Abstract
Architectural proteins are essential epigenetic regulators that play a critical role in organizing chromatin and controlling gene expression. CTCF (CCCTC-binding factor) is a key architectural protein responsible for maintaining the intricate 3D structure of chromatin. Because of its multivalent properties and plasticity to bind various sequences, CTCF is similar to a Swiss knife for genome organization. Despite the importance of this protein, its mechanisms of action are not fully elucidated. It has been hypothesized that its versatility is achieved through interaction with multiple partners, forming a complex network that regulates chromatin folding within the nucleus. In this review, we delve into CTCF’s interactions with other molecules involved in epigenetic processes, particularly histone and DNA demethylases, as well as several long non-coding RNAs (lncRNAs) that are able to recruit CTCF. Our review highlights the importance of CTCF partners to shed light on chromatin regulation and pave the way for future exploration of the mechanisms that enable the finely-tuned role of CTCF as a master regulator of chromatin.
Keywords:
CTCF; epigenetics; chromatin regulation; histone; demethylases; lncRNAs; TET; KDM; BORIS; CTCF-s 1. Introduction
Chromatin, a macromolecular complex of DNA, RNA, and proteins, provides a framework for the packaging of genetic material within the cell nucleus. Its organization plays a crucial role in gene expression and is regulated by a diverse array of protein complexes in response to a dynamic code of histone posttranslational modifications and DNA modifications [1]. CTCF (CCCTC-binding factor) is a crucial architectural protein believed to play a critical role in maintaining chromatin organization through its interactions with various protein complexes [2]. Among other functions, CTCF is a versatile protein known to participate in various processes related to the chromatin structure, including insulation [3], alternative splicing [4,5,6], transcriptional activation [7], and chromatin loop formation [8]. It is not clear how CTCF has such a dynamic range of functions; however, the response to this question may lie in the context-dependent interactions of CTCF with several protein partners.
Epigenetic complexes, which regulate histone post-translational modifications and DNA methylation, usually contain enzymes that chemically modify the amino-terminal ends of histones, forming a code that determines the chromatin state through a system of writing, reading, and erasing complexes [9,10,11]. The mechanisms by which epigenetic components are recruited to specific regions of the genome have not been fully understood, mainly due to the lack of DNA binding domains in most proteins with epigenetic functions [12]. This is why CTCF is a fundamental protein since it could be the bridge between many epigenetic factors and the DNA [13]. The importance of CTCF protein–protein interactions is highlighted by BORIS (Brother of the Regulator of Imprinted Sites), the paralogous protein of CTCF. The similarity of DNA-binding domains between BORIS and CTCF suggests they share similar targets in the genome [14]; however, due to the low degree of conservation between their terminal domains, it is believed that they interact with different cofactors, which cause them to have opposite consequences in gene expression and chromatin structure [15,16,17].
In addition, long non-coding RNAs (lncRNAs) have been described as crucial factors in chromatin architecture [18]. Recent evidence indicates that CTCF interacts with several lncRNAs that modulate its recruitment and binding to the DNA. Depletion of CTCF RNA binding domains impairs chromatin loop formation and alters transcriptional profiles [19,20]. Moreover, lncRNAs serve as a scaffold for the interaction of CTCF with other proteins in the form of RNA bridges [21] or could even cause it to detach from its DNA binding sites [22]. Without a doubt, CTCF depends on its interactions with other proteins and nucleic acids to exert a wide range of functions. In this review, we aim to shed light on the role of CTCF partners in shaping the 3D organization and gene regulation of chromatin, specifically those with epigenetic function.
