Lessons from Using Genetically Engineered Mouse Models of MYC-Induced Lymphoma
Abstract
:1. Introduction
2. MYC and the Origin of the Eµ-Myc Mouse Model
3. B-Cell Lymphomas from Eµ-Myc Transgenic Mice Arise in a Competent Immune System and Are Highly Heterogeneous
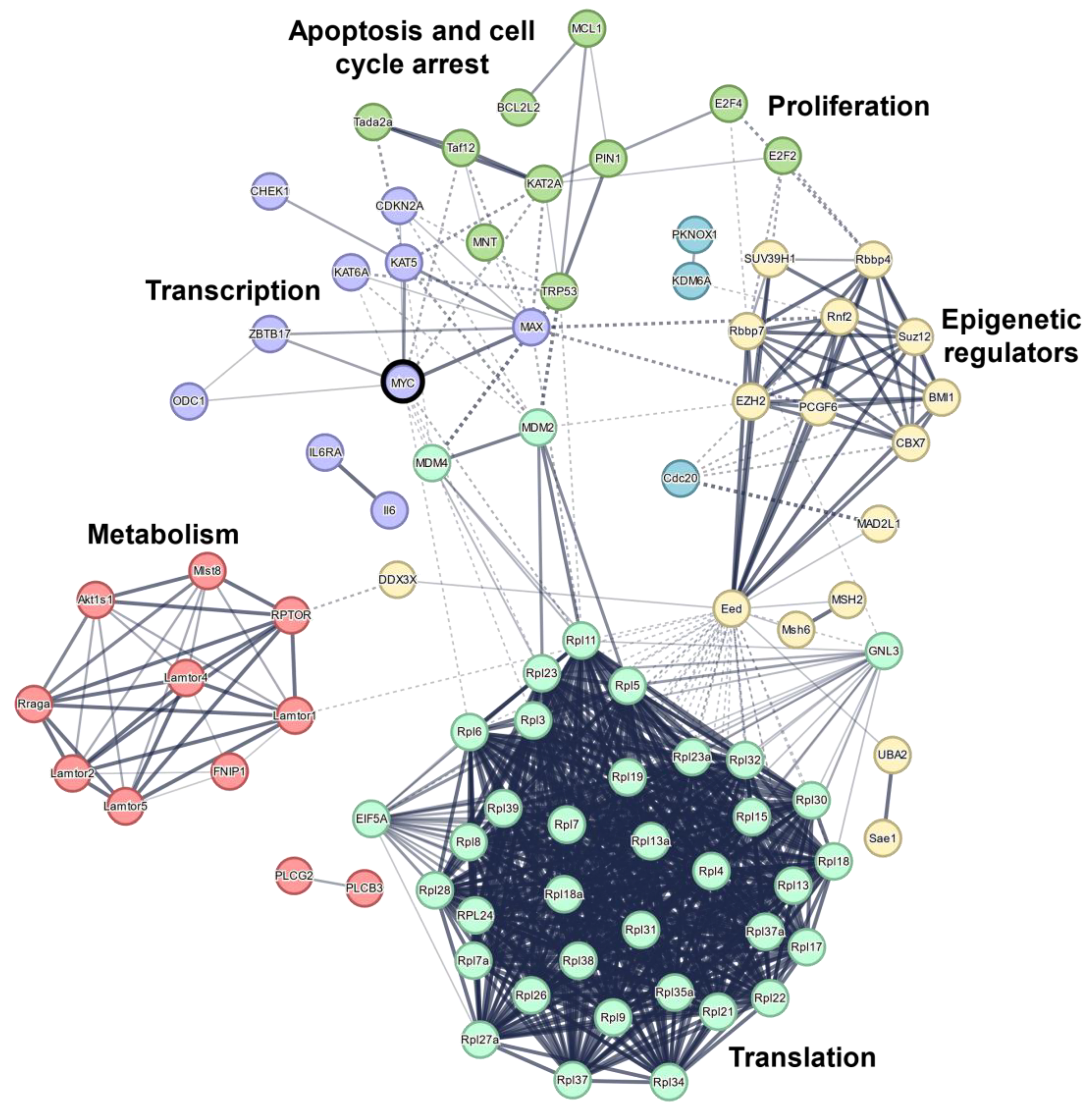
4. Critical Genes for MYC-Induced Lymphomagenesis Share Common Pathways
4.1. Epigenetic Modifiers Cooperate with MYC to Maintain Uncontrolled Proliferation
4.2. Impairing Direct MYC Interaction Partners Is Most Effective in Prolonging Survival
4.3. MYC-Induced Apoptosis Might Be Triggered by Transcriptional Stress and DNA Damage
4.4. Why Is Targeting MYC So Effective but Difficult to Realize?
5. Outlook
| Name | Gene | Function | Model | Survival | [%] | Ref. |
|---|---|---|---|---|---|---|
| µMT (IgM heavy chain) | Ighm | Receptor | Full KO | * CTRL: 120 d, KO: 80 d | 66.67 | [45] |
| 4E-BP1 | Eif4ebp1 | Translation | Dox-inducible KO | * CTRL: 90 d, KO: 145 d | 161.1 | [139] |
| A1/BFL-1 | Bcl2a1a | Apoptosis | (a) Full KO (b) Transplantation of tamoxifen-inducible KO cells (c) Constitutive miR-shRNA (KD) | (a) CTRL: 92 d, KO: 94 d (b) Vehicle: 17 d, Tamoxifen: 23 d (c) CTRL: 103 d, KD: 109 d | (a) 102.2 (b) 135.3 (c) 105.8 | [140,141] |
| AID | Aicda | DNA damage and repair | Full KO | (a) no effect (b) CTRL: 112 d, KO: 130 d | (a) 100 (b) 116.1 | [140,141] |
| ALOX12 | Alox12 | Metabolism | Full heterozygous KO | CTRL: 220 d, +/−: 70 d | 31.8 | [137] |
| AMD1 | Amd1 | Metabolism | Transplanted shRNA transduced FL cells | * CTRL: 112 d, KO: 70 d | 62.5 | [142] |
| EIF5A | Eif5a | Translation | Transplanted shRNA transduced FL cells | * CTRL: 112 d, KO: 56 d | 50 | [142] |
| AMPKα1 | Prkaa1 | Signaling | Full KO | CTRL: 10 wks, KO: 7 wks | 70 | [51] |
| APAF1 | Apaf1 | Apoptosis | Transplanted FL cells from Eµ-Myc full KO mice | No effect | 100 | [143] |
| ATF2 | Atf2 | Transcription factor | CD19-cre, B-cell-specific KO | No effect | 100 | [144] |
| ATF4 | Atf4 | Transcription factor | Tamoxifen-inducible | * Vehicle: 40 d, Tamoxifen: 80 d | 200 | [145] |
| ATF7 | Atf7 | Transcription factor | CD19-cre, B-cell-specific KO | WT: 105 d, KO: 135 d | 128.6 | [144] |
| BAD | Bad | Apoptosis | Full KO | CTRL: 138 d, WT: 78 d | 56.5 | [146] |
| BAX | Bax | Apoptosis | Full KO | CTRL: 21.7 wks WT: 12.6 wks | 58.1 | [146] |
| BCL-2 | Bcl2 | Apoptosis | (a) Full heterozygous KO (b) Transplanted FL cells from Eµ-Myc full KO mice | (a) CTRL: 116 d, KO: 154 d (b) No effect | (a) 132.8 (b) 100 | [147,148] |
| BCL-W | Bcl2l2 | Apoptosis | Full KO | CTRL: 90 d, KO: 298.5 s | 331.7 | [106] |
| BCL-x | Bcl2l1 | Apoptosis | (a) Full heterozygous KO (b) Transplantation of tamoxifen-inducible KO cells | (a) CTRL: 116 d, KO: 174 d (b) CTRL: 19 d, KO: 25 d | (a) 150 (b) 131.6 | [107,148] |
| BIF-1 | Sh3glb1 | Apoptosis | Full KO | CTRL: 107 d, KO: 65 d | 60.7 | [149] |
| BIK | Bik | Apoptosis | Full KO | No effect | 100 | [150] |
| BIM | Bcl2l11 | Apoptosis | (a) Full KO (b) Mb1-cre, B-cell-specific KO | (a) CTRL: 15 wks, KO: 8.2 wk (b) CTRL: 72 d, KO: 113 d | (a) 54.7 (b) 63.7 | [109,110] |
| BMF | Bmf | Apoptosis | Full KO | CTRL: 138 d, KO: 87 d | 63 | [151] |
| BMI1 | Bmi1 | Epigenetic regulator | (a,b) Full heterozygous KO (c) Transplanted overexpressing FL cells | (a) * CTRL: 150 d, KO: >300 d (b) * CTRL: 100 d, KO: >250 d (c) CTRL: >300 d, OE: 74 d | (a) >200 (b) >250 (c) 24.7 | [152,153,154] |
| BOK | Bok | Apoptosis | Full KO | CTRL: 107 d; KO: 121 d | 113.1 | [155] |
| BTK/TEC | Btk Tec | Signaling | Full heterozygous KO: BTK+/− TEC+/− | CTRL: 100 d, KO: 60 d | 60 | [156] |
| BUB1 | Bub1 | PTM | Overexpression of point mutant (T85) | CTRL: 21 wks, MUT: 13 wks | 61.9 | [157] |
| CAML | Caml | Signaling | Subcutaneous transplant of tamoxifen-inducible full KO | Vehicle: 7 d, Tamoxifen: >25 d | >357 | [158] |
| Caspase 9 | Casp9 | Apoptosis | FL transplantation of full KO cells | CTRL: 57 wk; KO: 54 wk | 94.7 | [143] |
| Caspase 2 | Casp2 | Apoptosis | Full KO | CTRL: 16 wks, KO: 8 wks | 50 | [159] |
| CBX7 | Cbx7 | Epigenetic regulator | FL cells with overexpression | CTRL: >300 d, OE: 43 d | <14.3 | [154] |
| CD19 | Cd19 | Receptor | Full KO | CTRL: 13.4 wks, KO: 24.3 wks | 181.3 | [43] |
| CDK4 | Cdk4 | pRB-axis | Full KO | CTRL: 18 wks, KO: 11 wks | 61.1 | [160] |
| CHK1 | Chek1 | DNA damage and repair | (a) Full heterozygous KO (b) Mb1-cre, B-cell-specific KO | (a) CTRL: 106 d, KO: 205 d (b) CTRL: 106 d, KO: >350 d | (a) 193 (b) >330 | [120] |
| CKS1 | Cks1b | pRB-axis | Full KO | CTRL: 91 d, KO: 268 d | 294.5 | [161] |
| CREBBP | Crebbp | Epigenetic regulator | AID-cre + immunization | * CTRL: 85 d, KO: 55 d | 64.7 | [162] |
| cREL | Rel | Transcription factor | Full KO | CTRL: 115 d, KO: 79 d | 68.7 | [163] |
| CSN6 | Cops6 | PTM | Full heterozygous KO | * CTRL: 100 d, KO: 190 d | 190 | [164] |
| CUL9 | Cul9 | PTM | Full KO | CTRL: 126.4 d, KO: 85.1 d | 67.3 | [165] |
| DDX3X | Ddx3x | Helicase | (a) CD19-cre, B-cell-specific KO (b) Vav-cre, B-cell-specific KO | (a) ♂: CTRL: 83 d, KO: 105 d; ♀: CTRL: 87 d, KO: 212 d (b) ♂: CTRL: 98 d, KO: >350 d ♀: CTRL: 110.5 d; KO: 83 d | (a)♂: 126.5; ♀: 243.7 (b)♂: >357.1; ♀: 75.1 | [62] |
| DICER | Dicer1 | Splicing | CD19-cre, B-cell-specific KO | WT: 194 d, KO: 351 d | 180.9 | [166] |
| DMP1 | Dmp1 | p53-axis | Full KO | * CTRL: 22 wks, KO: 13 wks | 59.1 | [167] |
| DNMT3B | Dnmt3b | Epigenetic regulator | Full heterozygous KO | * CTRL: 125 d, KO: 75 d | 60 | [168] |
| DPY30 | Dpy30 | Epigenetic regulator | Full heterozygous KO | CTRL: 121 d, KO: 180.5 d | 149.2 | [169] |
| E2F1 | E2f1 | pRB-axis | (a,b) Full KO | (a) * CTRL: 24 wks, KO: 16 wks (b) No effect | (a) 150 (b) 100 | [78,170] |
| E2F2 | E2f2 | pRB-axis | Full KO | WT: 126 d, KO: 63 d | 50 | [78] |
| E2F3 | E2f3 | pRB-axis | Full KO | No effect | 100 | [78] |
| E2F4 | E2f4 | pRB-axis | Full KO | CTRL: 110 d, KO: 375 d | 340.9 | [78] |
| E6AP | Ube3a | PTM | Full heterozygous KO | CTRL: 103 d, KO: 153 d | 148.5 | [52] |
| EZH2 | Ezh2 | Epigenetic regulator | (a) GOF mutant (b) Transplanted shRNA transduced FL cells | (a) CTRL: 137.5 d, MUT: 51 d (b) CTRL: 220 d, KD:55 d | (a) 37.1 (b) 25 | [86,153] |
| FNIP1 | Fnip1 | Metabolism | Full KO | * CTRL: 110 d, KO: >300 d | >272.7 | [171] |
| FOXO | Foxo4 | TF | Dominant negative mutant, transplanted transduced FL cells | * CTRL: >250 d, MUT: 50 d | <20 | [172] |
| GCN2 | Eif2ak4 | Translation | Transplanted tamoxifen-inducible lymphoma cells | No effect | 100 | [145] |
| GCN5 | Kat2a | Epigenetic regulator | CD19-cre, B-cell-specific KO | CTRL: 21 wks, KO: 58.4 wks | 278.1 | [83] |
| H2A.X | H2ax | Epigenetic regulator | Full KO | No effect | 100 | [25] |
| HDAC1 | Hdac1 | Epigenetic regulator | Mb1-cre, B-cell-specific KO | CTRL: 161 d, KO: 170 d | 105.6 | [173] |
| HDAC2 | Hdac2 | Epigenetic regulator | Mb1-cre, B-cell-specific KO | CTRL: 161 d, KO: 164 d | 101.9 | [173] |
| IBTK | Ibtk | Signaling | Full KO | CTRL: 90 d, KO: 150 d | 166.7 | [174] |
| ID2 | Id2 | TF | Full KO | No effect | 100 | [175] |
| IL6R (gp130) | Il6ra | Receptor | FL xenograft with CD19-cre deleted cells | CTRL: 277 d, KO: 20 d | 7.2 | [176] |
| IL7R | Il7r | Receptor | (a) LOF (no activation of survival mechanism) (b) Transplanted cells | (a) CTRL: 15.5 wks, MUT: 66.5 wks (b) No effect | (a) 429 (b) 100 | [177] |
| INK4A/P16 | Cdkn2a | p53-axis | Full heterozygous KO | * CTRL: 150 d, KO: 45 d | 30 | [152] |
| INK4C/P18 | Cdkn2c | p53-axis | Full KO | No effect | 100 | [175] |
| KLRK1 | Klrk1 | Receptor | Full KO | WT: 22 wks, KO: 15 wks | 68.2 | [178] |
| KSR1 | Ksr1 | Signaling | Full KO | CTRL: 95 d, KO: 138 d | 145.3 | [179] |
| L24 | Rpl24 | Translation | Full heterozygous KO | * CTRL: 100 d, KO: 210 d | 210 | [180] |
| L38 | Rpl38 | Epigenetic regulator | Full heterozygous KO | * CTRL: 70 d, KO: 110 d | 157.1 | [180] |
| LGL | Llgl1 | Cytoskeleton | Full KO | No effect | 100 | [181] |
| MAD2 | Mad2l1 | Spindle assembly | Transplanted HSCs with overexpression | * CTRL: >350 d, OE: 60 d | <17.1 | [182] |
| MAX | Max | TF | Mb1-cre, B-cell-specific KO | CTRL: 97 d, KO: 300 d | 309.3 | [92] |
| MCL1 | Mcl1 | Apoptosis | (a) CD19-cre, B-cell-specific KO (b) Rag1-cre, heterozygous KO (c) Transplanted tamoxifen-inducible lymphoma cells (d) Transgene (H2K promoter) (e) Transgene (VavP promoter) | (a) CTRL: 91 d, KO: 123 d (b) CTRL: 129 d, KO: 346 d (c) WT: 19 d, KO: 35 d (d) WT: 134 d, OE: 72 d (e) WT: 94 d, OE: 30.5 d | (a) 135.2 (b) 268.2 (c) 184.2 (d) 53.7 (e) 32.4 | [107,108,183,184] |
| MDM2 | Mdm2 | p53-axis | (a) Full heterozygous KO (b) Point mutation (LOF) C305F | (a) CTRL: 20.6 wks, KO: 44.3 wks (b) CTRL: 20.7 wks MUT: 11.6 wks | (a) 215 (b) 56.0 | [69,74] |
| MDM4 | Mdm4 | p53-axis | (a) Transgene (b) Full heterozygous KO | (a) CTRL: 31 wks, OE: 34 wks (b) * CTRL: 350 d, KO: >400 d | 109.7 >114.3 | [185,186] |
| MDMX | Mdmx | Deleted in mice | Point mutation W201S/W202G | * CTRL: 170 d, MUT: 80 d | 47.1 | [73] |
| MGA | Mga | TF | CD19-cre, B-cell-specific KO | CTRL: 97 d, KO: 87 d | 89.7 | [187] |
| MHCII | H2 | Receptor | Full KO + immunization | No effect | 100 | [162] |
| MIF | Mif | Cytokine | Full KO | CTRL: 2.67 months, KO: 3.67 months | 137.5 | [188] |
| miR146a | Mir146 | microRNA | Full KO | CTRL: 104.5 d, KO: 82.5 d | 78.9 | [189] |
| miR-17-92 | Mir17hg | microRNA | (a) Transplanted overexpressing FL cells (b) Transplanted tamoxifen-inducible KO cells | (a) * CTRL: >200 d, OE: 125 d (b) * CTRL: 20 d, KO: 33 d | <62.5 165 | [190,191] |
