Genome- and Transcriptome-Wide Association Studies Identify Susceptibility Genes and Pathways for Periodontitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Periodontitis GWAS Data
2.2. Cross-Tissue TWAS Analysis Using UTMOST
2.3. Single-Tissue TWAS Analysis Using FUSION
2.4. Expression Analysis of Candidate Genes in the Gingival Tissues of Periodontitis Patients
2.5. Gene Annotation and Enrichment Analyses
3. Results
3.1. Novel Risk Locus Discovered by Meta-Analysis
3.2. Cross-Tissue Transcriptome-Wide Significant Genes
3.3. Single-Tissue Transcriptome-Wide Significant Genes
3.4. Significant Upregulation of EZH1 in Periodontitis
3.5. Functional Annotation of EZH1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shimizu, S.; Momozawa, Y.; Takahashi, A.; Nagasawa, T.; Ashikawa, K.; Terada, Y.; Izumi, Y.; Kobayashi, H.; Tsuji, M.; Kubo, M.; et al. A genome-wide association study of periodontitis in a Japanese population. J. Dent. Res. 2015, 94, 555–561. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S149–S161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. 1), S173–S182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, T.; Lamster, I.B.; Levin, L. Current Concepts in the Management of Periodontitis. Int. Dent. J. 2021, 71, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Gera, I.; Vari, M. Genetic background of periodontitis. Part II. Genetic polymorphism in periodontal disease. A review of literature. Fogorv. Szle. 2009, 102, 131–140. [Google Scholar]
- Michalowicz, B.S.; Diehl, S.R.; Gunsolley, J.C.; Sparks, B.S.; Brooks, C.N.; Koertge, T.E.; Califano, J.V.; Burmeister, J.A.; Schenkein, H.A. Evidence of a substantial genetic basis for risk of adult periodontitis. J. Periodontol. 2000, 71, 1699–1707. [Google Scholar] [CrossRef]