2. CTCF Is a Multifaceted Protein
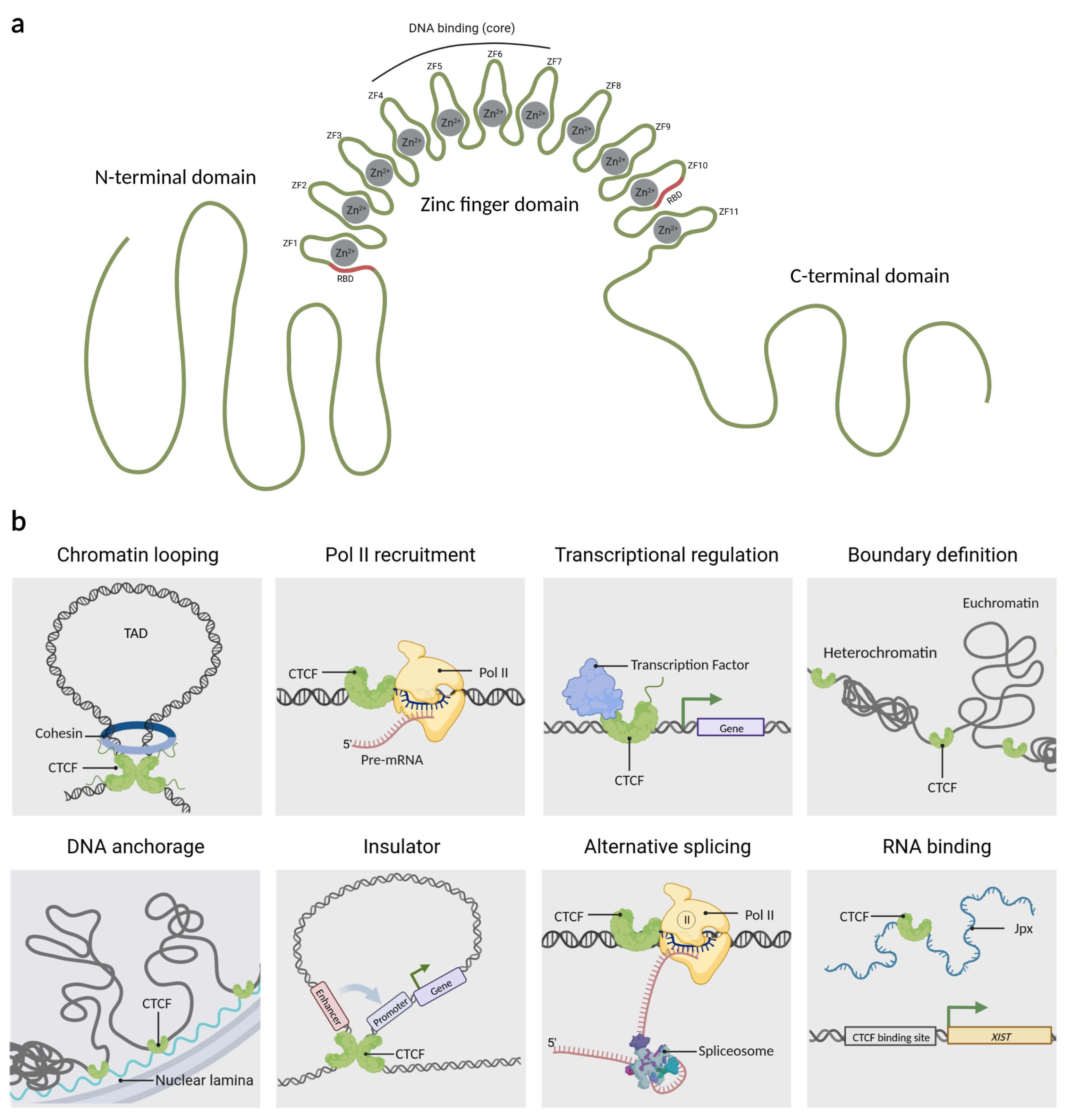
Originally, CTCF was described in chickens as a protein that binds to a region upstream of the c-myc promoter. Because that binding site has three regularly spaced repetitions of the sequence CCCTC, the protein was named CCCTC-binding factor or CTCF [23]. Later, it was found that CTCF is a ubiquitously expressed and highly conserved protein in vertebrates [14,24]. CTCF consists of 727 amino acids (aa) distributed in three domains; a zinc finger DNA-binding domain flanked by the intrinsically disordered N- and C-terminal regions (Figure 1a). The DNA binding domain of CTCF has 11 zinc fingers (ZF) which allow it to interact dynamically with the DNA [25,26,27]. CTCF uses different combinations of its ZF to recognize and bind to a variety of DNA sequences, which is why it is considered a multivalent protein [28,29]. However, around 80% of its target sequences contain the core motif 5’-CCACCAGGTGG-3’ that is recognized by ZFs 4 to 7. Unconserved flanking sequences can be recognized by ZF 1–2 or ZF 8–11, which helps to stabilize the CTCF-DNA complex [30,31,32]. A peculiarity of CTCF is that ZF1 and ZF10 have an RNA binding domain (RBD) which is used to interact with several lncRNAs, providing extra anchorage points for the protein [19,31].