| MIZ-1 | Zbtb17 | TF | Mb1-cre, B-cell-specific KO | * CTRL: 110 d, KO: 350 d | 318.2 | [50] |
| MNT | Mnt | TF | (a) Full heterozygous KO (b) Rag1-cre, KO | (a) CTRL: 17 wks KO: 28 wks (b) CTRL: 86 d, KO: 463 d | (a) 538.4 (b) 164.7 | [192,193] |
| MOZ | Kat6a | Epigenetic regulator | Full heterozygous KO | CTRL: 105 d, KO: 411 d | 391.4 | [84] |
| MPL | Mpl | Receptor | Full KO | CTRL: 87 d, KO: 76.5 | 87.9 | [194] |
| MSH2 | Msh2 | DNA damage and repair | (a) Full KO (b) Mutation (G674A) | (a) * CTRL: 100 d, KO: 40 d (b) * CTRL: 100 d, MUT: 40 d | (a) 40 (b) 40 | [45] |
| MTAP | Mtap | Metabolism | Full heterozygous KO | CTRL: 130 d, KO: 87 d | 66.9 | [195] |
| MTBP | Mtbp | p53-axis | Full heterozygous KO | CTRL: 135 d, KO: 270 d | 200 | [196] |
| MYSM1 | Mysm1 | PTM | Tamoxifen-inducible full KO | * CTRL: >150 d, KO: 80 d | <53.3 | [197] |
| NFKB1/P105 | Nfkb1 | TF | Full KO | No effect | 100 | [198] |
| NFKB2/P100 | Nfkb2 | TF | Full KO | CTRL: 205 d, KO: 171 d | 83.4 | [199] |
| NOXA | Pmaip1 | Apoptosis | Full KO | No effect | 100 | [111] |
| Nucleostemin | Gnl3 | Signaling | Full heterozygous KO | * CTRL: 100 d, KO: 260 d | 260 | [200] |
| ODC | Odc1 | Metabolism | Full heterozygous KO | CTRL: 110 d, KO: 320 d | 290.9 | [201] |
| OGG1 | Ogg1 | DNA damage and repair | Full KO | No effect | 100 | [202] |
| p19/ARF | Cdkn2a | p53-axis | (a) Full heterozygous KO (b) Full KO (c) Full KO | (a) * CTRL:135 d, KO: 35 d (b) CTRL: 20.7 wks, KO: 10.1 wks (c) CTRL: 89 d, KO: 73 d | (a) 25.9 (b) 48.8 (c) 82.0 | [71,72,74] |
| p27 | Cdkn1b | p53-axis | Full KO | CTRL: 120 d, KO: 80 d | 66.7 | [203] |
| p38 | Mapk14 | Signaling | Heterozygous mutation (T180A, T182F) | CTRL: 77 d, MUT: 85 | 110.4 | [204] |
| p53 | Trp53 | p53-axis | (a) Full heterozygous KO (b) Full heterozygous KO (c) Full heterozygous KO (d) Full KO (e) Full heterozygous KO (f) Point mutation LOF (G515C) | (a) CTRL: 20.6 wks, KO: 5.6 wks (b) * CTRL: 137.5 d, KO: 37.5 d (c) * CTRL: 100 d, KO: 30 d (d) CTRL: 89 d, KO: 40 d (e) CTRL: 138 d, KO: 35 d (f) CTRL: 138 d, MUT: 62 d | (a) 27.2 (b) 27.3 (c) 30 (d) 44.9 (e) 25.4 (f) 44.9 | [69,70,71,72,111] |
| P73 | Trp73 | TF | Full KO | No effect | 100 | [205] |
| PARP1 | Parp1 | Apoptosis | Full KO | CTRL: 127 d, KO: 90 d | 70.1 | [206] |
| PARP2 | Parp2 | Apoptosis | Full KO | CTRL: 127 d, KO: 326 d | 257 | [206] |
| PARP14 | Parp14 | Metabolism | Full KO | * CTRL: 13 wks, KO: 20 wks | 153.8 | [207] |
| PCGF6 | Pcgf6 | Epigenetic regulator | CD19-cre, B-cell-specific KO | CTRL: 203 d, KO: 65 d | 32.0 | [187] |
| PFP | Prf1 | Apoptosis | Full KO | CTRL: 135 d, KO: 139 d | 103 | [208] |
| PIN1 | Pin1 | Isomerase | Full KO | CTRL: 108 d, KO: 431 d | 399.1 | [101] |
| PLCβ3 | Plcb3 | Signaling | Full heterozygous KO | * CTRL: >365 d, KO: 100 d | <27.4 | [209] |
| PLCγ2 | Plcg2 | Signaling | Full KO | * CTRL 20 wks, KO: 10 wks | 50 | [210] |
| PML | Pml | Apoptosis, Signaling | Full heterozygous KO | CTRL: 103 d, KO: 153 d | 149 | [52] |
| PRDM11 | Prdm11 | Epigenetic regulator | Full KO | CTRL: 113 d, KO: 94 d | 83.2 | [211] |
| PRDM15 | Prdm15 | Transcription | Tamoxifen-inducible KO | CTRL: 107 d, KO: 332 d | 310.3 | [212] |
| PREP1 | Pknox1 | TF | Tamoxifen-inducible heterozygous KO | CTRL: 58 wks, KO: 23 wks | 39.7 | [213] |
| PRMT5 | Prmt5 | RNA/Splicing | Tamoxifen-inducible heterozygous KO | * CTRL: 90 d, KO: 175 d | 194.4 | [17] |
| PUMA | Bbc3 | Apoptosis | (a) Full KO (b) Full KO | (a) CTRL: 100 d, KO: 66 d (b) * CTRL: 15 wks, KO: 11 wks | (a) 66 (b) 73.3 | [111,214] |
| RAC1 | Rac1 | Signaling | Transplanted, transduced cells | * CTRL: 18 d, KD: 28 d | 155.6 | [215] |
| RAG1 | Rag1 | DNA damage and repair | Full KO | * CTRL 110 d, KO: 90 d | 81.8 | [141] |
| RAIDD | Cradd | Apoptosis | Full KO | * CTRL: 120 d, KO: 110 | 91.7 | [216] |
| RAP1 | Terf2ip | Signaling | Full KO | * CTRL: 15 wks, KO: 12 wks | 80 | [217] |
| RAPTOR | Rptor | Metabolism | CD2-cre | * CTRL: 18 wks, KO: >55 wks | >305.6 | [218] |
| pRB | Rb1 | pRB-axis | Full heterozygous KO | * CTRL: 135 d, KO: 125 d | 92.6 | [71] |
| RIPK3 | Ripk3 | Signaling | Full KO | CTRL: 118 d, KO: 97 d | 82.2 | [219] |
| RUNX1 | Runx1 | TF | Mx1-cre + pIpC; heterozygous for p53 | No effect | 100 | [220] |
| SAE2 | Uba2 | PTM | Transplanted, transduced lymphoma cells | * CTRL: 35 d, KD: >100 d | >285.7 | [221] |
| Scribble | Scrib | Scaffold | Transplanted FL cells | * CTRL: 175 d, KO: 280 d | 160 | [222] |
| Septin 4 | Septin4 | Cytoskeleton | Full KO | * CTRL: 270 d, KO: 100 d | 37.0 | [223] |
| Sirtuin 4 | Sirt4 | Epigenetic regulator | Full KO | CTRL: 195 d, KO: 139 d | 71.3 | [224] |
| SKP2 | Skp2 | PTM | Full KO | CTRL: 100 d, KO: 150 d | 150 | [225] |
| SMARCAL1 | Smarcal1 | Helicase | Full KO | CTRL: 187 d, KO: 224 d | 119.8 | [116] |
| SMYD2 | Smyd2 | Epigenetic regulator | CD19-cre, B-cell-specific KO | * CTRL: 150 d, KO: 175 d | 116.7 | [226] |
| SUV39H1 | Suv39h1 | Epigenetic regulator | Full KO | * CTRL: 125 d, KO: 60 d | 48 | [90] |
| SUZ12 | Suz12 | Epigenetic regulator | Heterozygous LOF mutation | CTRL: 103 d, MUT: 72 d | 69.9 | [153] |
| TCRα | Trac | Receptor | Full KO | No effect | 100 | [45] |
| TCRΔ | Trdc | Receptor | Full KO | No effect | 100 | [45] |
| TEL2 | Etv7 | Deleted in mice | Transplanted, transduced cells (overexpression) | CTRL: >16 wks, OE: 13 wks | 81.3 | [227] |
| TIP60 | Kat5 | Epigenetic regulator | Full heterozygous KO | * CTRL: 52 wks, KO: 12 wks | 23.1 | [121] |
| TIS11B | Zfp36l1 | Transcription | Eµ-Tis11b (overexpression) | * CTRL: 140 d, OE: 100 d | 71.4 | [228] |
| TRAIL-R | Tnfrsf10b | Apoptosis | Full KO | CTRL: 119 d, KO: 82 d | 68.9 | [229] |
| Tristetraprolin | Zfp36 | Transcription | Eµ-TTP (overexpression) | (a) CTRL: 103.5 d, OE: 194 d (b) CTRL: 121 d, OE: 277 d | (a) 187.4 (b) 228.9 | [228] |
| UCH-L1 | Uchl1 | Ubiquitin system | (a) Transgene (b) Full KO | (a) * CTRL: >60 wks, TG: 45 wks (b) * CTRL: >60 wks, KO: >60 wks | (a) <75 (b) 100 | [230] |
| UNG1 | Ung | Repair | Full KO | * CTRL: 110 d, KO: 85 d | 77.3 | [202] |
| UTX | Kdm6a | Epigenetic regulator | CD19-cre, B-cell-specific KO | * ♂: CTRL: 145 d, KO: 120 d; ♀: CTRL: >200 d, KO: 70 d | ♂: 82.8 ♀: <35 | [231] |
| WIP1 | Ppm1d | Signaling | Full KO | CTRL: 77 d, KO: 138 d | 179.2 | [204] |
| WRN | Wrn | Helicase | Mutation in helicase domain | CTRL: 115 d, KO: 151 d | 131.3 | [232] |
| XPO1 | Xpo1 | Nuclear export | Point mutation (E571K), tamoxifen-inducible | CTRL: 35 d, KO: 28 d | 80 | [233] |
| ZMAT3 | Zmat3 | Transcription | Full KO | CTRL: 125 d, KO: 93 d | 74.4 | [234] |
| ZRANB3 | Zranb3 | Helicase | Full KO | CTRL: 104 d, KO: 138 d | 132.7 | [116] |
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the Path to Cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, P.; Medeiros, L.J. The Impact of MYC Rearrangements and “Double Hit” Abnormalities in Diffuse Large B-Cell Lymphoma. Curr. Hematol. Malig. Rep. 2013, 8, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Brown, N.; Song, J.Y.; Yang, L.; Skrabek, P.; Nasr, M.R.; Wong, J.T.; Bedell, V.; Murata-Collins, J.; Kochan, L.; et al. Relative Frequency and Clinicopathologic Characteristics of MYC-Rearranged Follicular Lymphoma. Hum. Pathol. 2021, 114, 19–27. [Google Scholar] [CrossRef]
- Molyneux, E.M.; Rochford, R.; Griffin, B.; Newton, R.; Jackson, G.; Menon, G.; Harrison, C.J.; Israels, T.; Bailey, S. Burkitt’s Lymphoma. Lancet 2012, 379, 1234–1244. [Google Scholar] [CrossRef] [Green Version]
- Ross, J.; Miron, C.E.; Plescia, J.; Laplante, P.; McBride, K.; Moitessier, N.; Möröy, T. Targeting MYC: From Understanding Its Biology to Drug Discovery. Eur. J. Med. Chem. 2021, 213, 113137. [Google Scholar] [CrossRef]
- Baudino, T.A.; Cleveland, J.L. The Max Network Gone Mad. Mol. Cell Biol. 2001, 21, 691–702. [Google Scholar] [CrossRef] [Green Version]
- Poole, C.J.; van Riggelen, J. MYC—Master Regulator of the Cancer Epigenome and Transcriptome. Genes 2017, 8, 142. [Google Scholar] [CrossRef]
- Dang, C.V.; Lee, W.M. Identification of the Human C-Myc Protein Nuclear Translocation Signal. Mol. Cell Biol. 1988, 8, 4048–4054. [Google Scholar] [CrossRef] [Green Version]
- Zeller, K.I.; Zhao, X.; Lee, C.W.H.; Chiu, K.P.; Yao, F.; Yustein, J.T.; Ooi, H.S.; Orlov, Y.L.; Shahab, A.; Yong, H.C.; et al. Global Mapping of C-Myc Binding Sites and Target Gene Networks in Human B Cells. Proc. Natl. Acad. Sci. USA 2006, 103, 17834–17839. [Google Scholar] [CrossRef]
- Li, Z.; Van Calcar, S.; Qu, C.; Cavenee, W.K.; Zhang, M.Q.; Ren, B. A Global Transcriptional Regulatory Role for C-Myc in Burkitt’s Lymphoma Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 8164–8169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabò, A.; Kress, T.R.; Pelizzola, M.; De Pretis, S.; Gorski, M.M.; Tesi, A.; Morelli, M.J.; Bora, P.; Doni, M.; Verrecchia, A.; et al. Selective Transcriptional Regulation by Myc in Cellular Growth Control and Lymphomagenesis. Nature 2014, 511, 488–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez-Sola, D.; Victora, G.D.; Ying, C.Y.; Phan, R.T.; Saito, M.; Nussenzweig, M.C.; Dalla-Favera, R. The Proto-Oncogene MYC Is Required for Selection in the Germinal Center and Cyclic Reentry. Nat. Immunol. 2012, 13, 1083–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calado, D.P.; Sasaki, Y.; Godinho, S.A.; Pellerin, A.; Köchert, K.; Sleckman, B.P.; De Alborán, I.M.; Janz, M.; Rodig, S.; Rajewsky, K. The Cell-Cycle Regulator c-Myc Is Essential for the Formation and Maintenance of Germinal Centers. Nat. Immunol. 2012, 13, 1092–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toboso-Navasa, A.; Gunawan, A.; Morlino, G.; Nakagawa, R.; Taddei, A.; Damry, D.; Patel, Y.; Chakravarty, P.; Janz, M.; Kassiotis, G.; et al. Restriction of Memory b Cell Differentiation at the Germinal Center b Cell Positive Selection Stage. J. Exp. Med. 2020, 217, e20191933. [Google Scholar] [CrossRef]