- Nibali, L.; Bayliss-Chapman, J.; Almofareh, S.A.; Zhou, Y.; Divaris, K.; Vieira, A.R. What Is the Heritability of Periodontitis? A Systematic Review. J. Dent. Res. 2019, 98, 632–641. [Google Scholar] [CrossRef]
- Loos, B.G.; Van Dyke, T.E. The role of inflammation and genetics in periodontal disease. Periodontol. 2000 2020, 83, 26–39. [Google Scholar] [CrossRef]
- Brodzikowska, A.; Gorski, B. Polymorphisms in Genes Involved in Inflammation and Periodontitis: A Narrative Review. Biomolecules 2022, 12, 552. [Google Scholar] [CrossRef]
- Ryder, M.I.; Couch, E.T.; Chaffee, B.W. Personalized periodontal treatment for the tobacco- and alcohol-using patient. Periodontol. 2000 2018, 78, 30–46. [Google Scholar] [CrossRef]
- Bartold, P.M. Lifestyle and periodontitis: The emergence of personalized periodontics. Periodontol. 2000 2018, 78, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef] [PubMed]
- Macarthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; Mcmahon, A.; Milano, A.; Morales, J.; et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.S.; Richter, G.M.; Nothnagel, M.; Manke, T.; Dommisch, H.; Jacobs, G.; Arlt, A.; Rosenstiel, P.; Noack, B.; Groessner-Schreiber, B.; et al. A genome-wide association study identifies GLT6D1 as a susceptibility locus for periodontitis. Hum. Mol. Genet. 2010, 19, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, L.; Navarra, C.O.; Pirastu, N.; Lenarda, R.D.; Gasparini, P.; Robino, A. A genome-wide association study identifies an association between variants in EFCAB4B gene and periodontal disease in an Italian isolated population. J. Periodontal Res. 2018, 53, 992–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munz, M.; Richter, G.M.; Loos, B.G.; Jepsen, S.; Divaris, K.; Offenbacher, S.; Teumer, A.; Holtfreter, B.; Kocher, T.; Bruckmann, C.; et al. Meta-analysis of genome-wide association studies of aggressive and chronic periodontitis identifies two novel risk loci. Eur. J. Hum. Genet. 