Human Placental Endothelial Cell and Trophoblast Heterogeneity and Differentiation Revealed by Single-Cell RNA Sequencing
Abstract
1. Background
2. Materials and Methods
2.1. Human Placenta Sample Collection
2.2. Single Cell Dissociation
2.3. scRNA-seq Data Processing, Quality Control, and Analysis
2.4. Cell Type Identification
2.5. Cluster Visualization
2.6. Analysis of Sub-Clusters within Endothelial Cells and Trophoblasts
2.7. Pseudotime Trajectory Analysis
2.8. RNA Velocity Analysis
2.9. Cell-Cycle Analysis
2.10. Metabolic Gene Expression Analysis
2.11. Gene Set Variation Analysis
2.12. Identification of TFs Using SCENIC
2.13. Cell-Cell Communication Analysis
2.14. Differentially Expressed Genes between Development Stages
2.15. Immunofluorescence
2.16. Cell Culture
2.17. Western Blot Analysis
2.18. RNA Isolation and Quantitative RT-PCR
2.19. Statistical Analysis
3. Results
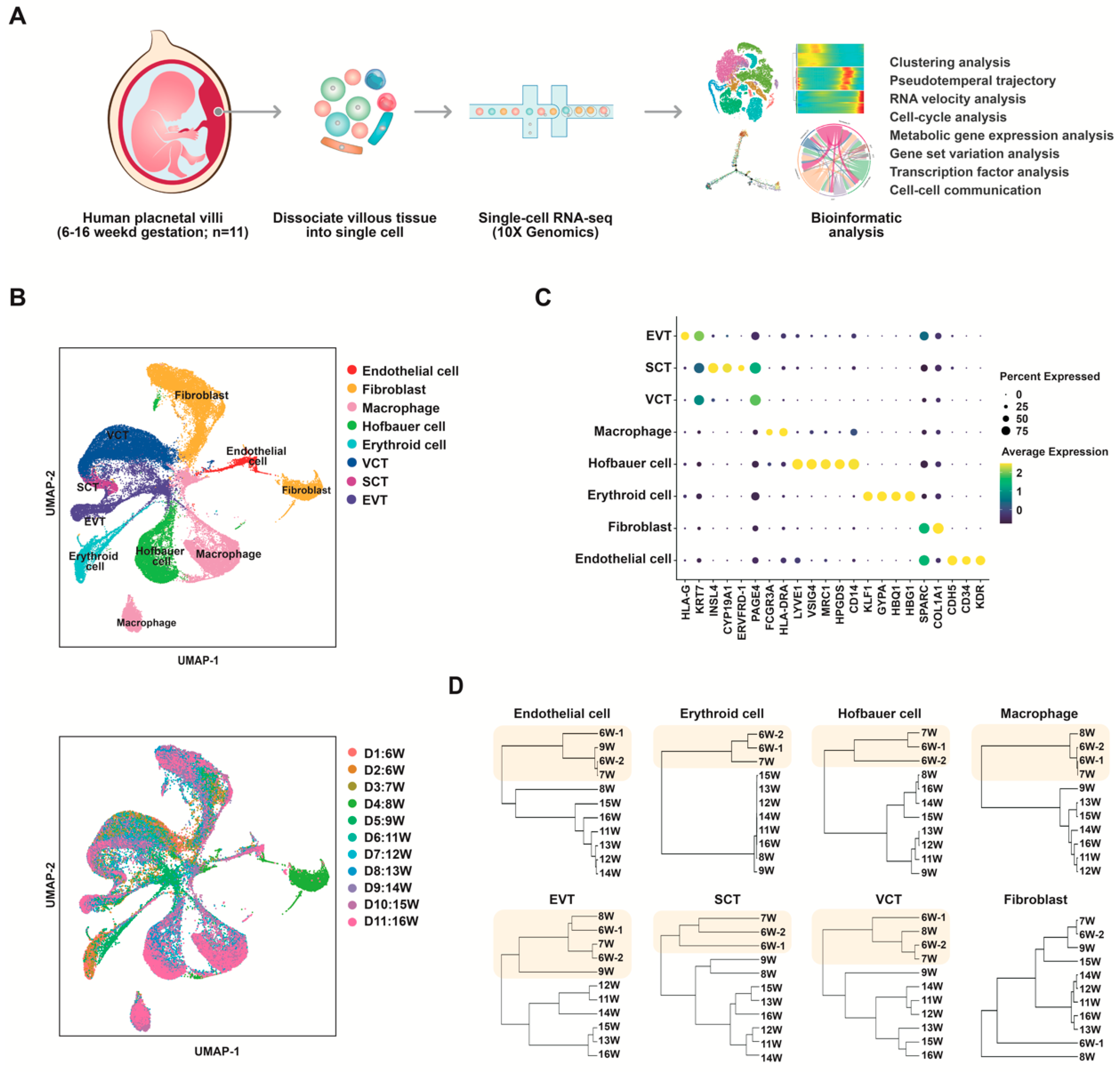
3.1. Single-Cell Transcriptome Profiling of Normal Human Placentas in the First and Second Trimesters
3.2. Identification and Characterization of Distinct Endothelial Clusters in Human Placenta
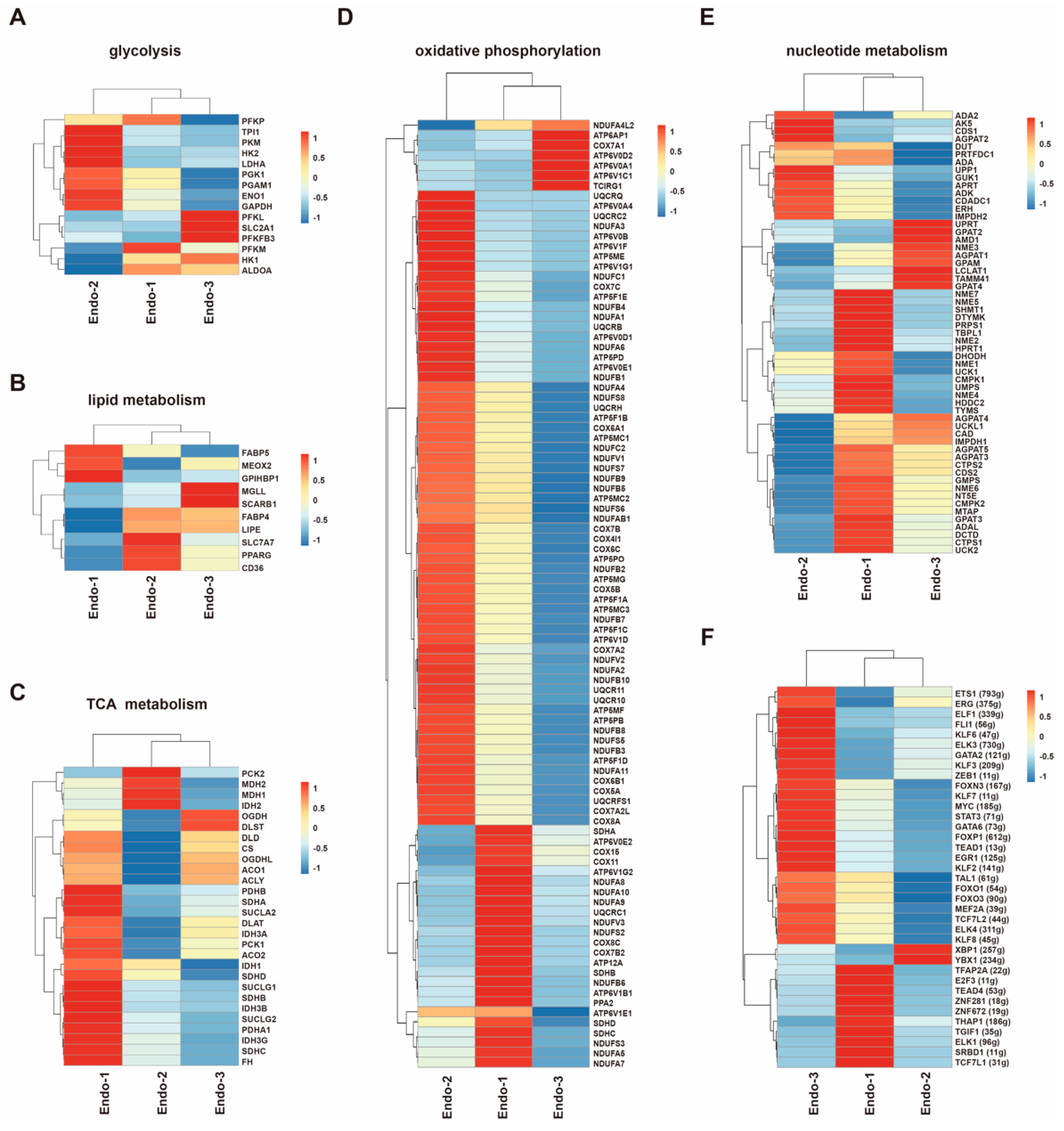
3.3. Metabolic Transcriptome Signatures in Different Placental Endothelial Clusters
3.4. Transcription Factor Regulatory Network in Different Placental Endothelial Clusters
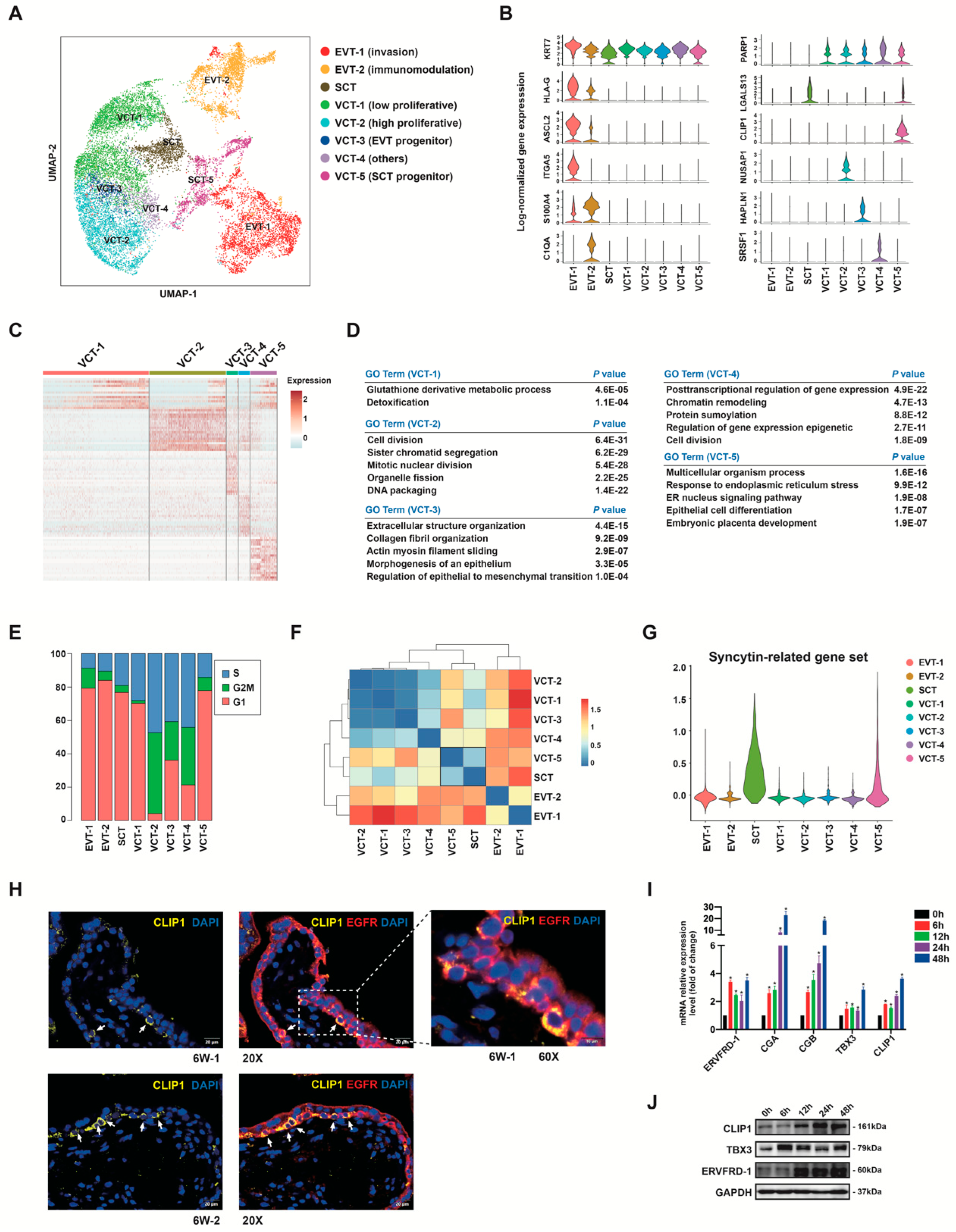
3.5. Subclusters of Placental Trophoblast Cells
3.6. Human Trophoblast Progenitors Contribute to EVT Differentiation
3.7. VCT to EVT Differentiation Analysis
3.8. Other Cell Types
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knofler, M.; Haider, S.; Saleh, L.; Pollheimer, J.; Gamage, T.; James, J. Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell. Mol. Life Sci. 2019, 76, 3479–3496. [Google Scholar] [CrossRef] [PubMed]
- Napso, T.; Yong, H.E.J.; Lopez-Tello, J.; Sferruzzi-Perri, A.N. The Role of Placental Hormones in Mediating Maternal Adaptations to Support Pregnancy and Lactation. Front. Physiol. 2018, 9, 1091. [Google Scholar] [CrossRef] [PubMed]
- Erlebacher, A. Immunology of the maternal-fetal interface. Annu. Rev. Immunol. 2013, 31, 387–411. [Google Scholar] [CrossRef] [PubMed]
- Maltepe, E.; Fisher, S.J. Placenta: The forgotten organ. Annu. Rev. Cell Dev. Biol. 2015, 31, 523–552. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.J.; Redman, C.W.; Roberts, J.M.; Moffett, A. Pre-eclampsia: Pathophysiology and clinical implications. BMJ 2019, 366, l2381. [Google Scholar] [CrossRef]