- Lin, C.Y.; Lovén, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional Amplification in Tumor Cells with Elevated C-Myc. Cell 2012, 151, 56–67. [Google Scholar] [CrossRef] [Green Version]
- Koh, C.M.; Bezzi, M.; Low, D.H.P.; Ang, W.X.; Teo, S.X.; Gay, F.P.H.; Al-Haddawi, M.; Tan, S.Y.; Osato, M.; Sabò, A.; et al. MYC Regulates the Core Pre-MRNA Splicing Machinery as an Essential Step in Lymphomagenesis. Nature 2015, 523, 96–100. [Google Scholar] [CrossRef]
- Iritani, B.M.; Eisenman, R.N. C-Myc Enhances Protein Synthesis and Cell Size during B Lymphocyte Development. Proc. Natl. Acad. Sci. USA 1999, 96, 13180–13185. [Google Scholar] [CrossRef] [Green Version]
- Kieffer-Kwon, K.R.; Nimura, K.; Rao, S.S.P.; Xu, J.; Jung, S.; Pekowska, A.; Dose, M.; Stevens, E.; Mathe, E.; Dong, P.; et al. Myc Regulates Chromatin Decompaction and Nuclear Architecture during B Cell Activation. Mol. Cell 2017, 67, 566–578.e10. [Google Scholar] [CrossRef]
- Adhikary, S.; Eilers, M. Transcriptional Regulation and Transformation by Myc Proteins. Nat. Rev. Mol. Cell Biol. 2005, 6, 635–645. [Google Scholar] [CrossRef]
- Lourenco, C.; Resetca, D.; Redel, C.; Lin, P.; MacDonald, A.S.; Ciaccio, R.; Kenney, T.M.G.; Wei, Y.; Andrews, D.W.; Sunnerhagen, M.; et al. MYC Protein Interactors in Gene Transcription and Cancer. Nat. Rev. Cancer 2021, 21, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Rickert, R.C. New Insights into Pre-BCR and BCR Signalling with Relevance to B Cell Malignancies. Nat. Rev. Immunol. 2013, 13, 578–591. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Harris, A.W.; Pinkert, C.A.; Corcoran, L.M.; Alexander, W.S.; Cory, S.; Palmiter, R.D.; Brinster, R.L. The C-Myc Oncogene Driven by Immunoglobulin Enhancers Induces Lymphoid Malignancy in Transgenic Mice. Nature 1985, 318, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Lefebure, M.; Tothill, R.W.; Kruse, E.; Hawkins, E.D.; Shortt, J.; Matthews, G.M.; Gregory, G.P.; Martin, B.P.; Kelly, M.J.; Todorovski, I.; et al. Genomic Characterisation of Eμ-Myc Mouse Lymphomas Identifies Bcor as a Myc Co-Operative Tumour-Suppressor Gene. Nat. Commun. 2017, 8, 14581. [Google Scholar] [CrossRef] [Green Version]
- Fusello, A.; Horowitz, J.; Yang-Iott, K.; Brady, B.L.; Yin, B.; Rowh, M.A.W.; Rappaport, E.; Bassing, C.H. Histone H2AX Suppresses Translocations in Lymphomas of Eμ-c-Myc Transgenic Mice That Contain a Germline Amplicon of Tumor-Promoting Genes. Cell Cycle 2013, 12, 2867–2875. [Google Scholar] [CrossRef] [Green Version]
- Eick, D.; Bornkamm, G.W. Expression of Normal and Translocated C-Myc Alleles in Burkitt’s Lymphoma Cells: Evidence for Different Regulation. EMBO J. 1989, 8, 1965–1972. [Google Scholar] [CrossRef]
- Bemark, M.; Neuberger, M.S. The C-MYC Allele That Is Translocated into the IgH Locus Undergoes Constitutive Hypermutation in a Burkitt’s Lymphoma Line. Oncogene 2000, 19, 3404–3410. [Google Scholar] [CrossRef] [Green Version]
- Sidman, C.L.; Denial, T.M.; Marshall, J.D.; Roths, J.B. Multiple Mechanisms of Tumorigenesis in Eµ-Myc Transgenic Mice. Cancer Res. 1993, 53, 1665–1669. [Google Scholar]
- Joshi, G.; Eberhardt, A.O.; Lange, L.; Winkler, R.; Hoffmann, S.; Kosan, C.; Bierhoff, H. Dichotomous Impact of Myc on Rrna Gene Activation and Silencing in b Cell Lymphomagenesis. Cancers 2020, 12, 3009. [Google Scholar] [CrossRef]
- Thürmer, M.; Gollowitzer, A.; Pein, H.; Neukirch, K.; Gelmez, E.; Waltl, L.; Wielsch, N.; Winkler, R.; Löser, K.; Grander, J.; et al. PI(18:1/18:1) Is a SCD1-Derived Lipokine That Limits Stress Signaling. Nat. Commun. 2022, 13, 2982. [Google Scholar] [CrossRef]
- Wiese, K.E.; Haikala, H.M.; von Eyss, B.; Wolf, E.; Esnault, C.; Rosenwald, A.; Treisman, R.; Klefström, J.; Eilers, M. Repression of SRF Target Genes Is Critical for Myc-Dependent Apoptosis of Epithelial Cells. EMBO J. 2015, 34, 1554–1571. [Google Scholar] [CrossRef] [PubMed]
- Sauvé, S.; Naud, J.F.; Lavigne, P. The Mechanism of Discrimination between Cognate and Non-Specific DNA by Dimeric b/HLH/LZ Transcription Factors. J. Mol. Biol. 2007, 365, 1163–1175. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wu, G.; Zhan, X.; Nolan, A.; Koh, C.; de Marzo, A.; Doan, H.M.; Fan, J.; Cheadle, C.; Fallahi, M.; et al. Cell-Type Independent MYC Target Genes Reveal a Primordial Signature Involved in Biomass Accumulation. PLoS ONE 2011, 6, e26057. [Google Scholar] [CrossRef] [PubMed]
- Knudson, A.G. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [Green Version]
- Eischen, C.M.; Weber, J.D.; Roussel, M.F.; Sherr, C.J.; Cleveland, J.L. Disruption of the ARF-Mdm2-P53 Tumor Suppressor Pathway in Myc-Induced Lymphomagenesis. Genes Dev. 1999, 13, 2658–2669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, C.; Berger, A.; Hoelzl, M.A.; Putz, E.M.; Frenzel, A.; Simma, O.; Moritz, N.; Hoelbl, A.; Kovacic, B.; Freissmuth, M.; et al. The Cooperating Mutation or “Second Hit” Determines the Immunologic Visibility toward MYC-Induced Murine Lymphomas. Blood 2011, 118, 4635–4645. [Google Scholar] [CrossRef] [Green Version]
- Vallespinós, M.; Fernández, D.; Rodríguez, L.; Alvaro-Blanco, J.; Baena, E.; Ortiz, M.; Dukovska, D.; Martínez, D.; Rojas, A.; Campanero, M.R.; et al. B Lymphocyte Commitment Program Is Driven by the Proto-Oncogene c-Myc. J. Immunol. 2011, 186, 6726–6736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croxford, J.L.; Li, M.; Tang, F.; Pan, M.F.; Huang, C.W.; Kamran, N.; Meow, C.; Phua, L.; Chng, W.J.; Ng, S.B.; et al. ATM-Dependent Spontaneous Regression of Early Em—Myc—Induced Murine B-Cell Leukemia Depends on Natural Killer and T Cells. Blood 2013, 121, 2512–2521. [Google Scholar] [CrossRef]
- Granato, A.; Hayashi, E.A.; Baptista, B.J.A.; Bellio, M.; Nobrega, A. IL-4 Regulates Bim Expression and Promotes B Cell Maturation in Synergy with BAFF Conferring Resistance to Cell Death at Negative Selection Checkpoints. J. Immunol. 2014, 192, 5761–5775. [Google Scholar] [CrossRef] [Green Version]
- Ouk, C.; Roland, L.; Gachard, N.; Poulain, S.; Oblet, C.; Rizzo, D.; Saintamand, A.; Lemasson, Q.; Carrion, C.; Thomas, M.; et al. Continuous MYD88 Activation Is Associated with Expansion and Then Transformation of IgM Differentiating Plasma Cells. Front. Immunol. 2021, 12, 641692. [Google Scholar] [CrossRef]
- Piskor, E.M.; Winkler, R.; Kosan, C. Analyzing Lymphoma Development and Progression Using HDACi in Mouse Models. Methods Mol. Biol. 2023, 2589, 3–15. [Google Scholar] [CrossRef]
- Hunter, J.E.; Butterworth, J.; Perkins, N.D.; Bateson, M.; Richardson, C.A. Using Body Temperature, Food and Water Consumption as Biomarkers of Disease Progression in Mice with Eμ-Myc Lymphoma. Br. J. Cancer 2014, 110, 928–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poe, J.C.; Minard-Colin, V.; Kountikov, E.I.; Haas, K.M.; Tedder, T.F. A C-Myc and Surface CD19 Signaling Amplification Loop Promotes B Cell Lymphoma Development and Progression in Mice. J. Immunol. 2012, 189, 2318–2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, R.; Mägdefrau, A.S.; Piskor, E.M.; Kleemann, M.; Beyer, M.; Linke, K.; Hansen, L.; Schaffer, A.M.; Hoffmann, M.E.; Poepsel, S.; et al. Targeting the MYC Interaction Network in B-Cell Lymphoma via Histone Deacetylase 6 Inhibition. Oncogene 2022, 41, 4560–4572. [Google Scholar] [CrossRef] [PubMed]
- Nepal, R.M.; Tong, L.; Kolaj, B.; Edelmann, W.; Martin, A. Msh2-Dependent DNA Repair Mitigates a Unique Susceptibility of B Cell Progenitors to c-Myc-Induced Lymphomas. Proc. Natl. Acad. Sci. USA 2009, 106, 18698–18703. [Google Scholar] [CrossRef] [Green Version]
- Hilmenyuk, T.; Ruckstuhl, C.A.; Hayoz, M.; Berchtold, C.; Nuoffer, J.M.; Solanki, S.; Keun, H.C.; Beavis, P.A.; Riether, C.; Ochsenbein, A.F. T Cell Inhibitory Mechanisms in a Model of Aggressive Non-Hodgkin’s Lymphoma. Oncoimmunology 2018, 7, e1365997. [Google Scholar] [CrossRef] [Green Version]
- Davila, M.L.; Kloss, C.C.; Gunset, G.; Sadelain, M. CD19 CAR-Targeted T Cells Induce Long-Term Remission and B Cell Aplasia in an Immunocompetent Mouse Model of B Cell Acute Lymphoblastic Leukemia. PLoS ONE 2013, 8, e61338. [Google Scholar] [CrossRef] [Green Version]
- Mattarollo, S.R.; West, A.C.; Steegh, K.; Duret, H.; Paget, C.; Martin, B.; Matthews, G.M.; Shortt, J.; Chesi, M.; Bergsagel, P.L.; et al. NKT Cell Adjuvant-Based Tumor Vaccine for Treatment of Myc Oncogene-Driven Mouse B-Cell Lymphoma. Blood 2012, 120, 3019–3029. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Doff, B.L.; Rearden, R.C.; Leggatt, G.R.; Mattarollo, S.R. NKT Cell-Targeted Vaccination plus Anti-4–1BB Antibody Generates Persistent CD8 T Cell Immunity against B Cell Lymphoma. Oncoimmunology 2015, 4, e990793. [Google Scholar] [CrossRef] [Green Version]
- Ross, J.; Rashkovan, M.; Fraszczak, J.; Joly-Beauparlant, C.; Vadnais, C.; Winkler, R.; Droit, A.; Kosan, C.; Moroy, T. Deletion of the MIZ-1 POZ Domain Increases Efficacy of Cytarabine Treatment in T- And B-ALL/Lymphoma Mouse Models. Cancer Res. 2019, 79, 4184–4195. [Google Scholar] [CrossRef]