2019, 27, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Munz, M.; Willenborg, C.; Richter, G.M.; Jockel-Schneider, Y.; Graetz, C.; Staufenbiel, I.; Wellmann, J.; Berger, K.; Krone, B.; Hoffmann, P.; et al. A genome-wide association study identifies nucleotide variants at SIGLEC5 and DEFA1A3 as risk loci for periodontitis. Hum. Mol. Genet. 2017, 26, 2577–2588. [Google Scholar] [CrossRef] [Green Version]
- Tam, V.; Patel, N.; Turcotte, M.; Bosse, Y.; Pare, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Gallagher, M.D.; Chen-Plotkin, A.S. The Post-GWAS Era: From Association to Function. Am. J. Hum. Genet. 2018, 102, 717–730. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Li, M.; Lu, Q.; Weng, H.; Wang, J.; Zekavat, S.M.; Yu, Z.; Li, B.; Gu, J.; Muchnik, S.; et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 2019, 51, 568–576. [Google Scholar] [CrossRef]
- Carithers, L.J.; Moore, H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank. 2015, 13, 307–308. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Barbeira, A.N.; Dickinson, S.P.; Bonazzola, R.; Zheng, J.; Wheeler, H.E.; Torres, J.M.; Torstenson, E.S.; Shah, K.P.; Garcia, T.; Edwards, T.L.; et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018, 9, 1825. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Wheeler, H.E.; Shah, K.P.; Mozaffari, S.V.; Aquino-Michaels, K.; Carroll, R.J.; Eyler, A.E.; Denny, J.C.; Nicolae, D.L.; Cox, N.J.; et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015, 47, 1091–1098. [Google Scholar] [CrossRef] [Green Version]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.; Jansen, R.; de Geus, E.J.; Boomsma, D.I.; Wright, F.A.; et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef] [Green Version]