- Haram, K.; Mortensen, J.H.; Myking, O.; Roald, B.; Magann, E.F.; Morrison, J.C. Early development of the human placenta and pregnancy complications. J. Matern. Fetal Neonatal. Med. 2020, 33, 3538–3545. [Google Scholar] [CrossRef]
- Brosens, I.; Pijnenborg, R.; Vercruysse, L.; Romero, R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 2011, 204, 193–201. [Google Scholar] [CrossRef]
- Perez-Garcia, V.; Fineberg, E.; Wilson, R.; Murray, A.; Mazzeo, C.I.; Tudor, C.; Sienerth, A.; White, J.K.; Tuck, E.; Ryder, E.J.; et al. Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 2018, 555, 463–468. [Google Scholar] [CrossRef]
- Aplin, J.D.; Myers, J.E.; Timms, K.; Westwood, M. Tracking placental development in health and disease. Nat. Rev. Endocrinol. 2020, 16, 479–494. [Google Scholar] [CrossRef]
- Rodesch, F.; Simon, P.; Donner, C.; Jauniaux, E. Oxygen measurements in endometrial and trophoblastic tissues during early pregnancy. Obstet. Gynecol. 1992, 80, 283–285. [Google Scholar]
- Caniggia, I.; Winter, J.; Lye, S.J.; Post, M. Oxygen and placental development during the first trimester: Implications for the pathophysiology of pre-eclampsia. Placenta 2000, 21, S25–S30. [Google Scholar] [CrossRef] [PubMed]
- Wulff, C.; Weigand, M.; Kreienberg, R.; Fraser, H.M. Angiogenesis during primate placentation in health and disease. Reproduction 2003, 126, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, L.P.; Caton, J.S.; Redmer, D.A.; Grazul-Bilska, A.T.; Vonnahme, K.A.; Borowicz, P.P.; Luther, J.S.; Wallace, J.M.; Wu, G.; Spencer, T.E. Evidence for altered placental blood flow and vascularity in compromised pregnancies. J. Physiol. 2006, 572, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Mayhew, T.M.; Charnock-Jones, D.S.; Kaufmann, P. Aspects of human fetoplacental vasculogenesis and angiogenesis. III. Changes in complicated pregnancies. Placenta 2004, 25, 127–139. [Google Scholar] [CrossRef]
- Redman, C.W.; Sargent, I.L. Latest advances in understanding preeclampsia. Science 2005, 308, 1592–1594. [Google Scholar] [CrossRef]
- Okae, H.; Toh, H.; Sato, T.; Hiura, H.; Takahashi, S.; Shirane, K.; Kabayama, Y.; Suyama, M.; Sasaki, H.; Arima, T. Derivation of Human Trophoblast Stem Cells. Cell Stem Cell 2018, 22, 50–63. [Google Scholar] [CrossRef]
- Haider, S.; Meinhardt, G.; Saleh, L.; Kunihs, V.; Gamperl, M.; Kaindl, U.; Ellinger, A.; Burkard, T.R.; Fiala, C.; Pollheimer, J.; et al. Self-Renewing Trophoblast Organoids Recapitulate the Developmental Program of the Early Human Placenta. Stem Cell Rep. 2018, 11, 537–551. [Google Scholar] [CrossRef]