- Faubert, B.; Boily, G.; Izreig, S.; Griss, T.; Samborska, B.; Dong, Z.; Dupuy, F.; Chambers, C.; Fuerth, B.J.; Viollet, B.; et al. AMPK Is a Negative Regulator of the Warburg Effect and Suppresses Tumor Growth in Vivo. Cell. Metab. 2013, 17, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Wolyniec, K.; Shortt, J.; de Stanchina, E.; Levav-Cohen, Y.; Alsheich-Bartok, O.; Louria-Hayon, I.; Corneille, V.; Kumar, B.; Woods, S.J.; Opat, S.; et al. E6AP Ubiquitin Ligase Regulates PML-Induced Senescence in Myc-Driven Lymphomagenesis. Blood 2012, 120, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Rempel, R.E.; Chang, J.T.; Yao, G.; Lagoo, A.S.; Potti, A.; Bild, A.; Nevins, J.R. Utilization of Pathway Signatures to Reveal Distinct Types of B Lymphoma in the Eμ-Myc Model and Human Diffuse Large B-Cell Lymphoma. Cancer Res. 2008, 68, 8525–8534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular Subtypes of Diffuse Large B Cell Lymphoma Are Associated with Distinct Pathogenic Mechanisms and Outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Wall, M.; Poortinga, G.; Stanley, K.L.; Lindemann, R.K.; Bots, M.; Chan, C.J.; Bywater, M.J.; Kinross, K.M.; Astle, M.V.; Waldeck, K.; et al. The MTORC1 Inhibitor Everolimus Prevents and Treats Eμ-Myc Lymphoma by Restoring Oncogene-Induced Senescence. Cancer Discov. 2013, 3, 82–95. [Google Scholar] [CrossRef] [Green Version]
- Witzig, T.E.; Reeder, C.B.; Laplant, B.R.; Gupta, M.; Johnston, P.B.; Micallef, I.N.; Porrata, L.F.; Ansell, S.M.; Colgan, J.P.; Jacobsen, E.D.; et al. A Phase II Trial of the Oral MTOR Inhibitor Everolimus in Relapsed Aggressive Lymphoma. Leukemia 2011, 25, 341–347. [Google Scholar] [CrossRef]
- Hartleben, G.; Müller, C.; Krämer, A.; Schimmel, H.; Zidek, L.M.; Dornblut, C.; Winkler, R.; Eichwald, S.; Kortman, G.; Kosan, C.; et al. Tuberous Sclerosis Complex Is Required for Tumor Maintenance in MYC-driven Burkitt’s Lymphoma. EMBO J. 2018, 37, e98589. [Google Scholar] [CrossRef]
- Xu, W.; Berning, P.; Erdmann, T.; Grau, M.; Bettazová, N.; Zapukhlyak, M.; Frontzek, F.; Kosnopfel, C.; Lenz, P.; Grondine, M.; et al. MTOR Inhibition Amplifies the Anti-Lymphoma Effect of PI3Kβ/δ Blockage in Diffuse Large B-Cell Lymphoma. Leukemia 2022. [Google Scholar] [CrossRef]
- Schleich, K.; Kase, J.; Dörr, J.R.; Trescher, S.; Bhattacharya, A.; Yu, Y.; Wailes, E.M.; Fan, D.N.Y.; Lohneis, P.; Milanovic, M.; et al. H3K9me3-Mediated Epigenetic Regulation of Senescence in Mice Predicts Outcome of Lymphoma Patients. Nat. Commun. 2020, 11, 3651. [Google Scholar] [CrossRef]
- Rickert, R.C.; Roes, J.; Rajewsky, K. B Lymphocyte-Specific, Cre-Mediated Mutagenesis in Mice. Nucleic Acids Res. 1997, 25, 1317–1318. [Google Scholar] [CrossRef]
- Hobeika, E.; Thiemann, S.; Storch, B.; Jumaa, H.; Nielsen, P.J.; Pelanda, R.; Reth, M. Testing Gene Function Early in the B Cell Lineage in Mb1-Cre Mice. Proc. Natl. Acad. Sci. USA 2006, 103, 13789–13794. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, M.; Beauchemin, H.; Fraszczak, J.; Ross, J.; Shooshtarizadeh, P.; Chen, R.; Möröy, T. The X-Linked Helicase DDX3X Is Required for Lymphoid Differentiation and MYC-Driven Lymphomagenesis. Cancer Res. 2022, 82, 3172–3186. [Google Scholar] [CrossRef] [PubMed]
- Crouch, E.E.; Li, Z.; Takizawa, M.; Fichtner-Feigl, S.; Gourzi, P.; Montaño, C.; Feigenbaum, L.; Wilson, P.; Janz, S.; Papavasiliou, F.N.; et al. Regulation of AID Expression in the Immune Response. J. Exp. Med. 2007, 204, 1145–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casola, S.; Cattoretti, G.; Uyttersprot, N.; Koralov, S.B.; Segal, J.; Hao, Z.; Waisman, A.; Egert, A.; Ghitza, D.; Rajewsky, K. Tracking Germinal Center B Cells Expressing Germ-Line Immunoglobulin Γ1 Transcripts by Conditional Gene Targeting. Proc. Natl. Acad. Sci. USA 2006, 103, 7396–7401. [Google Scholar] [CrossRef] [Green Version]
- Kraus, M.; Alimzhanov, M.B.; Rajewsky, N.; Rajewsky, K. Survival of Resting Mature B Lymphocytes Depends on BCR Signaling via the Igα/β Heterodimer. Cell 2004, 117, 787–800. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Grishagin, I.; Wang, Y.; Zhao, T.; Greene, J.; Obenauer, J.C.; Ngan, D.; Nguyen, D.T.; Guha, R.; Jadhav, A.; et al. The NCATS BioPlanet—An Integrated Platform for Exploring the Universe of Cellular Signaling Pathways for Toxicology, Systems Biology, and Chemical Genomics. Front. Pharm. 2019, 10, 445. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. Revigo Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Alt, J.R.; Greiner, T.C.; Cleveland, J.L.; Eischen, C.M. Mdm2 Haplo-Insufficiency Profoundly Inhibits Myc-Induced Lymphomagenesis. EMBO J. 2003, 22, 1442–1450. [Google Scholar] [CrossRef] [Green Version]
- Post, S.M.; Quintás-Cardama, A.; Terzian, T.; Smith, C.; Eischen, C.M.; Lozano, G. P53-Dependent Senescence Delays E-Myc-Induced B-Cell Lymphomagenesis. Oncogene 2010, 29, 1260–1269. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, C.A.; McCurrach, M.E.; de Stanchina, E.; Wallace-Brodeur, R.R.; Lowe, S.W. INK4a/ARF Mutations Accelerate Lymphomagenesis and Promote Chemoresistance by Disabling P53. Genes Dev. 1999, 13, 2670–2677. [Google Scholar] [CrossRef] [PubMed]
- Yetil, A.; Anchang, B.; Gouw, A.M.; Adam, S.J.; Zabuawala, T.; Parameswaran, R.; van Riggelen, J.; Plevritis, S.; Felsher, D.W. P19ARF Is a Critical Mediator of Both Cellular Senescence and an Innate Immune Response Associated with MYC Inactivation in Mouse Model of Acute Leukemia. Oncotarget 2015, 6, 3563–3577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Chen, L.; Yang, L.; Xie, X.; Gan, L.; Cleveland, J.L.; Chen, J. MDMX Acidic Domain Inhibits P53 DNA Binding in Vivo and Regulates Tumorigenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E3368–E3377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Carlson, N.R.; Dong, J.; Zhang, Y. Oncogenic C-Myc-Induced Lymphomagenesis Is Inhibited Non-Redundantly by the P19Arf-Mdm2-P53 and RP-Mdm2-P53 Pathways. Oncogene 2015, 34, 5709–5717. [Google Scholar] [CrossRef] [Green Version]
- Donehower, L.A.; Harvey, M.; Slagle, B.L.; McArthur, M.J.; Montgomery, C.A.; Butel, J.S.; Bradley, A. Mice Deficient for P53 Are Developmentally Normal but Susceptible to Spontaneous Tumours. Nature 1992, 356, 215–221. [Google Scholar] [CrossRef]
- Rowh, M.A.W.; Demicco, A.; Horowitz, J.E.; Yin, B.; Yang-Iott, K.S.; Fusello, A.M.; Hobeika, E.; Reth, M.; Bassing, C.H. Tp53 Deletion in B Lineage Cells Predisposes Mice to Lymphomas with Oncogenic Translocations. Oncogene 2011, 30, 4757–4764. [Google Scholar] [CrossRef] [Green Version]
- Gostissa, M.; Bianco, J.M.; Malkin, D.J.; Kutok, J.L.; Rodig, S.J.; Morse, H.C.; Bassing, C.H.; Alt, F.W. Conditional Inactivation of P53 in Mature B Cells Promotes Generation of Nongerminal Center-Derived B-Cell Lymphomas. Proc. Natl. Acad. Sci. USA 2013, 110, 2934–2939. [Google Scholar] [CrossRef] [Green Version]
- Rempel, R.E.; Mori, S.; Gasparetto, M.; Glozak, M.A.; Andrechek, E.R.; Adler, S.B.; Laakso, N.M.; Lagoo, A.S.; Storms, R.; Smith, C.; et al. A Role for E2F Activities in Determining the Fate of Myc-Induced Lymphomagenesis. PLoS Genet. 2009, 5, e1000640. [Google Scholar] [CrossRef] [Green Version]
- Kent, L.N.; Leone, G. The Broken Cycle: E2F Dysfunction in Cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Bouchard, C.; Dittrich, O.; Kiermaier, A.; Dohmann, K.; Menkel, A.; Eilers, M.; Lüscher, B. Regulation of Cyclin D2 Gene Expression by the Myc/Max/Mad Network: Myc-Dependent TRRAP Recruitment and Histone Acetylation at the Cyclin D2 Promoter. Genes Dev. 2001, 15, 2042–2047. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Lisanti, M.P.; Liao, D.J. Reviewing Once More the C-Myc and Ras Collaboration: Converging at the Cyclin D1-CDK4 Complex and Challenging Basic Concepts of Cancer Biology. Cell Cycle 2011, 10, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.-C.; Chang, W.-C.; Shieh, S.-Y.; Tarn, W.-Y. DDX3 Regulates Cell Growth through Translational Control of Cyclin E1. Mol. Cell Biol. 2010, 30, 5444–5453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farria, A.T.; Plummer, J.B.; Salinger, A.P.; Shen, J.; Lin, K.; Lu, Y.; McBride, K.M.; Koutelou, E.; Dent, S.Y.R. Transcriptional Activation of MYC-Induced Genes by GCN5 Promotes B-Cell Lymphomagenesis. Cancer Res. 2020, 80, 5543–5553. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, B.N.; Lee, S.C.W.; El-Saafin, F.; Vanyai, H.K.; Hu, Y.; Pang, S.H.M.; Grabow, S.; Strasser, A.; Nutt, S.L.; Alexander, W.S.; et al. MOZ Regulates B-Cell Progenitors and, Consequently, Moz Haploinsufficiency Dramatically Retards MYC-Induced Lymphoma Development. Blood 2015, 125, 1910–1921. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Leaver, D.J.; Hermans, S.J.; Kelly, G.L.; Brennan, M.S.; Downer, N.L.; Nguyen, N.; Wichmann, J.; McRae, H.M.; Yang, Y.; et al. Inhibitors of Histone Acetyltransferases KAT6A/B Induce Senescence and Arrest Tumour Growth. Nature 2018, 560, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Berg, T.; Thoene, S.; Yap, D.; Wee, T.; Schoeler, N.; Rosten, P.; Lim, E.; Bilenky, M.; Mungall, A.J.; Oellerich, T.; et al. A Transgenic Mouse Model Demonstrating the Oncogenic Role of Mutations in the Polycomb-Group Gene EZH2 in Lymphomagenesis. Blood 2014, 123, 3914–3924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef] [PubMed]
- Vandel, L.; Nicolas, E.; Vaute, O.; Ferreira, R.; Ait-Si-Ali, S.; Trouche, D. Transcriptional Repression by the Retinoblastoma Protein through the Recruitment of a Histone Methyltransferase. Mol. Cell Biol. 2001, 21, 6484–6494. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, S.J.; Schneider, R.; Bauer, U.M.; Bannister, A.J.; Morrison, A.; O’Carroll, D.; Firestein, R.; Cleary, M.; Jenuwein, T.; Herrera, R.E.; et al. Rb Targets Histone H3 Methylation and HP1 to Promoters. Nature 2001, 412, 561–565. [Google Scholar] [CrossRef]