- Battle, A.; Brown, C.D.; Engelhardt, B.E.; Montgomery, S.B. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Fontenla, C.; Carracedo, A. UTMOST, a single and cross-tissue TWAS (Transcriptome Wide Association Study), reveals new ASD (Autism Spectrum Disorder) associated genes. Transl. Psychiatry 2021, 11, 256. [Google Scholar] [CrossRef]
- Zhu, M.; Fan, J.; Zhang, C.; Xu, J.; Yin, R.; Zhang, E.; Wang, Y.; Ji, M.; Sun, Q.; Dai, J.; et al. A cross-tissue transcriptome-wide association study identifies novel susceptibility genes for lung cancer in Chinese populations. Hum. Mol. Genet. 2021, 30, 1666–1676. [Google Scholar] [CrossRef]
- Shungin, D.; Haworth, S.; Divaris, K.; Agler, C.S.; Kamatani, Y.; Keun, L.M.; Grinde, K.; Hindy, G.; Alaraudanjoki, V.; Pesonen, P.; et al. Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat. Commun. 2019, 10, 2773. [Google Scholar] [CrossRef] [Green Version]
- Kurki, M.I.; Karjalainen, J.; Palta, P.; Donner, K. FinnGen: Unique genetic insights from combining isolated population and national health register data. medRxiv 2022. [Google Scholar] [CrossRef]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demmer, R.T.; Behle, J.H.; Wolf, D.L.; Handfield, M.; Kebschull, M.; Celenti, R.; Pavlidis, P.; Papapanou, P.N. Transcriptomes in healthy and diseased gingival tissues. J. Periodontol. 2008, 79, 2112–2124. [Google Scholar] [CrossRef] [PubMed]
- Kebschull, M.; Demmer, R.T.; Grun, B.; Guarnieri, P.; Pavlidis, P.; Papapanou, P.N. Gingival tissue transcriptomes identify distinct periodontitis phenotypes. J. Dent. Res. 2014, 93, 459–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, A.P.; King, B.L.; Mockus, S.; Murphy, C.G.; Saraceni-Richards, C.; Rosenstein, M.; Wiegers, T.; Mattingly, C.J. The Comparative Toxicogenomics Database: Update 2011. Nucleic Acids Res. 2011, 39, D1067–D1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021, 2, 100141. [Google Scholar] [CrossRef]