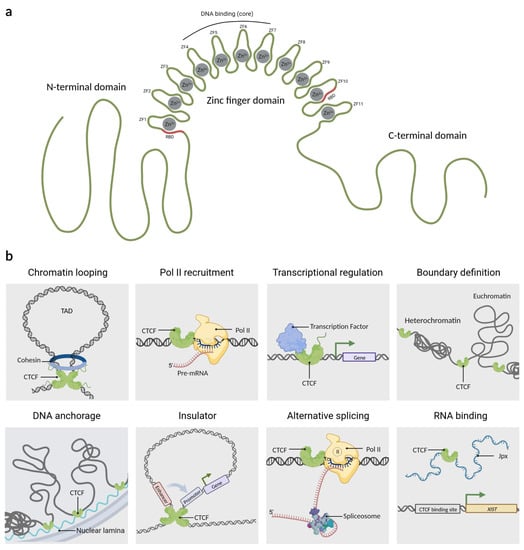
Figure 1.
The architectonic factor CTCF. (a) CTCF is an 82-kDa protein that contains three domains: an N-terminal region, a C-terminal region, and a central domain of 11 zinc fingers. Moreover, CTCF uses the zinc finger domain cooperatively to bind to DNA. RBD:RNA binding domain, ZF: zinc finger. (b) Overview of the wide arrange of CTCF mechanisms of action as: Chromatin looping, RNA Polymerase II (Pol II) recruitment, transcriptional regulation, boundary definition, DNA anchorage, insulator, alternative splicing, and RNA binding, among others. TAD: topologically associated domain. Created with BioRender.com (accessed on 23 April 2023).
CTCF has tens of thousands of genomic binding sites, some of which are conserved between species and tissues [33]. CTCF actions are dependent on its binding site location; which are mainly located in intergenic regions, although they could also be present in regulatory regions such as enhancers, gene promoters, and within gene bodies [34,35,36]. The main functions of CTCF include maintaining topologically associated domains (TADs), acting as a barrier to the spread of heterochromatic structures, and defining the boundaries between euchromatin and heterochromatin, for this reason, CTCF has been coined as an architectural protein [37,38,39,40]. CTCF also regulates DNA anchorage to cellular structures such as the nuclear lamina [37,38], acts as a protein insulator by controlling the interactions between enhancers and promoters [41], and can function as a scaffold protein for transcription factors [42,43,44] and epigenetic factors [45]. Based on the location of the CTCF in other genomic sites, it has also been demonstrated to be involved in processes such as alternative splicing by pausing RNA Polymerase II (RNAP II) binding to alternative exons, thus providing the required temporal context for co-transcriptional spliceosome formation at weak upstream splice sites [4]. CTCF also interacts with lncRNAs which is important for the transcriptional regulation of genes such as Xist, a lncRNA responsible for X chromosome inactivation. For this reason, CTCF has been considered a very versatile protein similar to a swiss army knife. A summary of its functions is shown in Figure 1b.