- Reimann, M.; Lee, S.; Loddenkemper, C.; Dörr, J.R.; Tabor, V.; Aichele, P.; Stein, H.; Dörken, B.; Jenuwein, T.; Schmitt, C.A. Tumor Stroma-Derived TGF-β Limits Myc-Driven Lymphomagenesis via Suv39h1-Dependent Senescence. Cancer Cell 2010, 17, 262–272. [Google Scholar] [CrossRef] [Green Version]
- Fernández-serrano, M.; Winkler, R.; Santos, J.C.; le Pannérer, M.M.; Buschbeck, M.; Roué, G. Histone Modifications and Their Targeting in Lymphoid Malignancies. Int. J. Mol. Sci. 2022, 23, 253. [Google Scholar] [CrossRef] [PubMed]
- Mathsyaraja, H.; Freie, B.; Cheng, P.F.; Babaeva, E.; Catchpole, J.T.; Janssens, D.; Henikoff, S.; Eisenman, R.N. Max Deletion Destabilizes MYC Protein and Abrogates Eµ-Myc Lymphomagenesis. Genes Dev. 2019, 33, 1252–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prochownik, E.V.; Vogt, P.K. Therapeutic Targeting of Myc. Genes Cancer 2010, 1, 650–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, A.G.; Parker, J.B.; Sagar, V.; Truica, M.I.; Soni, P.N.; Han, H.; Schiltz, G.E.; Abdulkadir, S.A.; Chakravarti, D. A MYC Inhibitor Selectively Alters the MYC and MAX Cistromes and Modulates the Epigenomic Landscape to Regulate Target Gene Expression. Sci. Adv. 2022, 8, eabh3635. [Google Scholar] [CrossRef] [PubMed]
- Castell, A.; Yan, Q.; Fawkner, K.; Bazzar, W.; Zhang, F.; Wickström, M.; Alzrigat, M.; Franco, M.; Krona, C.; Cameron, D.P.; et al. MYCMI-7: A Small MYC-Binding Compound That Inhibits MYC: MAX Interaction and Tumor Growth in a MYC-Dependent Manner. Cancer Res. Commun. 2022, 2, 182–201. [Google Scholar] [CrossRef]
- Salghetti, S.E.; Kim, S.Y.; Tansey, W.P. Destruction of Myc by Ubiquitin-Mediated Proteolysis: Cancer-Associated and Transforming Mutations Stabilize Myc. EMBO J. 1999, 18, 717–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sears, R.; Leone, G.; DeGregori, J.; Nevins, J.R. Ras Enhances Myc Protein Stability. Mol. Cell 1999, 3, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Gregory, M.A.; Hann, S.R. C-Myc Proteolysis by the Ubiquitin-Proteasome Pathway: Stabilization of c-Myc in Burkitt’s Lymphoma Cells. Mol Cell Biol. 2000, 20, 2423–2435. [Google Scholar] [CrossRef] [Green Version]
- Sears, R.; Nuckolls, F.; Haura, E.; Taya, Y.; Tamai, K.; Nevins, J.R. Multiple Ras-Dependent Phosphorylation Pathways Regulate Myc Protein Stability. Genes Dev. 2000, 14, 2501–2514. [Google Scholar] [CrossRef] [Green Version]
- Cohn, G.M.; Liefwalker, D.F.; Langer, E.M.; Sears, R.C. PIN1 Provides Dynamic Control of MYC in Response to Extrinsic Signals. Front. Cell Dev. Biol. 2020, 8, 224. [Google Scholar] [CrossRef]
- D’Artista, L.; Bisso, A.; Piontini, A.; Doni, M.; Verrecchia, A.; Kress, T.R.; Morelli, M.J.; del Sal, G.; Amati, B.; Campaner, S. Pin1 Is Required for Sustained B Cell Proliferation upon Oncogenic Activation of Myc. Oncotarget 2016, 7, 21786–21798. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Alexandrova, N.; Niklinski, J.; Bliskovsky, V.; Otterson, G.A.; Blake, M.; Kaye, F.J.; Zajac-Kaye, M. The N-Terminal Domain of c-Myc Associates with Alpha-Tubulin and Microtubules in Vivo and in Vitro. Mol. Cell Biol. 1995, 15, 5188–5195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuyama, A.; Shimazu, T.; Sumida, Y.; Saito, A.; Yoshimatsu, Y.; Seigneurin-Berny, D.; Osada, H.; Komatsu, Y.; Nishino, N.; Khochbin, S.; et al. In Vivo Destabilization of Dynamic Microtubules by HDAC6-Mediated Deacetylation. EMBO J. 2002, 21, 6820–6831. [Google Scholar] [CrossRef] [Green Version]
- Miyake, Y.; Keusch, J.J.; Wang, L.; Saito, M.; Hess, D.; Wang, X.; Melancon, B.J.; Helquist, P.; Gut, H.; Matthias, P. Structural Insights into HDAC6 Tubulin Deacetylation and Its Selective Inhibition. Nat. Chem. Biol. 2016, 12, 748–754. [Google Scholar] [CrossRef]
- Adams, C.M.; Kim, A.S.; Mitra, R.; Choi, J.K.; Gong, J.Z.; Eischen, C.M. BCL-W Has a Fundamental Role in B Cell Survival and Lymphomagenesis. J. Clin. Investig. 2017, 127, 635–650. [Google Scholar] [CrossRef] [Green Version]
- Kelly, G.L.; Grabow, S.; Glaser, S.P.; Fitzsimmons, L.; Aubrey, B.J.; Okamoto, T.; Valente, L.J.; Robati, M.; Tai, L.; Douglas Fairlie, W.; et al. Targeting of MCL-1 Kills MYC-Driven Mouse and Human Lymphomas Even When They Bear Mutations in P53. Genes Dev. 2014, 28, 58–70. [Google Scholar] [CrossRef] [Green Version]
- Grabow, S.; Kelly, G.L.; Delbridge, A.R.D.; Kelly, P.N.; Bouillet, P.; Adams, J.M.; Strasser, A. Critical B-Lymphoid Cell Intrinsic Role of Endogenous MCL-1 in c-MYC-Induced Lymphomagenesis. Cell Death Dis. 2016, 7, e2132. [Google Scholar] [CrossRef]
- Egle, A.; Harris, A.W.; Bouillet, P.; Cory, S. Bim Is a Suppressor of Myc-Induced Mouse B Cell Leukemia. Proc. Natl. Acad. Sci. USA 2004, 101, 6164–6169. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; King, A.; Bouillet, P.; Tarlinton, D.M.; Strasser, A.; Heierhorst, J. Proapoptotic BIM Impacts B Lymphoid Homeostasis by Limiting the Survival of Mature B Cells in a Cell-Autonomous Manner. Front. Immunol. 2018, 8, 592. [Google Scholar] [CrossRef]
- Michalak, E.M.; Jansen, E.S.; Happo, L.; Cragg, M.S.; Tai, L.; Smyth, G.K.; Strasser, A.; Adams, J.M.; Scott, C.L. Puma and to a Lesser Extent Noxa Are Suppressors of Myc-Induced Lymphomagenesis. Cell Death Differ. 2009, 16, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.H.; McMahon, S.B. Targeting of Miz-1 Is Essential for Myc-Mediated Apoptosis. J. Biol. Chem. 2006, 281, 3283–3289. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.H.; McMahon, S.B. BCL2 Is a Downstream Effector of MIZ-1 Essential for Blocking c-MYC-Induced Apoptosis. J. Biol. Chem. 2007, 282, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Baluapuri, A.; Wolf, E.; Eilers, M. Target Gene-Independent Functions of MYC Oncoproteins. Nat. Rev. Mol. Cell Biol. 2020, 21, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Murga, M.; Campaner, S.; Lopez-Contreras, A.J.; Toledo, L.I.; Soria, R.; Montaña, M.F.; D’Artista, L.; Schleker, T.; Guerra, C.; Garcia, E.; et al. Exploiting Oncogene-Induced Replicative Stress for the Selective Killing of Myc-Driven Tumors. Nat. Struct. Mol. Biol. 2011, 18, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Puccetti, M.V.; Adams, C.M.; Kushinsky, S.; Eischen, C.M. SMARCAL1 and ZrAnB3 Protect Replication Forks from MYC-Induced DNA Replication Stress. Cancer Res. 2019, 79, 1612–1623. [Google Scholar] [CrossRef]
- Hamperl, S.; Cimprich, K.A. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell 2016, 167, 1455–1467. [Google Scholar] [CrossRef] [Green Version]
- Sollier, J.; Cimprich, K.A. Breaking Bad: R-Loops and Genome Integrity. Trends Cell Biol. 2015, 25, 514–522. [Google Scholar] [CrossRef] [Green Version]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774–786.e19. [Google Scholar] [CrossRef] [Green Version]
- Schuler, F.; Weiss, J.G.; Lindner, S.E.; Lohmüller, M.; Herzog, S.; Spiegl, S.F.; Menke, P.; Geley, S.; Labi, V.; Villunger, A. Checkpoint Kinase 1 Is Essential for Normal B Cell Development and Lymphomagenesis. Nat. Commun. 2017, 8, 1697. [Google Scholar] [CrossRef] [Green Version]
- Gorrini, C.; Squatrito, M.; Luise, C.; Syed, N.; Perna, D.; Wark, L.; Martinato, F.; Sardella, D.; Verrecchia, A.; Bennett, S.; et al. Tip60 Is a Haplo-Insufficient Tumour Suppressor Required for an Oncogene-Induced DNA Damage Response. Nature 2007, 448, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.H.; Du, Y.; Ard, P.G.; Phillips, C.; Carella, B.; Chen, C.-J.; Rakowski, C.; Chatterjee, C.; Lieberman, P.M.; Lane, W.S.; et al. The C-MYC Oncoprotein Is a Substrate of the Acetyltransferases HGCN5/PCAF and TIP60. Mol. Cell Biol. 2004, 24, 10826–10834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Zhang, H.; Kajino, K.; Greene, M.I. BRCA1 Binds C-Myc and Inhibits Its Transcriptional and Transforming Activity in Cells. Oncogene 1998, 17, 1939–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herold, S.; Kalb, J.; Büchel, G.; Ade, C.P.; Baluapuri, A.; Xu, J.; Koster, J.; Solvie, D.; Carstensen, A.; Klotz, C.; et al. Recruitment of BRCA1 Limits MYCN-Driven Accumulation of Stalled RNA Polymerase. Nature 2019, 567, 545–549. [Google Scholar] [CrossRef]
- Choi, P.S.; Van Riggelen, J.; Gentles, A.J.; Bachireddy, P.; Rakhra, K.; Adam, S.J.; Plevritis, S.K.; Felsher, D.W. Lymphomas That Recur after MYC Suppression Continue to Exhibit Oncogene Addiction. Proc. Natl. Acad. Sci. USA 2011, 108, 17432–17437. [Google Scholar] [CrossRef] [Green Version]
- Van Riggelen, J.; Müller, J.; Otto, T.; Beuger, V.; Yetil, A.; Choi, P.S.; Kosan, C.; Möröy, T.; Felsher, D.W.; Eilers, M. The Interaction between Myc and Miz1 Is Required to Antagonize TGFβ-Dependent Autocrine Signaling during Lymphoma Formation and Maintenance. Genes Dev. 2010, 24, 1281–1294. [Google Scholar] [CrossRef] [Green Version]
- Muthalagu, N.; Monteverde, T.; Raffo-Iraolagoitia, X.; Wiesheu, R.; Whyte, D.; Hedley, A.; Laing, S.; Kruspig, B.; Upstill-Goddard, R.; Shaw, R.; et al. Repression of the Type i Interferon Pathway Underlies MYC-and KRAS-Dependent Evasion of NK and B Cells in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2020, 10, 872–887. [Google Scholar] [CrossRef] [Green Version]