- Masumoto, R.; Kitagaki, J.; Fujihara, C.; Matsumoto, M.; Miyauchi, S.; Asano, Y.; Imai, A.; Kobayashi, K.; Nakaya, A.; Yamashita, M.; et al. Identification of genetic risk factors of aggressive periodontitis using genomewide association studies in association with those of chronic periodontitis. J. Periodontal Res. 2019, 54, 199–206. [Google Scholar] [CrossRef]
- Wang, X.; Ma, F.; Jia, P. LncRNA AWPPH overexpression predicts the recurrence of periodontitis. Biosci. Rep. 2019, 39, BSR20190636. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Chi, C. lncRNA FGD5-AS1 and miR-130a Can Be Used for Prognosis Analysis of Patients with Chronic Periodontitis. Biomed Res. Int. 2021, 2021, 8544914. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, L.; Yan, X.; Shan, Y.; Liu, L.; Zhou, J.; Kuang, Q.; Li, M.; Long, H.; Lai, W. Identification of immune-related lncRNAs in periodontitis reveals regulation network of gene-lncRNA-pathway-immunocyte. Int. Immunopharmacol. 2020, 84, 106600. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, R.B.; Coletta, R.D.; Silverio, K.G.; Benevides, L.; Casati, M.Z.; Da, S.J.; Nociti, F.J. Impact of smoking on inflammation: Overview of molecular mechanisms. Inflamm. Res. 2011, 60, 409–424. [Google Scholar] [CrossRef]
- Walther, K.A.; Gonzales, J.R.; Groger, S.; Ehmke, B.; Kaner, D.; Lorenz, K.; Eickholz, P.; Kocher, T.; Kim, T.S.; Schlagenhauf, U.; et al. The Role of Polymorphisms at the Interleukin-1, Interleukin-4, GATA-3 and Cyclooxygenase-2 Genes in Non-Surgical Periodontal Therapy. Int. J. Mol. Sci. 2022, 23, 7266. [Google Scholar] [CrossRef]