- Turco, M.Y.; Gardner, L.; Kay, R.G.; Hamilton, R.S.; Prater, M.; Hollinshead, M.S.; McWhinnie, A.; Esposito, L.; Fernando, R.; Skelton, H.; et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 2018, 564, 263–267. [Google Scholar] [CrossRef]
- Io, S.; Kabata, M.; Iemura, Y.; Semi, K.; Morone, N.; Minagawa, A.; Wang, B.; Okamoto, I.; Nakamura, T.; Kojima, Y.; et al. Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell 2021, 28, 1023–1039.e13. [Google Scholar] [CrossRef]
- Dong, C.; Beltcheva, M.; Gontarz, P.; Zhang, B.; Popli, P.; Fischer, L.A.; Khan, S.A.; Park, K.M.; Yoon, E.J.; Xing, X.; et al. Derivation of trophoblast stem cells from naive human pluripotent stem cells. eLife 2020, 9, e52504. [Google Scholar] [CrossRef]
- Liu, X.; Ouyang, J.F.; Rossello, F.J.; Tan, J.P.; Davidson, K.C.; Valdes, D.S.; Schroder, J.; Sun, Y.B.Y.; Chen, J.; Knaupp, A.S.; et al. Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 2020, 586, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Cindrova-Davies, T.; Sferruzzi-Perri, A.N. Human placental development and function. Semin. Cell Dev. Biol. 2022, 131, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.E.; Stephenson, E.; Polanski, K.; Goncalves, A.; et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Suryawanshi, H.; Morozov, P.; Straus, A.; Sahasrabudhe, N.; Max, K.E.A.; Garzia, A.; Kustagi, M.; Tuschl, T.; Williams, Z. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018, 4, eaau4788. [Google Scholar] [CrossRef]
- Tsang, J.C.H.; Vong, J.S.L.; Ji, L.; Poon, L.C.Y.; Jiang, P.; Lui, K.O.; Ni, Y.B.; To, K.F.; Cheng, Y.K.Y.; Chiu, R.W.K.; et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc. Natl. Acad. Sci. USA 2017, 114, E7786–E7795. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, H.; Yang, Y.; Guo, F.; Yu, B.; Su, Z. Trophoblast Cell Subtypes and Dysfunction in the Placenta of Individuals with Preeclampsia Revealed by SingleCell RNA Sequencing. Mol. Cells 2022, 45, 317–328. [Google Scholar] [CrossRef]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Sendler, E.D.; Xu, Y.; Garcia-Flores, V.; Leng, Y.; Luca, F.; Hassan, S.S.; Gomez-Lopez, N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. eLife 2019, 8, e52004. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, F.; Peng, Y.; Chen, R.; Zhou, W.; Wang, H.; OuYang, J.; Yu, B.; Xu, Z. Transcriptomic Profiling of Human Placenta in Gestational Diabetes Mellitus at the Single-Cell Level. Front. Endocrinol. 2021, 12, 679582. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, X.; Wang, R.; Lu, X.; Dang, Y.L.; Wang, H.; Lin, H.Y.; Zhu, C.; Ge, H.; Cross, J.C.; et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018, 28, 819–832. [Google Scholar] [CrossRef]