- Bartha, Á.; Győrffy, B. TNMplot.Com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Rivas, M.A.; Melnick, A.M. Role of Chromosomal Architecture in Germinal Center B Cells and Lymphomagenesis. Curr. Opin. Hematol. 2019, 26, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Canela, A.; Maman, Y.; Jung, S.; Wong, N.; Callen, E.; Day, A.; Kieffer-Kwon, K.R.; Pekowska, A.; Zhang, H.; Rao, S.S.P.; et al. Genome Organization Drives Chromosome Fragility. Cell 2017, 170, 507–521.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hnisz, D.; Weintrau, A.S.; Day, D.S.; Valton, A.L.; Bak, R.O.; Li, C.H.; Goldmann, J.; Lajoie, B.R.; Fan, Z.P.; Sigova, A.A.; et al. Activation of Proto-Oncogenes by Disruption of Chromosome Neighborhoods. Science (1979) 2016, 351, 1454–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; McBride, K.M.; Hensley, S.; Lu, Y.; Chedin, F.; Bedford, M.T. Arginine Methylation Facilitates the Recruitment of TOP3B to Chromatin to Prevent R Loop Accumulation. Mol. Cell 2014, 53, 484–497. [Google Scholar] [CrossRef] [Green Version]
- Hall, Z.; Ament, Z.; Wilson, C.H.; Burkhart, D.L.; Ashmore, T.; Koulman, A.; Littlewood, T.; Evan, G.I.; Griffin, J.L. MYC Expression Drives Aberrant Lipid Metabolism in Lung Cancer. Cancer Res. 2016, 76, 4608–4618. [Google Scholar] [CrossRef] [Green Version]
- Eberlin, L.S.; Gabay, M.; Fan, A.C.; Gouw, A.M.; Tibshirani, R.J.; Felsher, D.W.; Zare, R.N. Alteration of the Lipid Profile in Lymphomas Induced by MYC Overexpression. Proc. Natl. Acad. Sci. USA 2014, 111, 10450–10455. [Google Scholar] [CrossRef] [Green Version]
- Chu, B.; Kon, N.; Chen, D.; Li, T.; Liu, T.; Jiang, L.; Song, S.; Tavana, O.; Gu, W. ALOX12 Is Required for P53-Mediated Tumour Suppression through a Distinct Ferroptosis Pathway. Nat. Cell Biol. 2019, 21, 579–591. [Google Scholar] [CrossRef]
- Hofmann, J.W.; Zhao, X.; De Cecco, M.; Peterson, A.L.; Pagliaroli, L.; Manivannan, J.; Hubbard, G.B.; Ikeno, Y.; Zhang, Y.; Feng, B.; et al. Reduced Expression of MYC Increases Longevity and Enhances Healthspan. Cell 2015, 160, 477–488. [Google Scholar] [CrossRef] [Green Version]
- Pourdehnad, M.; Truitt, M.L.; Siddiqi, I.N.; Ducker, G.S.; Shokat, K.M.; Ruggero, D. Myc and MTOR Converge on a Common Node in Protein Synthesis Control That Confers Synthetic Lethality in Myc-Driven Cancers. Proc. Natl. Acad. Sci. USA 2013, 110, 11988–11993. [Google Scholar] [CrossRef] [Green Version]
- Kotani, A.; Kakazu, N.; Tsuruyama, T.; Okazaki, I.M.; Muramatsu, M.; Kinoshita, K.; Nagaoka, H.; Yabe, D.; Honjo, T. Activation-Induced Cytidine Deaminase (AID) Promotes B Cell Lymphomagenesis in Emu-Cmyc Transgenic Mice. Proc. Natl. Acad. Sci. USA 2007, 104, 1616–1620. [Google Scholar] [CrossRef] [Green Version]
- Nepal, R.M.; Zaheen, A.; Basit, W.; Li, L.; Berger, S.A.; Martin, A. AID and RAG1 Do Not Contribute to Lymphomagenesis in Eμ C-Myc Transgenic Mice. Oncogene 2008, 27, 4752–4756. [Google Scholar] [CrossRef] [PubMed]
- Scuoppo, C.; Miething, C.; Lindqvist, L.; Reyes, J.; Ruse, C.; Appelmann, I.; Yoon, S.; Krasnitz, A.; Teruya-Feldstein, J.; Pappin, D.; et al. A Tumour Suppressor Network Relying on the Polyamine-Hypusine Axis. Nature 2012, 487, 244–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, C.L.; Schuler, M.; Marsden, V.S.; Egle, A.; Pellegrini, M.; Nesic, D.; Gerondakis, S.; Nutt, S.L.; Green, D.R.; Strasser, A. Apaf-1 and Caspase-9 Do Not Act as Tumor Suppressors in Myc-Induced Lymphomagenesis or Mouse Embryo Fibroblast Transformation. J. Cell Biol. 2004, 164, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Walczynski, J.; Lyons, S.; Jones, N.; Breitwieser, W. Sensitisation of C-MYC-Induced B-Lymphoma Cells to Apoptosis by ATF2. Oncogene 2014, 33, 1027–1036. [Google Scholar] [CrossRef]
- Tameire, F.; Verginadis, I.I.; Leli, N.M.; Polte, C.; Conn, C.S.; Ojha, R.; Salas Salinas, C.; Chinga, F.; Monroy, A.M.; Fu, W.; et al. ATF4 Couples MYC-Dependent Translational Activity to Bioenergetic Demands during Tumour Progression. Nat. Cell Biol. 2019, 21, 889–899. [Google Scholar] [CrossRef]
- Eischen, C.M.; Roussel, M.F.; Korsmeyer, S.J.; Cleveland, J.L. Bax Loss Impairs Myc-Induced Apoptosis and Circumvents the Selection of P53 Mutations during Myc-Mediated Lymphomagenesis. Mol. Cell Biol. 2001, 21, 7653–7662. [Google Scholar] [CrossRef] [Green Version]
- Kelly, P.N.; Puthalakath, H.; Adams, J.M.; Strasser, A. Endogenous Bcl-2 Is Not Required for the Development of Eμ-Myc-Induced B-Cell Lymphoma. Blood 2007, 109, 4907–4913. [Google Scholar] [CrossRef] [Green Version]
- Kelly, P.N.; Grabow, S.; Delbridge, A.R.D.; Strasser, A.; Adams, J.M. Endogenous Bcl-XL Is Essential for Myc-Driven Lymphomagenesis in Mice. Blood 2011, 118, 6380–6386. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Hori, T.; Cooper, T.K.; Liao, J.; Desai, N.; Serfass, J.M.; Young, M.M.; Park, S.; Izu, Y.; Wang, H.G. Bif-1 Haploinsufficiency Promotes Chromosomal Instability and Accelerates Myc-Driven Lymphomagenesis via Suppression of Mitophagy. Blood 2013, 121, 1622–1632. [Google Scholar] [CrossRef] [Green Version]
- Happo, L.; Phipson, B.; Smyth, G.K.; Strasser, A.; Scott, C.L. Neither Loss of Bik Alone, nor Combined Loss of Bik and Noxa, Accelerate Murine Lymphoma Development or Render Lymphoma Cells Resistant to DNA Damaging Drugs. Cell Death Dis. 2012, 3, e306. [Google Scholar] [CrossRef] [Green Version]
- Frenzel, A.; Labi, V.; Chmelewskij, W.; Ploner, C.; Geley, S.; Fiegl, H.; Tzankov, A.; Villunger, A. Suppression of B-Cell Lymphomagenesis by the BH3-Only Proteins Bmf and Bad. Blood 2010, 115, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.J.L.; Scheijen, B.; Voncken, J.W.; Kieboom, K.; Berns, A.; van Lohuizen, M. Bmi-1 Collaborates with c-Myc in Tumorigenesis by Inhibiting c-Myc- Induced Apoptosis via INK4a/ARF. Genes Dev. 1999, 13, 2678–2690. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.W.; Phipson, B.; Hyland, C.D.; Leong, H.S.; Allan, R.S.; Lun, A.; Hilton, D.J.; Nutt, S.L.; Blewitt, M.E.; Smyth, G.K.; et al. Polycomb Repressive Complex 2 (PRC2) Suppresses Em-Myc Lymphoma. Blood 2013, 122, 2654–2664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, C.L.; Gil, J.; Hernando, E.; Teruya-Feldstein, J.; Narita, M.; Martínez, D.; Visakorpi, T.; Mu, D.; Cordon-Cardo, C.; Peters, G.; et al. Role of the Chromobox Protein CBX7 in Lymphomagenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 5389–5394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, F.; Voss, A.; Kerr, J.B.; O’Reilly, L.A.; Tai, L.; Echeverry, N.; Bouillet, P.; Strasser, A.; Kaufmann, T. BCL-2 Family Member BOK Is Widely Expressed but Its Loss Has Only Minimal Impact in Mice. Cell Death Differ. 2012, 19, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Habib, T.; Park, H.; Tsang, M.; de Alborán, I.M.; Nicks, A.; Wilson, L.; Knoepfler, P.S.; Andrews, S.; Rawlings, D.J.; Eisenman, R.N.; et al. Myc Stimulates B Lymphocyte Differentiation and Amplifies Calcium Signaling. J. Cell Biol. 2007, 179, 717–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricke, R.M.; Jeganathan, K.B.; van Deursen, J.M. Bub1 Overexpression Induces Aneuploidy and Tumor Formation through Aurora B Kinase Hyperactivation. J. Cell Biol. 2011, 193, 1049–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shing, J.C.; Lindquist, L.D.; Borgese, N.; Bram, R.J. CAML Mediates Survival of Myc-Induced Lymphoma Cells Independent of Tail-Anchored Protein Insertion. Cell Death Discov. 2017, 3, 16098. [Google Scholar] [CrossRef] [Green Version]
- Ho, L.H.; Taylor, R.; Dorstyn, L.; Cakouros, D.; Bouillet, P.; Kumar, S. A Tumor Suppressor Function for Caspase-2. Proc. Natl. Acad. Sci. USA 2009, 106, 5336–5341. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Wu, Y.; Feng, X.; Shen, R.; Wang, J.H.; Fallahi, M.; Li, W.; Yang, C.; Hankey, W.; Zhao, W.; et al. CDK4 Deficiency Promotes Genomic Instability and Enhances Myc-Driven Lymphomagenesis. J. Clin. Investig. 2014, 124, 1672–1684. [Google Scholar] [CrossRef] [Green Version]
- Keller, U.B.; Old, J.B.; Dorsey, F.C.; Nilsson, J.A.; Nilsson, L.; MacLean, K.H.; Chung, L.; Yang, C.; Spruck, C.; Boyd, K.; et al. Myc Targets Cks1 to Provoke the Suppression of P27Kip1, Proliferation and Lymphomagenesis. EMBO J. 2007, 26, 2562–2574. [Google Scholar] [CrossRef] [PubMed]
- Hashwah, H.; Schmid, C.A.; Kasser, S.; Bertram, K.; Stelling, A.; Manz, M.G.; Müller, A. Inactivation of CREBBP Expands the Germinal Center B Cell Compartment, down-Regulates MHCII Expression and Promotes DLBCL Growth. Proc. Natl. Acad. Sci. USA 2017, 114, 9701–9706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, J.E.; Butterworth, J.A.; Zhao, B.; Sellier, H.; Campbell, K.J.; Thomas, H.D.; Bacon, C.M.; Cockell, S.J.; Gewurz, B.E.; Perkins, N.D. The NF-ΚB Subunit c-Rel Regulates Bach2 Tumour Suppressor Expression in B-Cell Lymphoma. Oncogene 2016, 35, 3476–3484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Shin, J.H.; Zhao, R.; Phan, L.; Wang, H.; Xue, Y.; Post, S.M.; Ho Choi, H.; Chen, J.S.; Wang, E.; et al. CSN6 Drives Carcinogenesis by Positively Regulating Myc Stability. Nat. Commun. 2014, 5, 5384. [Google Scholar] [CrossRef] [Green Version]
- Pei, X.H.; Bai, F.; Li, Z.; Smith, M.D.; Whitewolf, G.; Jin, R.; Xiong, Y. Cytoplasmic CUL9/PARC Ubiquitin Ligase Is a Tumor Suppressor and Promotes P53-Dependent Apoptosis. Cancer Res. 2011, 71, 2969–2977. [Google Scholar] [CrossRef] [Green Version]
- Arrate, M.P.; Vincent, T.; Odvody, J.; Kar, R.; Jones, S.N.; Eischen, C.M. MicroRNA Biogenesis Is Required for Myc-Induced b-Cell Lymphoma Development and Survival. Cancer Res. 2010, 70, 6083–6092. [Google Scholar] [CrossRef] [Green Version]