3. BORIS and CTCF-s Highlight the Importance of CTCF Protein–Protein Interactions
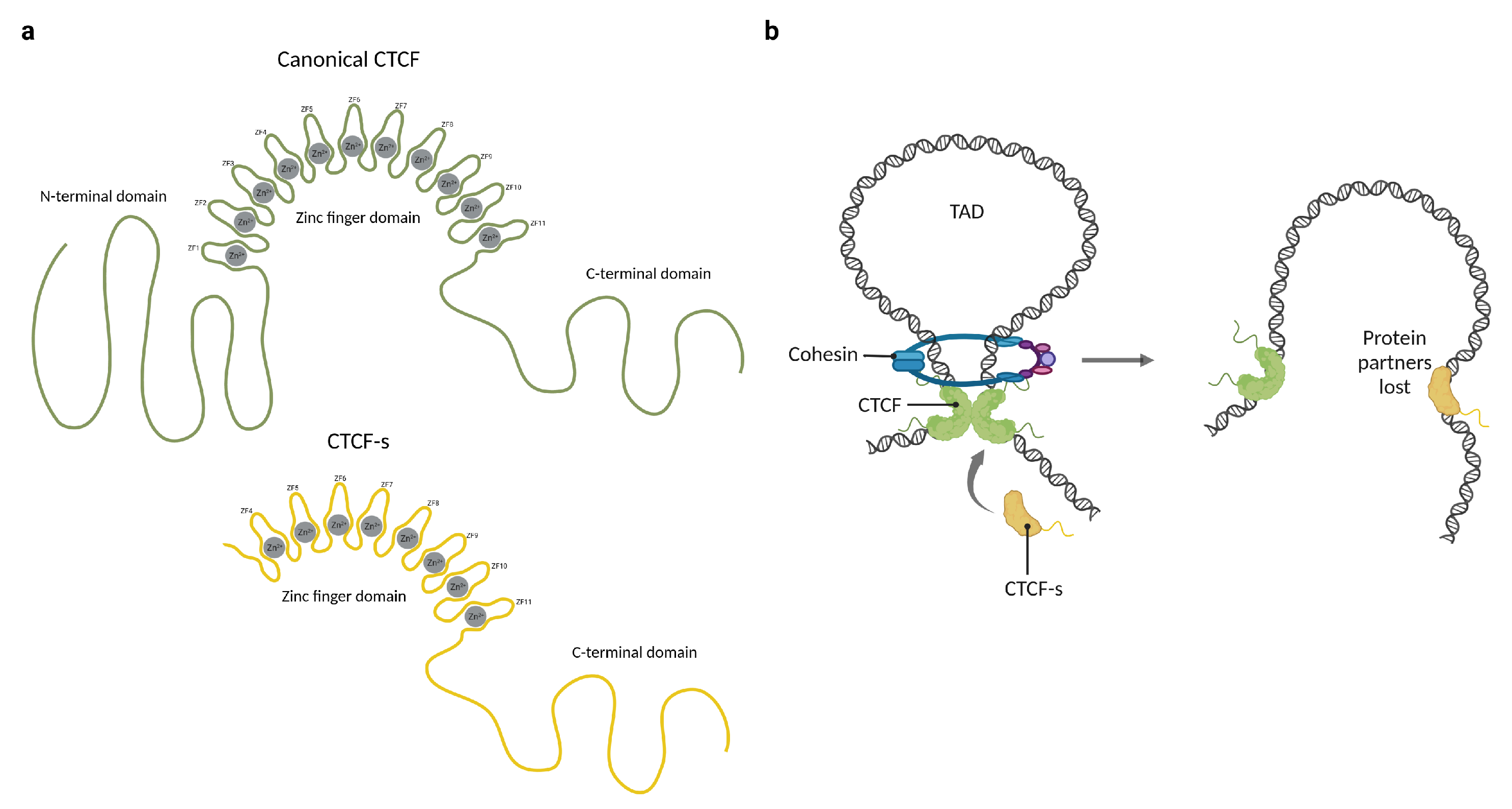
The mechanisms underlying CTCF functions are not yet fully understood, but it is probable that most of them depend on interactions with other proteins. One of the better-characterized CTCF protein–protein interactions is cohesin retention. The cohesin ring is a multi-protein complex involved in the formation of chromatin loops [46]. The mechanism of loop extrusion by cohesin involves the translocation of the complex along chromatin fibers, progressively extruding chromatin loops until it encounters a barrier that prevents further movement; such a barrier is frequently a CTCF dimer. In humans, CTCF interacts with SA1-SCC1 subunits of cohesins through its N-terminal domain, fixating the ring in place and establishing topologically associated domains [8,47,48]. Based on this mechanism of action, it has been proposed that upon binding to the DNA, the unbound ZFs and the terminal regions of CTCF might serve as a platform for interaction with other proteins. This hypothesis is supported by the discovery of a shorter isoform known as CTCF-short (CTCF-s), which lacks the N-terminal domain and the first three zinc fingers (Figure 2a). Because they share the core DNA binding domain, CTCF-s competes for the canonical CTCF binding sites and interferes with CTCF–cohesin interactions, causing a disruption in the long-range connection between enhancers and promoters (Figure 2b). Overexpression of CTCF-s leads to increased cell apoptosis in HeLa-S3 cells, but the physiological role of this isoform and its impact on CTCF interactions with other proteins remain uncertain [49].
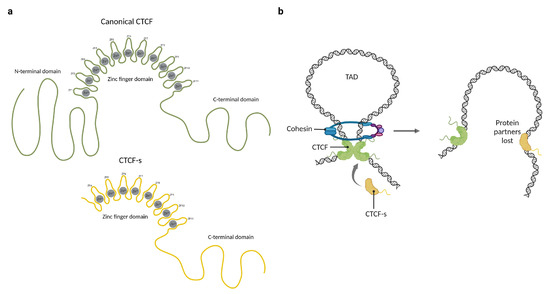
Figure 2.