- Li, W.; Zheng, Q.; Meng, H.; Chen, D. Integration of genome-wide association study and expression quantitative trait loci data identifies AIM2 as a risk gene of periodontitis. J. Clin. Periodontol. 2020, 47, 583–593. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Q.; Ding, Y.; Li, X.; Zhao, D.; Zhao, K.; Guo, Z.; Cao, X. Histone lysine methyltransferase Ezh1 promotes TLR-triggered inflammatory cytokine production by suppressing Tollip. J. Immunol. 2015, 194, 2838–2846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, W.; Wang, Q.; Chen, Q. The cytokine network involved in the host immune response to periodontitis. Int. J. Oral Sci. 2019, 11, 30. [Google Scholar] [CrossRef] [Green Version]
- Apolinario, V.G.; Aparecida, R.A.; Figueiredo, C.K.; Ferreira, O.L.; Tanaka, S.K.; Reis, M.M.; Sprone, R.M.; Goncalves, D.A.A.; Taba, M.J. Specific inhibition of IL-6 receptor attenuates inflammatory bone loss in experimental periodontitis. J. Periodontol. 2021, 92, 1460–1469. [Google Scholar] [CrossRef]
- Hintermann, E.; Haake, S.K.; Christen, U.; Sharabi, A.; Quaranta, V. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: A strategy for cell adhesion and migration disabling. Infect. Immun. 2002, 70, 5846–5856. [Google Scholar] [CrossRef] [Green Version]
- Yokoe, S.; Hasuike, A.; Watanabe, N.; Tanaka, H.; Karahashi, H.; Wakuda, S.; Takeichi, O.; Kawato, T.; Takai, H.; Ogata, Y.; et al. Epstein-Barr Virus Promotes the Production of Inflammatory Cytokines in Gingival Fibroblasts and RANKL-Induced Osteoclast Differentiation in RAW264.7 Cells. Int. J. Mol. Sci. 2022, 23, 809. [Google Scholar] [CrossRef]
- Imai, K.; Ogata, Y. How Does Epstein-Barr Virus Contribute to Chronic Periodontitis? Int. J. Mol. Sci. 2020, 21, 1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mcphee, C.K.; Baehrecke, E.H. Autophagy shows its animal side. Cell 2010, 141, 922–924. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Y.; Li, W. Autophagy and its significance in periodontal disease. J. Periodontal Res. 2021, 56, 18–26. [Google Scholar] [CrossRef]
