur. J. Immunol. 2017, 47, 1414–1426. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Song, Y.; Wang, Y.; Zhou, X. Functional analysis of a novel βV1 gene identified in a geminivirus betasatellite. Sci. China Life Sci. 2020, 63, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Tao, X.; Xie, Y.; Fauquet, C.M.; Zhou, X. A DNA β associated with Tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 2004, 78, 13966–13974. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, P.; Ma, Y.; Yao, X.; Sun, Y.; Huang, X.; Jin, J.; Zhang, Y.; Zhu, C.; Fang, R.; et al. A whitefly effector Bsp9 targets host immunity regulator WRKY33 to promote performance. Phil. Trans. R. Soc. B 2019, 374, 20180313. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Zhao, P.; Wang, D.; Mubin, M.; Fang, R.; Ye, J. Diverse Begomoviruses Evolutionarily Hijack Plant Terpenoid-Based Defense to Promote Whitefly Performance. Cells 2023, 12, 149. https://doi.org/10.3390/cells12010149
Wang N, Zhao P, Wang D, Mubin M, Fang R, Ye J. Diverse Begomoviruses Evolutionarily Hijack Plant Terpenoid-Based Defense to Promote Whitefly Performance. Cells. 2023; 12(1):149. https://doi.org/10.3390/cells12010149
Chicago/Turabian StyleWang, Ning, Pingzhi Zhao, Duan Wang, Muhammad Mubin, Rongxiang Fang, and Jian Ye. 2023. "Diverse Begomoviruses Evolutionarily Hijack Plant Terpenoid-Based Defense to Promote Whitefly Performance" Cells 12, no. 1: 149. https://doi.org/10.3390/cells12010149
APA StyleWang, N., Zhao, P., Wang, D., Mubin, M., Fang, R., & Ye, J. (2023). Diverse Begomoviruses Evolutionarily Hijack Plant Terpenoid-Based Defense to Promote Whitefly Performance. Cells, 12(1), 149. https://doi.org/10.3390/cells12010149