The participation of CTCF and CTCF-short (CTCF-s) in the formation of chromatin loops. (a) Representation of the domain distribution of CTCF and CTCF-short (CTCF-s). (b) CTCF physically binds to itself to form homodimers which promote chromatin loop formation through cohesin ring protein. CTCF-s competes with CTCF to alter the chromatin architecture and loop formation, mainly because CTCF-s is unable to interact with the cohesin ring. Created with BioRender.com (accessed on 23 April 2023).
Similarly, CTCF has a paralogous gene called CTCF-Like (CTCFL), which encodes the protein Brother of the Regulator of Imprinted Sites (BORIS). It is believed that CTCFL originated from a duplication event at some point before the evolution of mammals [14]. Unlike CTCF, BORIS is a protein that under physiological circumstances is only expressed in the testis, where it is required for spermatogenesis [50]. Nonetheless, BORIS has gained notoriety recently as a promising drug target because it is aberrantly expressed in several neoplasms and has been related to poor outcomes in cancer patients [51,52].
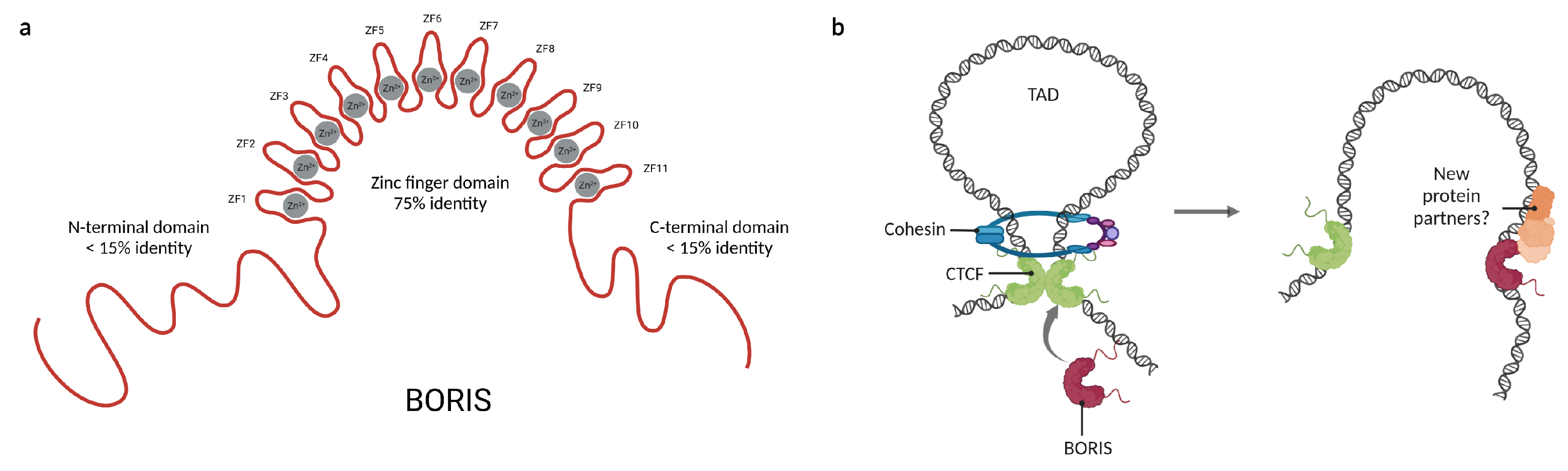
CTCF and BORIS share 75% of identity, mainly in their DNA binding domains, suggesting that they might compete for similar binding sites in the genome. Indeed, BORIS has been described to bind to a large subset of CTCF binding sites; however, there are a few differences in the target regions of both proteins [53]. While CTCF binds preferentially to intronic and intergenic regions, BORIS binds mainly to promoters [54,55]. Because the N- and C-terminal domains of BORIS are not conserved (Figure 3a), it has been suggested that BORIS may share binding sites with CTCF, but after binding will recruit different protein partners, interfering with the main functions of CTCF. In this regard, it has been reported that BORIS expression affects transcriptional regulation and the establishment of chromatin loops since BORIS alone is insufficient to recruit the cohesin complex, which is indispensable for CTCF-mediated chromatin loop formation [53,56].