- Inoue, K.; Zindy, F.; Randle, D.H.; Rehg, J.E.; Sherr, C.J. Dmp1 Is Haplo-Insufficient for Tumor Suppression and Modifies the Frequencies of Arf and P53 Mutations in Myc-Induced Lymphomas. Genes Dev. 2001, 15, 2934–2939. [Google Scholar] [CrossRef] [Green Version]
- Vasanthakumar, A.; Lepore, J.B.; Zegarek, M.H.; Kocherginsky, M.; Singh, M.; Davis, E.M.; Link, P.A.; Anastasi, J.; le Beau, M.M.; Karpf, A.R.; et al. Dnmt3b Is a Haploinsufficient Tumor Suppressor Gene in Myc-Induced Lymphomagenesis. Blood 2013, 121, 2059–2063. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Shah, K.; Busby, T.; Giles, K.; Khodadadi-Jamayran, A.; Li, W.; Jiang, H. Hijacking a Key Chromatin Modulator Creates Epigenetic Vulnerability for MYC-Driven Cancer. J. Clin. Investig. 2018, 128, 3605–3618. [Google Scholar] [CrossRef] [Green Version]
- Baudino, T.A.; Maclean, K.H.; Brennan, J.; Parganas, E.; Yang, C.; Aslanian, A.; Lees, J.A.; Sherr, C.J.; Roussel, M.F.; Cleveland, J.L. Myc-Mediated Proliferation and Lymphomagenesis, but Not Apoptosis, Are Compromised by E2f1 Loss. Mol. Cell 2003, 11, 905–914. [Google Scholar] [CrossRef]
- Park, H.; Staehling, K.; Tsang, M.; Appleby, M.W.; Brunkow, M.E.; Margineantu, D.; Hockenbery, D.M.; Habib, T.; Liggitt, H.D.; Carlson, G.; et al. Disruption of Fnip1 Reveals a Metabolic Checkpoint Controlling B Lymphocyte Development. Immunity 2012, 36, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, C.; Lee, S.; Paulus-Hock, V.; Loddenkemper, C.; Eilers, M.; Schmitt, C.A. FoxO Transcription Factors Suppress Myc-Driven Lymphomagenesis via Direct Activation of Arf. Genes Dev. 2007, 21, 2775–2787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillonel, V.; Reichert, N.; Cao, C.; Heideman, M.R.; Yamaguchi, T.; Matthias, G.; Tzankov, A.; Matthias, P. Histone Deacetylase 1 Plays a Predominant Pro-Oncogenic Role in Eμ-Myc Driven B Cell Lymphoma. Sci. Rep. 2016, 6, 37772. [Google Scholar] [CrossRef] [Green Version]
- Vecchio, E.; Golino, G.; Pisano, A.; Albano, F.; Falcone, C.; Ceglia, S.; Iaccino, E.; Mimmi, S.; Fiume, G.; Giurato, G.; et al. IBTK Contributes to B-Cell Lymphomagenesis in Eμ-Myc Transgenic Mice Conferring Resistance to Apoptosis. Cell Death Dis. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, L.M.; Keller, U.B.; Yang, C.; Nilsson, J.A.; Cleveland, J.L.; Roussel, M.F. Ink4c Is Dispensable for Tumor Suppression in Myc-Induced B-Cell Lymphomagenesis. Oncogene 2007, 26, 2833–2839. [Google Scholar] [CrossRef] [Green Version]
- Scherger, A.K.; Al-Maarri, M.; Maurer, H.C.; Schick, M.; Maurer, S.; Öllinger, R.; Gonzalez-Menendez, I.; Martella, M.; Thaler, M.; Pechloff, K.; et al. Activated Gp130 Signaling Selectively Targets B Cell Differentiation to Induce Mature Lymphoma and Plasmacytoma. JCI Insight 2019, 4, e128435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osborne, L.C.; Duthie, K.A.; Seo, J.H.; Gascoyne, R.D.; Abraham, N. Selective Ablation of the YxxM Motif of IL-7Rα Suppresses Lymphomagenesis but Maintains Lymphocyte Development. Oncogene 2010, 29, 3854–3864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerra, N.; Tan, Y.X.; Joncker, N.T.; Choy, A.; Gallardo, F.; Xiong, N.; Knoblaugh, S.; Cado, D.; Greenberg, N.R.; Raulet, D.H. NKG2D-Deficient Mice Are Defective in Tumor Surveillance in Models of Spontaneous Malignancy. Immunity 2008, 28, 571–580. [Google Scholar] [CrossRef] [Green Version]
- Gramling, M.W.; Eischen, C.M. Suppression of Ras/Mapk Pathway Signaling Inhibits Myc-Induced Lymphomagenesis. Cell Death Differ. 2012, 19, 1220–1227. [Google Scholar] [CrossRef] [Green Version]
- Barna, M.; Pusic, A.; Zollo, O.; Costa, M.; Kondrashov, N.; Rego, E.; Rao, P.H.; Ruggero, D. Suppression of Myc Oncogenic Activity by Ribosomal Protein Haploinsufficiency. Nature 2008, 456, 971–975. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, E.D.; Oliaro, J.; Ramsbottom, K.M.; Ting, S.B.; Sacirbegovic, F.; Harvey, M.; Kinwell, T.; Ghysdael, J.; Johnstone, R.W.; Humbert, P.O.; et al. Lethal Giant Larvae 1 Tumour Suppressor Activity Is Not Conserved in Models of Mammalian T and B Cell Leukaemia. PLoS ONE 2014, 9, e87376. [Google Scholar] [CrossRef]
- Sotillo, R.; Hernando, E.; Díaz-Rodríguez, E.; Teruya-Feldstein, J.; Cordón-Cardo, C.; Lowe, S.W.; Benezra, R. Mad2 Overexpression Promotes Aneuploidy and Tumorigenesis in Mice. Cancer Cell 2007, 11, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunelle, J.K.; Ryan, J.; Yecies, D.; Opferman, J.T.; Letai, A. MCL-1-Dependent Leukemia Cells Are More Sensitive to Chemotherapy than BCL-2-Dependent Counterparts. J. Cell Biol. 2009, 187, 429–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, K.J.; Bath, M.L.; Turner, M.L.; Vandenberg, C.J.; Bouillet, P.; Metcalf, D.; Scott, C.L.; Cory, S. Elevated Mcl-1 Perturbs Lymphopoiesis, Promotes Transformation of Hematopoietic Stem/Progenitor Cells, and Enhances Drug Resistance. Blood 2010, 116, 3197–3207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Clercq, S.; Gembarska, A.; Denecker, G.; Maetens, M.; Naessens, M.; Haigh, K.; Haigh, J.J.; Marine, J.-C. Widespread Overexpression of Epitope-Tagged Mdm4 Does Not Accelerate Tumor Formation In vivo. Mol. Cell Biol. 2010, 30, 5394–5405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terzian, T.; Wang, Y.; van Pelt, C.S.; Box, N.F.; Travis, E.L.; Lozano, G. Haploinsufficiency of Mdm2 and Mdm4 in Tumorigenesis and Development. Mol. Cell Biol. 2007, 27, 5479–5485. [Google Scholar] [CrossRef] [Green Version]
- Tanaskovic, N.; Dalsass, M.; Filipuzzi, M.; Ceccotti, G.; Verrecchia, A.; Nicoli, P.; Doni, M.; Olivero, D.; Pasini, D.; Koseki, H.; et al. Polycomb Group Ring Finger Protein 6 Suppresses Myc-Induced Lymphomagenesis. Life Sci. Alliance 2022, 5, e202101344. [Google Scholar] [CrossRef] [PubMed]
- Talos, F.; Mena, P.; Fingerle-Rowson, G.; Moll, U.; Petrenko, O. MIF Loss Impairs Myc-Induced Lymphomagenesis. Cell Death Differ. 2005, 12, 1319–1328. [Google Scholar] [CrossRef] [Green Version]
- Contreras, J.R.; Palanichamy, J.K.; Tran, T.M.; Fernando, T.R.; Rodriguez-Malave, N.I.; Goswami, N.; Arboleda, V.A.; Casero, D.; Rao, D.S. MicroRNA-146a Modulates B-Cell Oncogenesis by Regulating Egr1. Oncotarget 2015, 6, 11023–11037. [Google Scholar] [CrossRef] [Green Version]
- Mu, P.; Han, Y.C.; Betel, D.; Yao, E.; Squatrito, M.; Ogrodowski, P.; de Stanchina, E.; D’Andrea, A.; Sander, C.; Ventura, A. Genetic Dissection of the MiR-17-92 Cluster of MicroRNAs in Myc-Induced B-Cell Lymphomas. Genes Dev. 2009, 23, 2806–2811. [Google Scholar] [CrossRef] [Green Version]
- Olive, V.; Sabio, E.; Bennett, M.J.; de Jong, C.S.; Biton, A.; McGann, J.C.; Greaney, S.K.; Sodir, N.M.; Zhou, A.Y.; Balakrishnan, A.; et al. A Component of the Mir-17-92 Polycistronic Oncomir Promotes Oncogene-Dependent Apoptosis. Elife 2013, 2, e00822. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.J.; Vandenberg, C.J.; Anstee, N.S.; Hurlin, P.J.; Cory, S. Mnt Modulates Myc-Driven Lymphomagenesis. Cell Death Differ. 2017, 24, 2117–2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.V.; Vandenberg, C.J.; Ng, A.P.; Robati, M.R.; Anstee, N.S.; Rimes, J.; Hawkins, E.D.; Cory, S. Development and Survival of MYC-Driven Lymphomas Require the MYC Antagonist MNT to Curb MYC-Induced Apoptosis. Blood 2020, 135, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Au, A.E.; Lebois, M.; Sim, S.A.; Cannon, P.; Corbin, J.; Gangatirkar, P.; Hyland, C.D.; Moujalled, D.; Rutgersson, A.; Yassinson, F.; et al. Altered B-Lymphopoiesis in Mice with Deregulated Thrombopoietin Signaling. Sci. Rep. 2017, 7, 14953. [Google Scholar] [CrossRef] [Green Version]
- Kadariya, Y.; Tang, B.; Wang, L.; Al-Saleem, T.; Hayakawa, K.; Slifker, M.J.; Kruger, W.D. Germline Mutations in Mtap Cooperate with Myc to Accelerate Tumorigenesis in Mice. PLoS ONE 2013, 8, e67635. [Google Scholar] [CrossRef]
- Odvody, J.; Vincent, T.; Arrate, M.P.; Grieb, B.; Wang, S.; Garriga, J.; Lozano, G.; Iwakuma, T.; Haines, D.S.; Eischen, C.M. A Deficiency in Mdm2 Binding Protein Inhibits Myc-Induced B-Cell Proliferation and Lymphomagenesis. Oncogene 2010, 29, 3287–3296. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.H.; Wang, H.C.; Fiore, A.; Förster, M.; Tung, L.T.; Belle, J.I.; Robert, F.; Pelletier, J.; Langlais, D.; Nijnik, A. Loss of MYSM1 Inhibits the Oncogenic Activity of CMYC in B Cell Lymphoma. J. Cell Mol. Med. 2021, 25, 7089–7094. [Google Scholar] [CrossRef]
- Keller, U.; Nilsson, J.A.; Maclean, K.H.; Old, J.B.; Cleveland, J.L. Nfkb1 Is Dispensable for Myc-Induced Lymphomagenesis. Oncogene 2005, 24, 6231–6240. [Google Scholar] [CrossRef] [Green Version]
- Keller, U.; Huber, J.; Nilsson, J.A.; Fallahi, M.; Hall, M.A.; Peschel, C.; Cleveland, J.L. Myc Suppression of Nfkb2 Accelerates Lymphomagenesis. BMC Cancer 2010, 10, 348. [Google Scholar] [CrossRef] [Green Version]
- Zwolinska, A.K.; Heagle Whiting, A.; Beekman, C.; Sedivy, J.M.; Marine, J.C. Suppression of Myc Oncogenic Activity by Nucleostemin Haploinsufficiency. Oncogene 2012, 31, 3311–3321. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, J.A.; Keller, U.B.; Baudino, T.A.; Yang, C.; Norton, S.; Old, J.A.; Nilsson, L.M.; Neale, G.; Kramer, D.L.; Porter, C.W.; et al. Targeting Ornithine Decarboxylase in Myc-Induced Lymphomagenesis Prevents Tumor Formation. Cancer Cell 2005, 7, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Green, B.; Martin, A.; Belcheva, A. Deficiency in the DNA Glycosylases UNG1 and OGG1 Does Not Potentiate C-Myc-Induced B-Cell Lymphomagenesis. Exp. Hematol. 2018, 61, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.P.; Berns, A. Loss of P27Kip1 but Not P21Cip1 Decreases Survival and Synergizes with MYC in Murine Lymphomagenesis. EMBO J. 2002, 21, 3739–3748. [Google Scholar] [CrossRef] [PubMed]