- Marsh, B.; Zhou, Y.; Kapidzic, M.; Fisher, S.; Blelloch, R. Regionally distinct trophoblast regulate barrier function and invasion in the human placenta. eLife 2022, 11, e78829. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Gao, M.; Wang, C.; Zhang, J.; Noel, P.; Yang, C.; Von Hoff, D.; Han, H.; Zhang, M.Q.; Lin, W. SuperCT: A supervised-learning framework for enhanced characterization of single-cell transcriptomic profiles. Nucleic Acids Res. 2019, 47, e48. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, R.; Zhou, Y.; Fei, L.; Sun, H.; Lai, S.; Saadatpour, A.; Zhou, Z.; Chen, H.; Ye, F.; et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 2018, 172, 1091–1107.e17. [Google Scholar] [CrossRef]
- Lukowski, S.W.; Patel, J.; Andersen, S.B.; Sim, S.L.; Wong, H.Y.; Tay, J.; Winkler, I.; Powell, J.E.; Khosrotehrani, K. Single-Cell Transcriptional Profiling of Aortic Endothelium Identifies a Hierarchy from Endovascular Progenitors to Differentiated Cells. Cell Rep. 2019, 27, 2748–2758. [Google Scholar] [CrossRef]
- Esnault, C.; Priet, S.; Ribet, D.; Vernochet, C.; Bruls, T.; Lavialle, C.; Weissenbach, J.; Heidmann, T. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc. Natl. Acad. Sci. USA 2008, 105, 17532–17537. [Google Scholar] [CrossRef]
- Vargas, A.; Moreau, J.; Landry, S.; LeBellego, F.; Toufaily, C.; Rassart, É.; Lafond, J.; Barbeau, B. Syncytin-2 Plays an Important Role in the Fusion of Human Trophoblast Cells. J. Mol. Biol. 2009, 392, 301–318. [Google Scholar] [CrossRef]
- Potgens, A.J.; Drewlo, S.; Kokozidou, M.; Kaufmann, P. Syncytin: The major regulator of trophoblast fusion? Recent developments and hypotheses on its action. Hum. Reprod. Update 2004, 10, 487–496. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lonnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Rohlenova, K.; Goveia, J.; Garcia-Caballero, M.; Subramanian, A.; Kalucka, J.; Treps, L.; Falkenberg, K.D.; de Rooij, L.; Zheng, Y.; Lin, L.; et al. Single-Cell RNA Sequencing Maps Endothelial Metabolic Plasticity in Pathological Angiogenesis. Cell Metab. 2020, 31, 862–877.e14. [Google Scholar] [CrossRef] [PubMed]