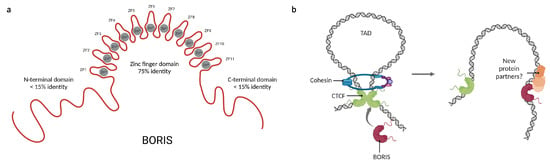
Figure 3.
Features and functions of Brother of the Regulator of imprinted sites (BORIS). (a) Representation of the domain distribution of BORIS and their percentage of identity with CTCF. (b) BORIS can alter chromatin loops by a competitive mechanism with CTCF and its inability to interact with the cohesin ring. Moreover, the recruitment of new protein partners by BORIS could explain the opposite behaviors of CTCF and BORIS. Created with BioRender.com (accessed on 23 April 2023).
Besides the impairment of chromatin loops, the differences between CTCF and BORIS terminal domains may affect which proteins are recruited upon binding (Figure 3b). Through a yeast two-hybrid assay, it was demonstrated that BORIS binds to a set of completely different protein partners than CTCF [57]. This explains the opposite consequences of their expression in cancer; while BORIS promotes cell proliferation and has been classified as an oncogene [58,59], CTCF is a known tumor suppressor [60]. Moreover, it has been observed that BORIS promotes the expression of some genes that are repressed by CTCF, such as hTERT [61], NY-ESO [17], and H19 [62]. So far, CDH8 and UBF are the only proteins known to bind both CTCF and BORIS [63,64]. A summary of the currently known BORIS protein partners is displayed in Table 1. The former reinforces the importance of CTCF protein–protein interactions for the maintenance of the 3D-chromatin structure and suggests that the terminal domains of these proteins serve as scaffolds for cofactor recruitment. Together, this suggests that the cellular functions of CTCF and BORIS could be defined by their interaction with other proteins.

Table 1.
Known protein–protein interactions of BORIS. Proteins that were experimentally validated to interact with CTCF as well are labeled in red.
4. CTCF Regulates the Chromatin Structure through Interactions with Several Epigenetic Factors
The chromatin status is dynamic and can be regulated by covalent modification of the amino-terminal ends of histones that protrude from the nucleosome and are accessible to enzymes that chemically modify them through a system of writing, reading, and erasing complexes [9]. These modifications correspond to a kind of code that works in conjunction with the DNA sequence to determine the state of the chromatin and establishes and stabilizes gene expression patterns [10]. Because of CTCF’s role as the master regulator of chromatin, it is highly probable that both its actions and DNA recruitment are dependent on the chromatin context. To better understand the interactions between CTCF and other proteins with epigenetic functions, we analyzed data from the literature, as well as the STRING database [69] and the Integrated Interactions Database [70] to find CTCF protein partners (Supplementary Table S1). While many of these partners are transcription factors that use CTCF as a scaffold to shape the chromatin structure [71], CTCF also interacts with other proteins that have epigenetic functions, such as DNA and histone demethylases [21,72,73]. The identification of CTCF protein partners involved in epigenetic processes may provide valuable insights into the complex regulatory mechanisms of chromatin organization and gene expression. To identify these proteins, we filtered our list of CTCF protein partners using the annotations available in the EpiFactors database [13]. The resulting CTCF epigenetic factor targets are shown in Figure 4.