- Jiang, M.; Li, Z.; Zhu, G. The role of autophagy in the pathogenesis of periodontal disease. Oral Dis. 2020, 26, 259–269. [Google Scholar] [CrossRef]
- Jiang, M.; Li, Z.; Zhu, G. The role of endoplasmic reticulum stress in the pathophysiology of periodontal disease. J. Periodontal Res. 2022, 57, 915–932. [Google Scholar] [CrossRef]
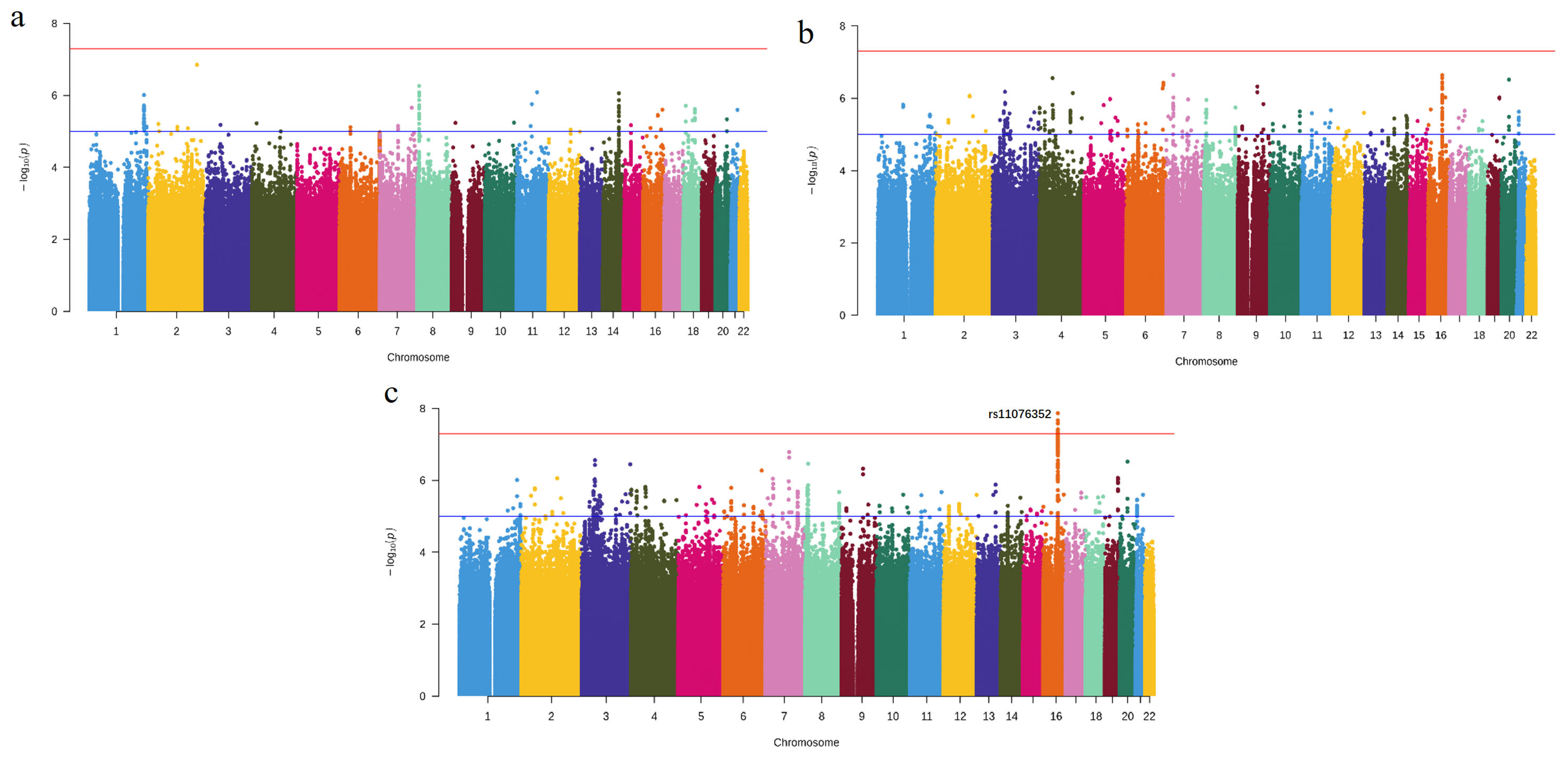
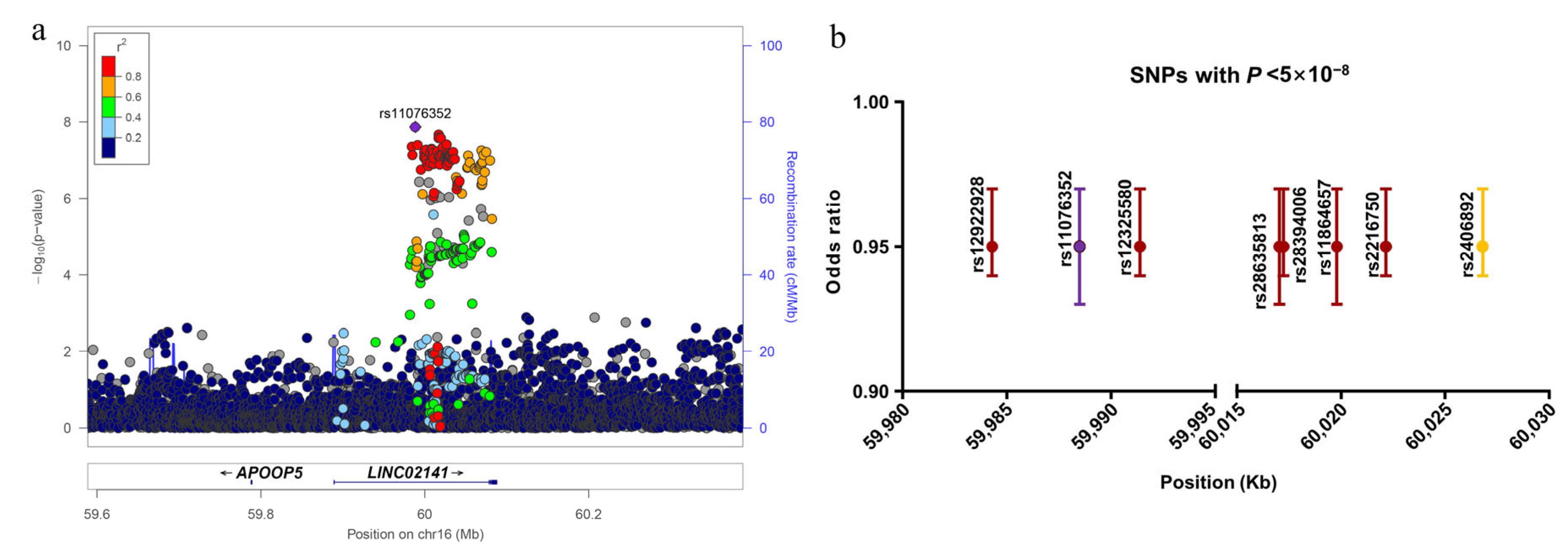
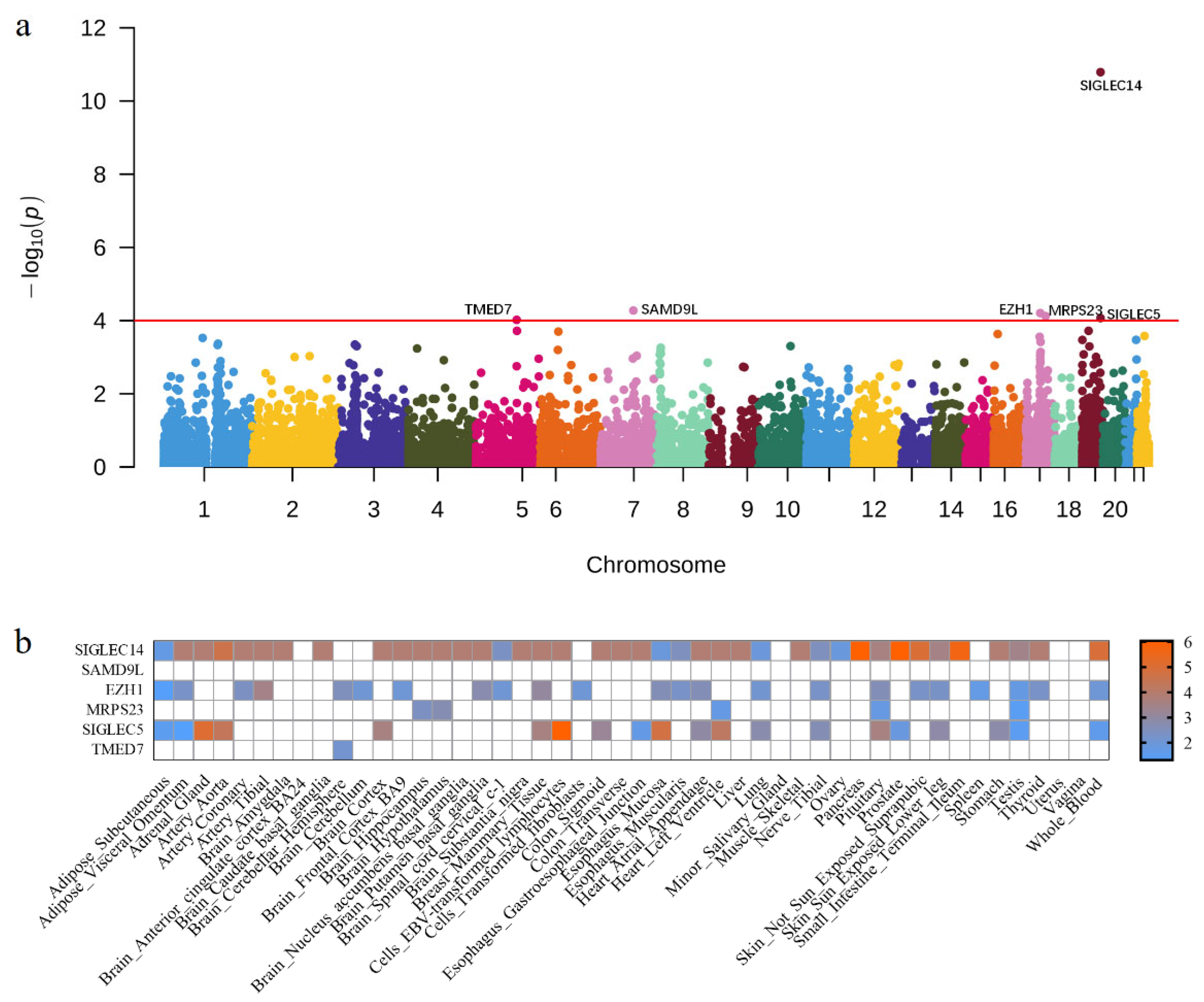
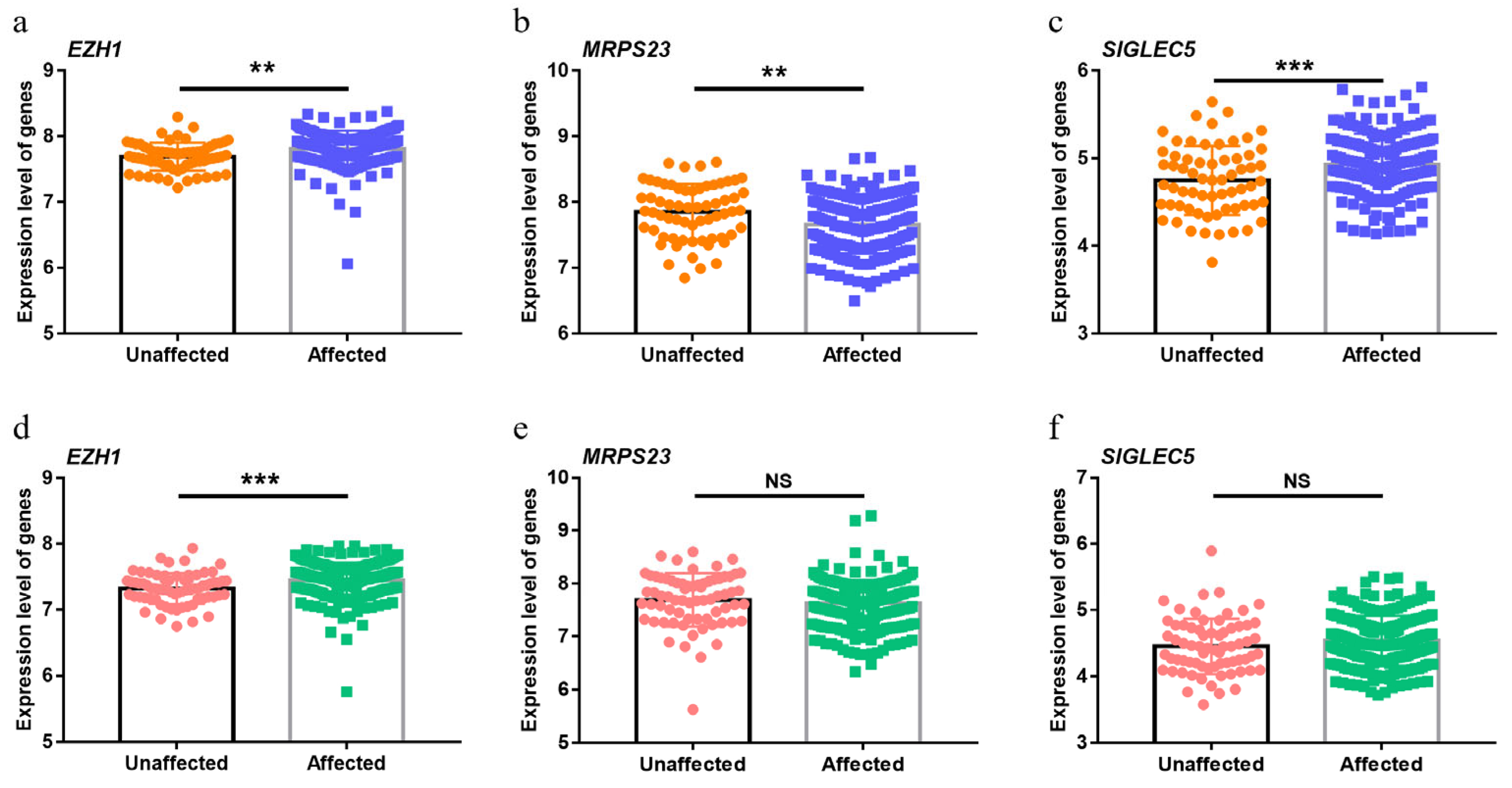

| Gene | Chr | BP | PUTMOST | PFUSION | n | Reported |
|---|---|---|---|---|---|---|
| SIGLEC14 | 19 | 52145806 | 1.63 × 10−11 | 9.34 × 10−7 | 40 | Yes |
| SAMD9L | 7 | 92759368 | 5.32 × 10−5 | NA | 0 | No |
| EZH1 | 17 | 40852293 | 6.24 × 10−5 | 3.83 × 10−3 | 23 | No |
| MRPS23 | 17 | 55916287 | 7.55 × 10−5 | 2.14 × 10−3 | 5 | No |
| SIGLEC5 | 19 | 52114756 | 8.54 × 10−5 | 9.34 × 10−7 | 20 | Yes |
| TMED7 | 5 | 114948905 | 9.56 × 10−5 | 7.53 × 10−3 | 1 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, G.; Cui, X.; Fan, L.; Pan, Y.; Wang, L. Genome- and Transcriptome-Wide Association Studies Identify Susceptibility Genes and Pathways for Periodontitis. Cells 2023, 12, 70. https://doi.org/10.3390/cells12010070
Zhu G, Cui X, Fan L, Pan Y, Wang L. Genome- and Transcriptome-Wide Association Studies Identify Susceptibility Genes and Pathways for Periodontitis. Cells. 2023; 12(1):70. https://doi.org/10.3390/cells12010070
Chicago/Turabian StyleZhu, Guirong, Xing Cui, Liwen Fan, Yongchu Pan, and Lin Wang. 2023. "Genome- and Transcriptome-Wide Association Studies Identify Susceptibility Genes and Pathways for Periodontitis" Cells 12, no. 1: 70. https://doi.org/10.3390/cells12010070
APA StyleZhu, G., Cui, X., Fan, L., Pan, Y., & Wang, L. (2023). Genome- and Transcriptome-Wide Association Studies Identify Susceptibility Genes and Pathways for Periodontitis. Cells, 12(1), 70. https://doi.org/10.3390/cells12010070