- Shreeram, S.; Weng, K.H.; Demidov, O.N.; Kek, C.; Yamaguchi, H.; Fornace, A.J.; Anderson, C.W.; Appella, E.; Bulavin, D.V. Regulation of ATM/P53-Dependent Suppression of Myc-Induced Lymphomas by Wip1 Phosphatase. J. Exp. Med. 2006, 203, 2793–2799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemajerova, A.; Petrenko, O.; Trümper, L.; Palacios, G.; Moll, U.M. Loss of P73 Promotes Dissemination of Myc-Induced B Cell Lymphomas in Mice. J. Clin. Investig. 2010, 120, 2070–2080. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Campos, M.A.; Lutfi, N.; Bonnin, S.; Martínez, C.; Velasco-Hernandez, T.; García-Hernández, V.; Martín-Caballero, J.; Ampurdanés, C.; Gimeno, R.; Colomo, L.; et al. Distinct Roles for PARP-1 and PARP-2 in c-Myc–Driven B-Cell Lymphoma in Mice. Blood 2022, 139, 228–239. [Google Scholar] [CrossRef]
- Cho, S.H.; Ahn, A.K.; Bhargava, P.; Lee, C.H.; Eischen, C.M.; McGuinness, O.; Boothby, M. Glycolytic Rate and Lymphomagenesis Depend on PARP14, an ADP Ribosyltransferase of the B Aggressive Lymphoma (BAL) Family. Proc. Natl. Acad. Sci. USA 2011, 108, 15972–15977. [Google Scholar] [CrossRef] [Green Version]
- Bolitho, P.; Street, S.E.A.; Westwood, J.A.; Edelmann, W.; MacGregor, D.; Waring, P.; Murray, W.K.; Godfrey, D.I.; Trapani, J.A.; Johnstone, R.W.; et al. Perforin-Mediated Suppression of B-Cell Lymphoma. Proc. Natl. Acad. Sci. USA 2009, 106, 2723–2728. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.; Hong, H.; Kawakami, Y.; Kato, Y.; Wu, D.; Yasudo, H.; Kimura, A.; Kubagawa, H.; Bertoli, L.F.; Davis, R.S.; et al. Tumor Suppression by Phospholipase C-Β3 via SHP-1-Mediated Dephosphorylation of Stat5. Cancer Cell 2009, 16, 161–171. [Google Scholar] [CrossRef] [Green Version]
- Wen, R.; Chen, Y.; Bai, L.; Fu, G.; Schuman, J.; Dai, X.; Zeng, H.; Yang, C.; Stephan, R.P.; Cleveland, J.L.; et al. Essential Role of Phospholipase Cγ2 in Early B-Cell Development and Myc-Mediated Lymphomagenesis. Mol. Cell Biol. 2006, 26, 9364–9376. [Google Scholar] [CrossRef] [Green Version]
- Fog, C.K.; Asmar, F.; Côme, C.; Jensen, K.T.; Johansen, J.V.; Kheir, T.B.; Jacobsen, L.; Friis, C.; Louw, A.; Rosgaard, L.; et al. Loss of PRDM11 Promotes MYC-Driven Lymphomagenesis. Blood 2015, 125, 1272–1281. [Google Scholar] [CrossRef] [PubMed]
- Mzoughi, S.; Fong, J.Y.; Papadopoli, D.; Koh, C.M.; Hulea, L.; Pigini, P.; di Tullio, F.; Andreacchio, G.; Hoppe, M.M.; Wollmann, H.; et al. PRDM15 Is a Key Regulator of Metabolism Critical to Sustain B-Cell Lymphomagenesis. Nat. Commun. 2020, 11, 3520. [Google Scholar] [CrossRef] [PubMed]
- Iotti, G.; Mejetta, S.; Modica, L.; Penkov, D.; Ponzoni, M.; Blasi, F. Reduction of Prep1 Levels Affects Differentiation of Normal and Malignant B Cells and Accelerates Myc Driven Lymphomagenesis. PLoS ONE 2012, 7, e48353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrison, S.P.; Jeffers, J.R.; Yang, C.; Nilsson, J.A.; Hall, M.A.; Rehg, J.E.; Yue, W.; Yu, J.; Zhang, L.; Onciu, M.; et al. Selection against PUMA Gene Expression in Myc-Driven B-Cell Lymphomagenesis. Mol. Cell Biol. 2008, 28, 5391–5402. [Google Scholar] [CrossRef] [Green Version]
- Meacham, C.E.; Ho, E.E.; Dubrovsky, E.; Gertler, F.B.; Hemann, M.T. In Vivo RNAi Screening Identifies Regulators of Actin Dynamics as Key Determinants of Lymphoma Progression. Nat. Genet. 2009, 41, 1133–1137. [Google Scholar] [CrossRef] [Green Version]
- Peintner, L.; Dorstyn, L.; Kumar, S.; Aneichyk, T.; Villunger, A.; Manzl, C. The Tumor-Modulatory Effects of Caspase-2 and Pidd1 Do Not Require the Scaffold Protein Raidd. Cell Death Differ. 2015, 22, 1803–1811. [Google Scholar] [CrossRef] [Green Version]
- Khattar, E.; Maung, K.Z.Y.; Chew, C.L.; Ghosh, A.; Mok, M.M.H.; Lee, P.; Zhang, J.; Chor, W.H.J.; Cildir, G.; Wang, C.Q.; et al. Rap1 Regulates Hematopoietic Stem Cell Survival and Affects Oncogenesis and Response to Chemotherapy. Nat. Commun. 2019, 10, 5349. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Yu, M.; Tan, H.; Li, Y.; Su, W.; Shi, H.; Dhungana, Y.; Guy, C.; Neale, G.; Cloer, C.; et al. Discrete Roles and Bifurcation of PTEN Signaling and MTORC1-Mediated Anabolic Metabolism Underlie IL-7-Driven B Lymphopoiesis. Sci. Adv. 2018, 4, eaar5701. [Google Scholar] [CrossRef] [Green Version]
- Thijssen, R.; Alvarez-Diaz, S.; Grace, C.; Gao, M.Y.; Segal, D.H.; Xu, Z.; Strasser, A.; Huang, D.C.S. Loss of RIPK3 Does Not Impact MYC-Driven Lymphomagenesis or Chemotherapeutic Drug-Induced Killing of Malignant Lymphoma Cells. Cell Death Differ. 2020, 27, 2531–2533. [Google Scholar] [CrossRef]
- Borland, G.; Kilbey, A.; Hay, J.; Gilroy, K.; Terry, A.; Mackay, N.; Bell, M.; McDonald, A.; Mills, K.; Cameron, E.; et al. Addiction to Runx1 Is Partially Attenuated by Loss of P53 in the Eμ-Myc Lymphoma Model. Oncotarget 2016, 7, 22973–22987. [Google Scholar] [CrossRef] [Green Version]
- Hoellein, A.; Fallahi, M.; Schoeffmann, S.; Steidle, S.; Schaub, F.X.; Rudelius, M.; Laitinen, I.; Nilsson, L.; Goga, A.; Peschel, C.; et al. Myc-Induced SUMOylation Is a Therapeutic Vulnerability for B-Cell Lymphoma. Blood 2014, 124, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, E.D.; Oliaro, J.; Ramsbottom, K.M.; Newbold, A.; Humbert, P.O.; Johnstone, R.W.; Russell, S.M. Scribble Acts as an Oncogene in Eμ-Myc-Driven Lymphoma. Oncogene 2016, 35, 1193–1197. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, M.; Kissel, H.; Brown, S.; Gorenc, T.; Schile, A.J.; Rafii, S.; Larisch, S.; Steller, H. Sept4/ARTS Is Required for Stem Cell Apoptosis and Tumor Suppression. Genes Dev. 2010, 24, 2282–2293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, S.M.; Lee, A.; Lee, J.; Haigis, M.C. SIRT4 Protein Suppresses Tumor Formation in Genetic Models of Myc-Induced B Cell Lymphoma. J. Biol. Chem. 2014, 289, 4135–4144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Old, J.B.; Kratzat, S.; Hoellein, A.; Graf, S.; Nilsson, J.A.; Nilsson, L.; Nakayama, K.I.; Peschel, C.; Cleveland, J.L.; Keller, U.B. Skp2 Directs Myc-Mediated Suppression of P27Kip1 yet Has Modest Effects on Myc-Driven Lymphomagenesis. Mol. Cancer Res. 2010, 8, 353–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagislar, S.; Sabò, A.; Kress, T.R.; Doni, M.; Nicoli, P.; Campaner, S.; Amati, B. Smyd2 Is a Myc-Regulated Gene Critical for MLL-AF9 Induced Leukemogenesis. Oncotarget 2016, 7, 66398–66415. [Google Scholar] [CrossRef] [Green Version]
- Cardone, M.; Kandilci, A.; Carella, C.; Nilsson, J.A.; Brennan, J.A.; Sirma, S.; Ozbek, U.; Boyd, K.; Cleveland, J.L.; Grosveld, G.C. The Novel ETS Factor TEL2 Cooperates with Myc in B Lymphomagenesis. Mol. Cell Biol. 2005, 25, 2395–2405. [Google Scholar] [CrossRef] [Green Version]
- Rounbehler, R.J.; Fallahi, M.; Yang, C.; Steeves, M.A.; Li, W.; Doherty, J.R.; Schaub, F.X.; Sanduja, S.; Dixon, D.A.; Blackshear, P.J.; et al. Tristetraprolin Impairs Myc-Induced Lymphoma and Abolishes the Malignant State. Cell 2012, 150, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Finnberg, N.; Klein-Szanto, A.J.P.; El-Deiry, W.S. TRAIL-R Deficiency in Mice Promotes Susceptibility to Chronic Inflammation and Tumorigenesis. J. Clin. Investig. 2008, 118, 111–123. [Google Scholar] [CrossRef]
- Hussain, S.; Bedekovics, T.; Liu, Q.; Hu, W.; Jeon, H.; Johnson, S.H.; Vasmatzis, G.; May, D.G.; Roux, K.J.; Galardy, P.J. UCH-L1 Bypasses MTOR to Promote Protein Biosynthesis and Is Required for MYC-Driven Lymphomagenesis in Mice. Blood 2018, 132, 2564–2574. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, Y.; Zheng, L.; Liu, M.; Chen, C.D.; Jiang, H. UTX Is an Escape from X-Inactivation Tumor-Suppressor in B Cell Lymphoma. Nat. Commun. 2018, 9, 2720. [Google Scholar] [CrossRef] [PubMed]
- Moser, R.; Toyoshima, M.; Robinson, K.; Gurley, K.E.; Howie, H.L.; Davison, J.; Morgan, M.; Kemp, C.J.; Grandori, C. MYC-Driven Tumorigenesis Is Inhibited by WRN Syndrome Gene Deficiency. Mol. Cancer Res. 2012, 10, 535–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.; Sendino, M.; Gorelick, A.N.; Pastore, A.; Chang, M.T.; Penson, A.V.; Gavrila, E.I.; Stewart, C.; Melnik, E.M.; Chavez, F.H.; et al. Altered Nuclear Export Signal Recognition as a Driver of Oncogenesis. Cancer Discov. 2019, 9, 1452–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Best, S.A.; Vandenberg, C.J.; Abad, E.; Whitehead, L.; Guiu, L.; Ding, S.; Brennan, M.S.; Strasser, A.; Herold, M.J.; Sutherland, K.D.; et al. Consequences of Zmat3 Loss in C-MYC- and Mutant KRAS-Driven Tumorigenesis. Cell Death Dis. 2020, 11, 877. [Google Scholar] [CrossRef] [PubMed]
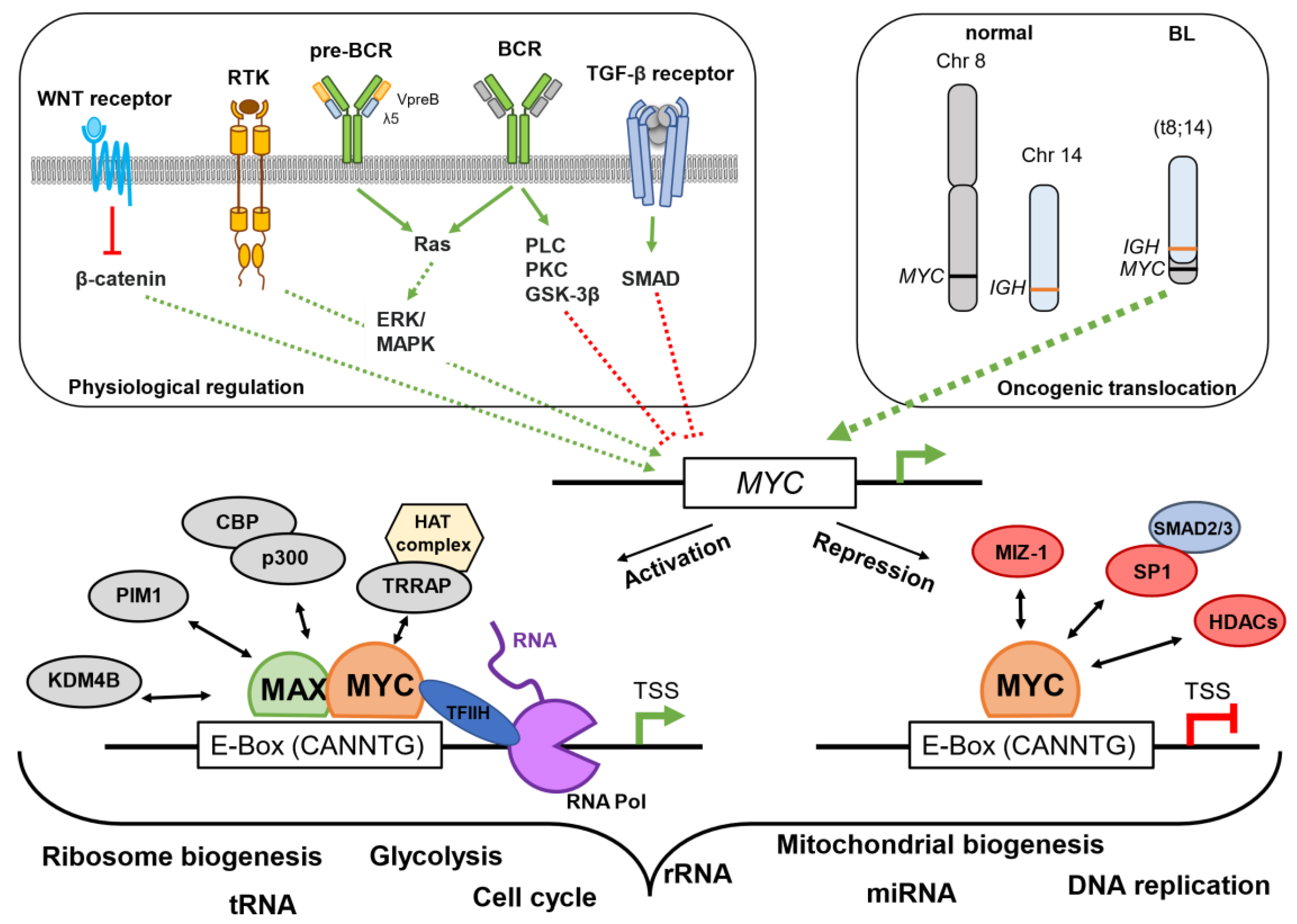


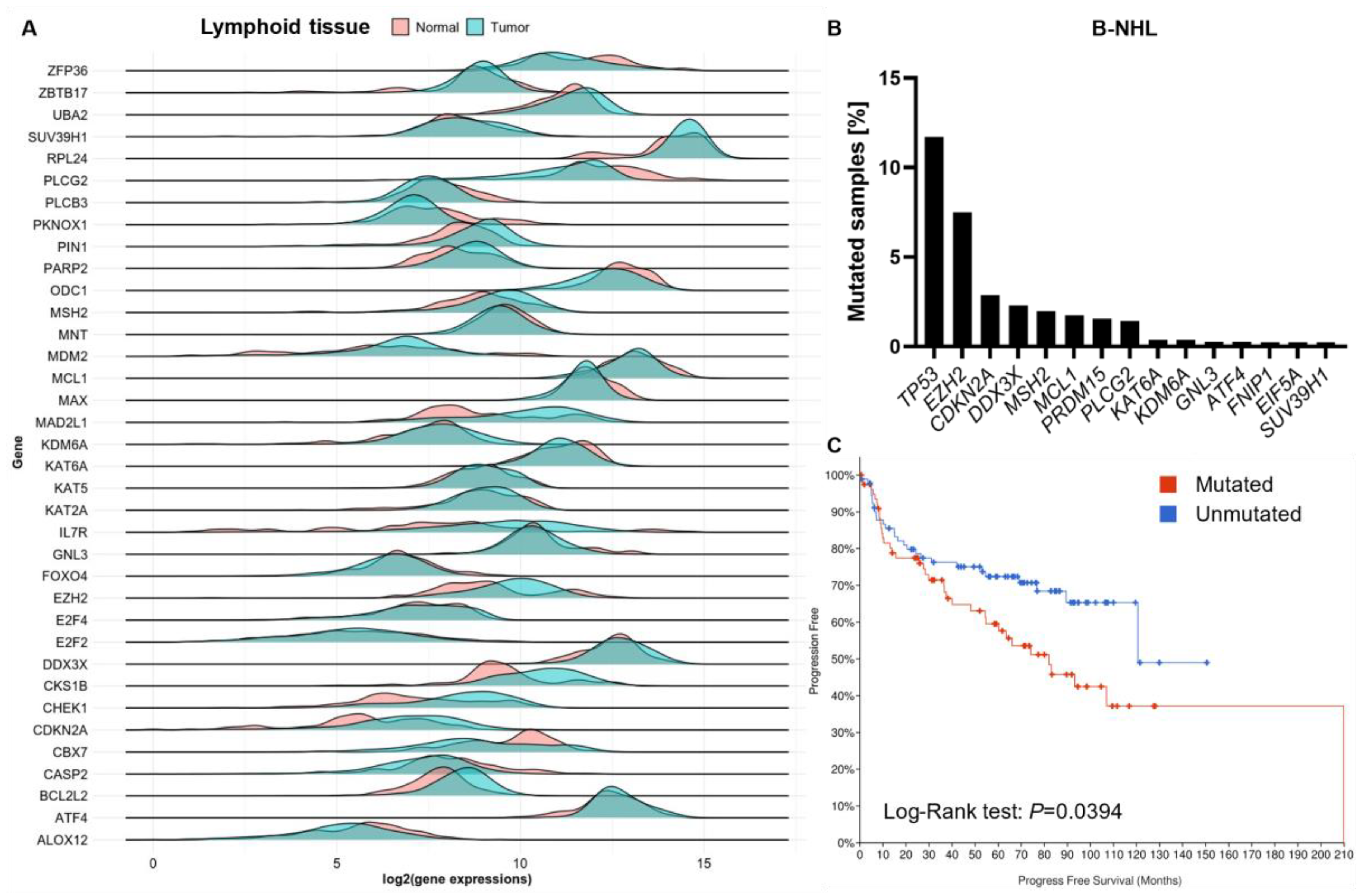
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winkler, R.; Piskor, E.-M.; Kosan, C. Lessons from Using Genetically Engineered Mouse Models of MYC-Induced Lymphoma. Cells 2023, 12, 37. https://doi.org/10.3390/cells12010037
Winkler R, Piskor E-M, Kosan C. Lessons from Using Genetically Engineered Mouse Models of MYC-Induced Lymphoma. Cells. 2023; 12(1):37. https://doi.org/10.3390/cells12010037
Chicago/Turabian StyleWinkler, René, Eva-Maria Piskor, and Christian Kosan. 2023. "Lessons from Using Genetically Engineered Mouse Models of MYC-Induced Lymphoma" Cells 12, no. 1: 37. https://doi.org/10.3390/cells12010037
APA StyleWinkler, R., Piskor, E.-M., & Kosan, C. (2023). Lessons from Using Genetically Engineered Mouse Models of MYC-Induced Lymphoma. Cells, 12(1), 37. https://doi.org/10.3390/cells12010037