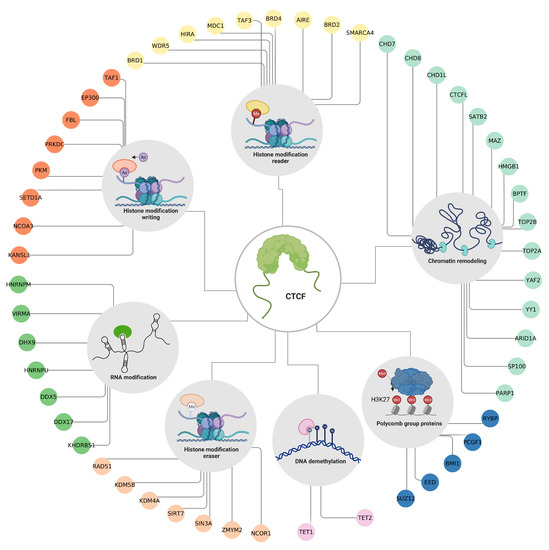
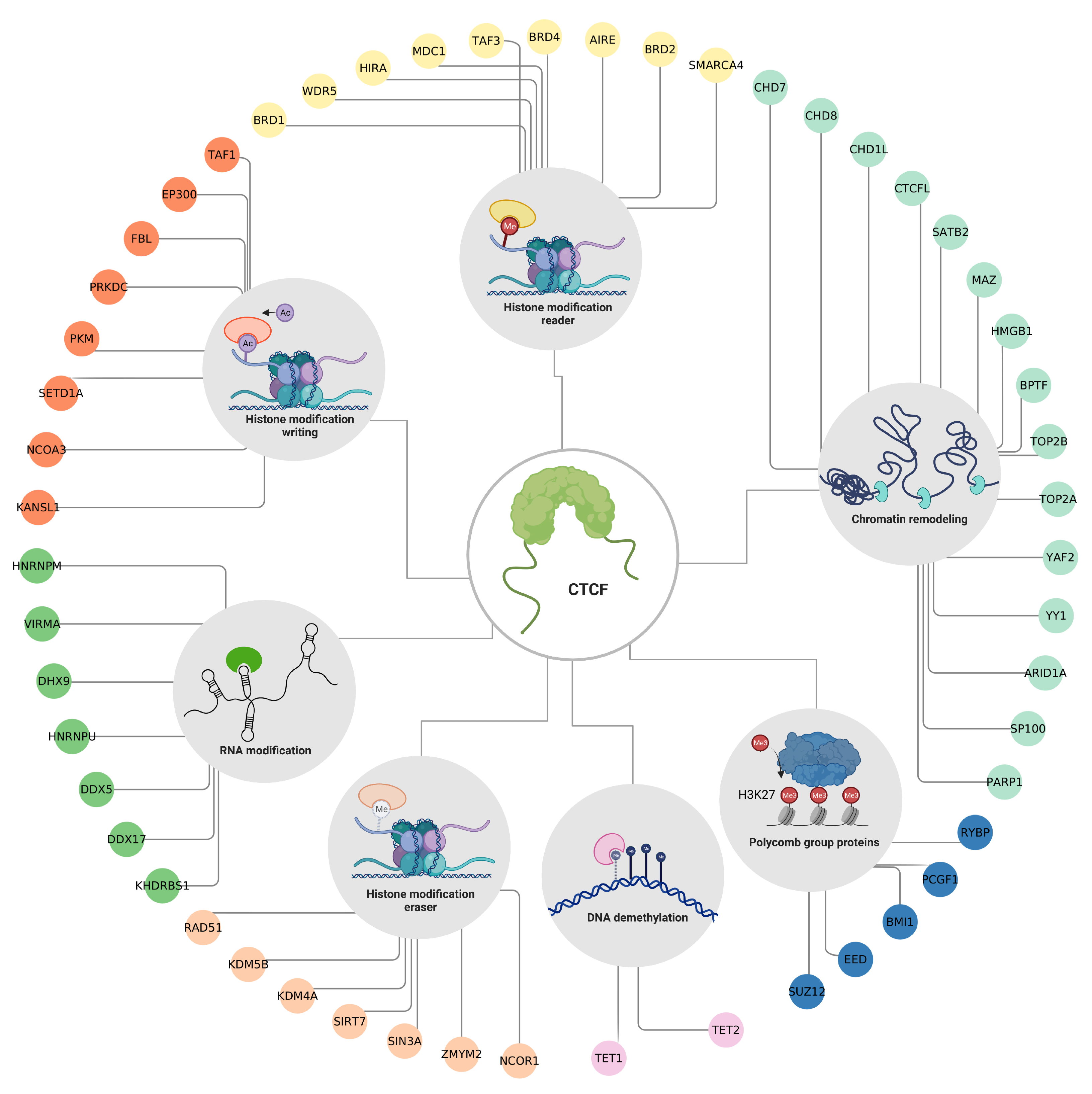
Figure 4.
Epigenetic factors that interact with CTCF. The protein–protein interactions between CTCF and other proteins with epigenetic functions. Colors are according to the EpiFactor category that each protein belongs to, as follows: histone modification reader in yellow, chromatin remodeling in mint, polycomb group proteins in blue navy, DNA demethylation in pink, histone modification eraser in salmon, RNA modification in green, and histone modification writing in orange. Created with BioRender.com (accessed on 23 April 2023).