- Kalucka, J.; de Rooij, L.; Goveia, J.; Rohlenova, K.; Dumas, S.J.; Meta, E.; Conchinha, N.V.; Taverna, F.; Teuwen, L.A.; Veys, K.; et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 2020, 180, 764–779.e20. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Aibar, S.; Gonzalez-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Apps, R.; Sharkey, A.; Gardner, L.; Male, V.; Trotter, M.; Miller, N.; North, R.; Founds, S.; Moffett, A. Genome-wide expression profile of first trimester villous and extravillous human trophoblast cells. Placenta 2011, 32, 33–43. [Google Scholar] [CrossRef]
- Lee, C.Q.; Gardner, L.; Turco, M.; Zhao, N.; Murray, M.J.; Coleman, N.; Rossant, J.; Hemberger, M.; Moffett, A. What Is Trophoblast? A Combination of Criteria Define Human First-Trimester Trophoblast. Stem Cell Rep. 2016, 6, 257–272. [Google Scholar] [CrossRef]
- Hoch, D.; Novakovic, B.; Cvitic, S.; Saffery, R.; Desoye, G.; Majali-Martinez, A. Sex matters: XIST and DDX3Y gene expression as a tool to determine fetal sex in human first trimester placenta. Placenta 2020, 97, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Laakkonen, P.; Waltari, M.; Holopainen, T.; Takahashi, T.; Pytowski, B.; Steiner, P.; Hicklin, D.; Persaud, K.; Tonra, J.R.; Witte, L.; et al. Vascular endothelial growth factor receptor 3 is involved in tumor angiogenesis and growth. Cancer Res. 2007, 67, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Rakocevic, J.; Kojic, S.; Orlic, D.; Stankovic, G.; Ostojic, M.; Petrovic, O.; Zaletel, I.; Puskas, N.; Todorovic, V.; Labudovic-Borovic, M. Co-expression of vascular and lymphatic endothelial cell markers on early endothelial cells present in aspirated coronary thrombi from patients with ST-elevation myocardial infarction. Exp. Mol. Pathol. 2016, 100, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, X.; Carmeliet, P. Hallmarks of Endothelial Cell Metabolism in Health and Disease. Cell Metab. 2019, 30, 414–433. [Google Scholar] [CrossRef] [PubMed]
- Vandekeere, S.; Dubois, C.; Kalucka, J.; Sullivan, M.R.; Garcia-Caballero, M.; Goveia, J.; Chen, R.; Diehl, F.F.; Bar-Lev, L.; Souffreau, J.; et al. Serine Synthesis via PHGDH Is Essential for Heme Production in Endothelial Cells. Cell Metab. 2018, 28, 573–587.e13. [Google Scholar] [CrossRef] [PubMed]
- Diebold, L.P.; Gil, H.J.; Gao, P.; Martinez, C.A.; Weinberg, S.E.; Chandel, N.S. Mitochondrial complex III is necessary for endothelial cell proliferation during angiogenesis. Nat. Metab. 2019, 1, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Cayrol, C.; Lacroix, C.; Mathe, C.; Ecochard, V.; Ceribelli, M.; Loreau, E.; Lazar, V.; Dessen, P.; Mantovani, R.; Aguilar, L.; et al. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 2007, 109, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Harel, S.; Sanchez, V.; Moamer, A.; Sanchez-Galan, J.E.; Abid Hussein, M.N.; Mayaki, D.; Blanchette, M.; Hussain, S.N.A. ETS1, ELK1, and ETV4 Transcription Factors Regulate Angiopoietin-1 Signaling and the Angiogenic Response in Endothelial Cells. Front. Physiol. 2021, 12, 683651. [Google Scholar] [CrossRef]
- Ong, Y.T.; Andrade, J.; Armbruster, M.; Shi, C.; Castro, M.; Costa, A.S.H.; Sugino, T.; Eelen, G.; Zimmermann, B.; Wilhelm, K.; et al. A YAP/TAZ-TEAD signalling module links endothelial nutrient acquisition to angiogenic growth. Nat. Metab. 2022, 4, 672–682. [Google Scholar] [CrossRef]
- Atkins, G.B.; Jain, M.K. Role of Kruppel-like transcription factors in endothelial biology. Circ. Res. 2007, 100, 1686–1695. [Google Scholar] [CrossRef]
- Dejana, E.; Taddei, A.; Randi, A.M. Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim. Biophys. Acta 2007, 1775, 298–312. [Google Scholar] [CrossRef] [PubMed]
- De Val, S.; Black, B.L. Transcriptional control of endothelial cell development. Dev. Cell 2009, 16, 180–195. [Google Scholar] [CrossRef] [PubMed]
- De Val, S. Key transcriptional regulators of early vascular development. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 1469–1475. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.W.; Zhou, Y.; Petrovic, J.; Fasolino, M.; Xu, L.; Shaffer, S.M.; Pear, W.S.; Vahedi, G.; Faryabi, R.B. TooManyCells identifies and visualizes relationships of single-cell clades. Nat. Methods 2020, 17, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Huppertz, B.; Gauster, M. Trophoblast fusion. Adv. Exp. Med. Biol. 2011, 713, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Cobos-Correa, A.; Trojanek, J.B.; Diemer, S.; Mall, M.A.; Schultz, C. Membrane-bound FRET probe visualizes MMP12 activity in pulmonary inflammation. Nat. Chem. Biol. 2009, 5, 628–630. [Google Scholar] [CrossRef]
- Knofler, M.; Pollheimer, J. IFPA Award in Placentology lecture: Molecular regulation of human trophoblast invasion. Placenta 2012, 33, S55–S62. [Google Scholar] [CrossRef]
- Haider, S.; Lackner, A.I.; Dietrich, B.; Kunihs, V.; Haslinger, P.; Meinhardt, G.; Maxian, T.; Saleh, L.; Fiala, C.; Pollheimer, J.; et al. Transforming growth factor-beta signaling governs the differentiation program of extravillous trophoblasts in the developing human placenta. Proc. Natl. Acad. Sci. USA 2022, 119, e2120667119. [Google Scholar] [CrossRef]
- Aplin, J.D.; Haigh, T.; Jones, C.J.; Church, H.J.; Vicovac, L. Development of cytotrophoblast columns from explanted first-trimester human placental villi: Role of fibronectin and integrin alpha5beta1. Biol. Reprod. 1999, 60, 828–838. [Google Scholar] [CrossRef]
- Damsky, C.H.; Librach, C.; Lim, K.H.; Fitzgerald, M.L.; McMaster, M.T.; Janatpour, M.; Zhou, Y.; Logan, S.K.; Fisher, S.J. Integrin switching regulates normal trophoblast invasion. Development 1994, 120, 3657–3666. [Google Scholar] [CrossRef]
- Wei, J.; Huang, X.J.; Huang, Y.; Xiong, M.Y.; Yao, X.Y.; Huang, Z.N.; Li, S.N.; Zhou, W.J.; Fang, D.L.; Deng, D.H.; et al. Key immune-related gene ITGB2 as a prognostic signature for acute myeloid leukemia. Ann. Transl. Med. 2021, 9, 1386. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.R.; Appios, A.; Zhao, X.; Dutkiewicz, R.; Donde, M.; Lee, C.Y.C.; Naidu, P.; Lee, C.; Cerveira, J.; Liu, B.; et al. Phenotypic and functional characterization of first-trimester human placental macrophages, Hofbauer cells. J. Exp. Med. 2021, 218, e20200891. [Google Scholar] [CrossRef] [PubMed]
- Murata, H.; Tanaka, S.; Okada, H. Immune Tolerance of the Human Decidua. J. Clin. Med. 2021, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Newbold, R.R.; Jefferson, W.N.; Grissom, S.F.; Padilla-Banks, E.; Snyder, R.J.; Lobenhofer, E.K. Developmental exposure to diethylstilbestrol alters uterine gene expression that may be associated with uterine neoplasia later in life. Mol. Carcinog. 2007, 46, 783–796. [Google Scholar] [CrossRef]
- Salsano, S.; Gonzalez-Martin, R.; Quinonero, A.; Lopez-Martin, S.; Gomez-Escribano, A.P.; Perez-Deben, S.; Yanez-Mo, M.; Dominguez, F. Novel nonclassic progesterone receptor PGRMC1 pulldown-precipitated proteins reveal a key role during human decidualization. Fertil. Steril. 2020, 113, 1050–1066.e7. [Google Scholar] [CrossRef]
- Risinger, J.I.; Allard, J.; Chandran, U.; Day, R.; Chandramouli, G.V.; Miller, C.; Zahn, C.; Oliver, J.; Litzi, T.; Marcus, C.; et al. Gene expression analysis of early stage endometrial cancers reveals unique transcripts associated with grade and histology but not depth of invasion. Front. Oncol. 2013, 3, 139. [Google Scholar] [CrossRef]
- De Bock, K.; Georgiadou, M.; Schoors, S.; Kuchnio, A.; Wong, B.W.; Cantelmo, A.R.; Quaegebeur, A.; Ghesquiere, B.; Cauwenberghs, S.; Eelen, G.; et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 2013, 154, 651–663. [Google Scholar] [CrossRef]
- Meinhardt, G.; Haider, S.; Kunihs, V.; Saleh, L.; Pollheimer, J.; Fiala, C.; Hetey, S.; Feher, Z.; Szilagyi, A.; Than, N.G.; et al. Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc. Natl. Acad. Sci. USA 2020, 117, 13562–13570. [Google Scholar] [CrossRef]
- Hemberger, M.; Udayashankar, R.; Tesar, P.; Moore, H.; Burton, G.J. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum. Mol. Genet. 2010, 19, 2456–2467. [Google Scholar] [CrossRef]
- Saha, B.; Ganguly, A.; Home, P.; Bhattacharya, B.; Ray, S.; Ghosh, A.; Rumi, M.A.K.; Marsh, C.; French, V.A.; Gunewardena, S.; et al. TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: An implication in early human pregnancy loss. Proc. Natl. Acad. Sci. USA 2020, 117, 17864–17875. [Google Scholar] [CrossRef]
- Li, Y.; Moretto-Zita, M.; Leon-Garcia, S.; Parast, M.M. p63 inhibits extravillous trophoblast migration and maintains cells in a cytotrophoblast stem cell-like state. Am. J. Pathol. 2014, 184, 3332–3343. [Google Scholar] [CrossRef] [PubMed]
- Jauniaux, E.; Watson, A.L.; Hempstock, J.; Bao, Y.P.; Skepper, J.N.; Burton, G.J. Onset of maternal arterial blood flow and placental oxidative stress. A possible factor in human early pregnancy failure. Am. J. Pathol. 2000, 157, 2111–2122. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Peng, H.; Hong, W.; Wei, Y.; Tian, H.; Huang, X.; Jia, L.; Zheng, J.; Duan, T.; He, Q.; et al. Human Placental Endothelial Cell and Trophoblast Heterogeneity and Differentiation Revealed by Single-Cell RNA Sequencing. Cells 2023, 12, 87. https://doi.org/10.3390/cells12010087
Li H, Peng H, Hong W, Wei Y, Tian H, Huang X, Jia L, Zheng J, Duan T, He Q, et al. Human Placental Endothelial Cell and Trophoblast Heterogeneity and Differentiation Revealed by Single-Cell RNA Sequencing. Cells. 2023; 12(1):87. https://doi.org/10.3390/cells12010087
Chicago/Turabian StyleLi, Han, Hao Peng, Wei Hong, Yingying Wei, Haojun Tian, Xiaojie Huang, Linyan Jia, Jing Zheng, Tao Duan, Qizhi He, and et al. 2023. "Human Placental Endothelial Cell and Trophoblast Heterogeneity and Differentiation Revealed by Single-Cell RNA Sequencing" Cells 12, no. 1: 87. https://doi.org/10.3390/cells12010087
APA StyleLi, H., Peng, H., Hong, W., Wei, Y., Tian, H., Huang, X., Jia, L., Zheng, J., Duan, T., He, Q., & Wang, K. (2023). Human Placental Endothelial Cell and Trophoblast Heterogeneity and Differentiation Revealed by Single-Cell RNA Sequencing. Cells, 12(1), 87. https://doi.org/10.3390/cells12010087