Among these interactions, many of the proteins participate in the shaping of the 3D conformation of the genome such as the DNA helicases CHD7 [74], CHD8 [63] and CHD1L [75], the topoisomerases TOP2A [76] and TOP2B [77], and the components of chromatin remodeling complexes such as ARID1A [78], YY1 [79], YAF2 [42] and BPTF [71]. The former suggests that CTCF works in combination with other remodeling cofactors to establish chromatin domains.
It is also worth noticing that CTCF interacts with several members of the Polycomb group (PcG). These proteins are part of a system that regulates post-translational modifiers of histones, and their action is generally associated with the transcriptional repression of tissue-specific genes. This group has two members, the Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2). PRC2 is the complex that acts as a writer, as it is responsible for mono-, di-, and trimethylated lysine 27 of histone 3 (H3K27me3). This mark is associated with silenced gene promoters and facultative heterochromatin. H3K27me3 is recognized by PRC1 (reader) that binds to chromatin, monoubiquitinates lysine 119 of histone H2A (H2AK119ub), and prevents transcription by blocking the recruitment of RNA polymerase II [80,81]. CTCF interacts with EED and SUZ12 which are members of the PRC2 complex; a couple of studies have proposed that CTCF could guide the PRC2 complex to gene promoters that are susceptible to repression through H3K27 methylation [82,83]. Furthermore, BMI1, PCGF1, and RYBP are members of the PRC1 complex. Although the biological significance of their interaction with CTCF remains unexplored, a study shows that these proteins may regulate the organization of CTCF-mediated chromatin interactions [84].
Besides PcG proteins and chromatin remodeling factors, CTCF’s relationship with proteins related to histone post-translational modifications are remarkable as well. CTCF interacts with proteins involved in the three stages of histone posttranslational modifications (writing, reading, and erasing). However, we would like to discuss further two particular cases that have not been broadly explored yet; histone and DNA demethylases.
5. CTCF as a Modulator of Histone Methylation
Histone methylation is a post-translational modification related to multiple biological functions. Methylation happens mainly in arginine (R) and lysine (K) residues. Arginines can be mono- or dimethylated, and this chemical modification generally potentiates the interaction with other enzymes that modify histone tails [85]. Moreover, lysine residues can be mono-, di-, or trimethylated; these histone marks are associated with either transcriptional activation or repression, depending on the lysine residue. As an example, di- and trimethylation at H3K4 is related to enhanced gene expression, whereas trimethylation at H3K9 and H3K27 is associated with transcriptional repression [10]. Because histone methylation is a covalent modification, it was initially assumed to be stable and irreversible. However, in 2004, the first histone lysine demethylase was characterized, and since then more than 20 enzymes have been described that can remove this covalent modification [86,87].
Currently, histone lysine demethylases (KDMs) are classified into two families based on their chemical mechanism of action: the amine oxidase-like and the oxygenase enzymes [88]. The amino oxidase-like family has two members: KDM1A; the first histone lysine demethylase described by Shi and colleagues in 2004; and KDM1B. These proteins have a common amine oxidase-like domain and are FAD-dependent [89]. KDM1 enzymes can remove mono- and dimethyl groups but cannot demethylate trimethylated lysines, due to their FAD-dependent catalytic mechanism [90]. The oxygenase family is the largest one, with more than 20 JmjC (Jumonji) domain-containing enzymes. These proteins enclose a Fe ion in their catalytic domain and use -ketoglutarate as a co-substrate [91]. This family is also divided into seven subfamilies (KDM2-8) according to the similarity of their catalytic domain and their substrate specificity [88].
In vitro studies have demonstrated that the simple binding of these enzymes to their substrates is sufficient for the demethylation reaction, suggesting that their recruitment must be tightly controlled in order to prevent aberrant demethylation [92,93,94]. It is not yet clear how the demethylases are directed to specific sites in the chromatin, especially since they lack DNA binding domains. One possible explanation could be that certain transcriptional factors and other chromatin-binding proteins might be responsible for the recruitment of these epigenetic components. KDMs activity could be regulated by protein–protein interactions allowing a dynamical interaction with the chromatin by taking advantage of the “reader” domains present in their binding partners [95,96]. Moreover, it has been suggested that the chromatin environment provides certain selectivity to demethylases since it controls the accessibility of these proteins to their target sites [97]. In addition, it is known that several transcription factors recruit histone demethylases upon binding to their target genes to promote a change in the chromatin state [98,99,100]. However, KDMs’ relationship with CTCF remains partially unexplored.
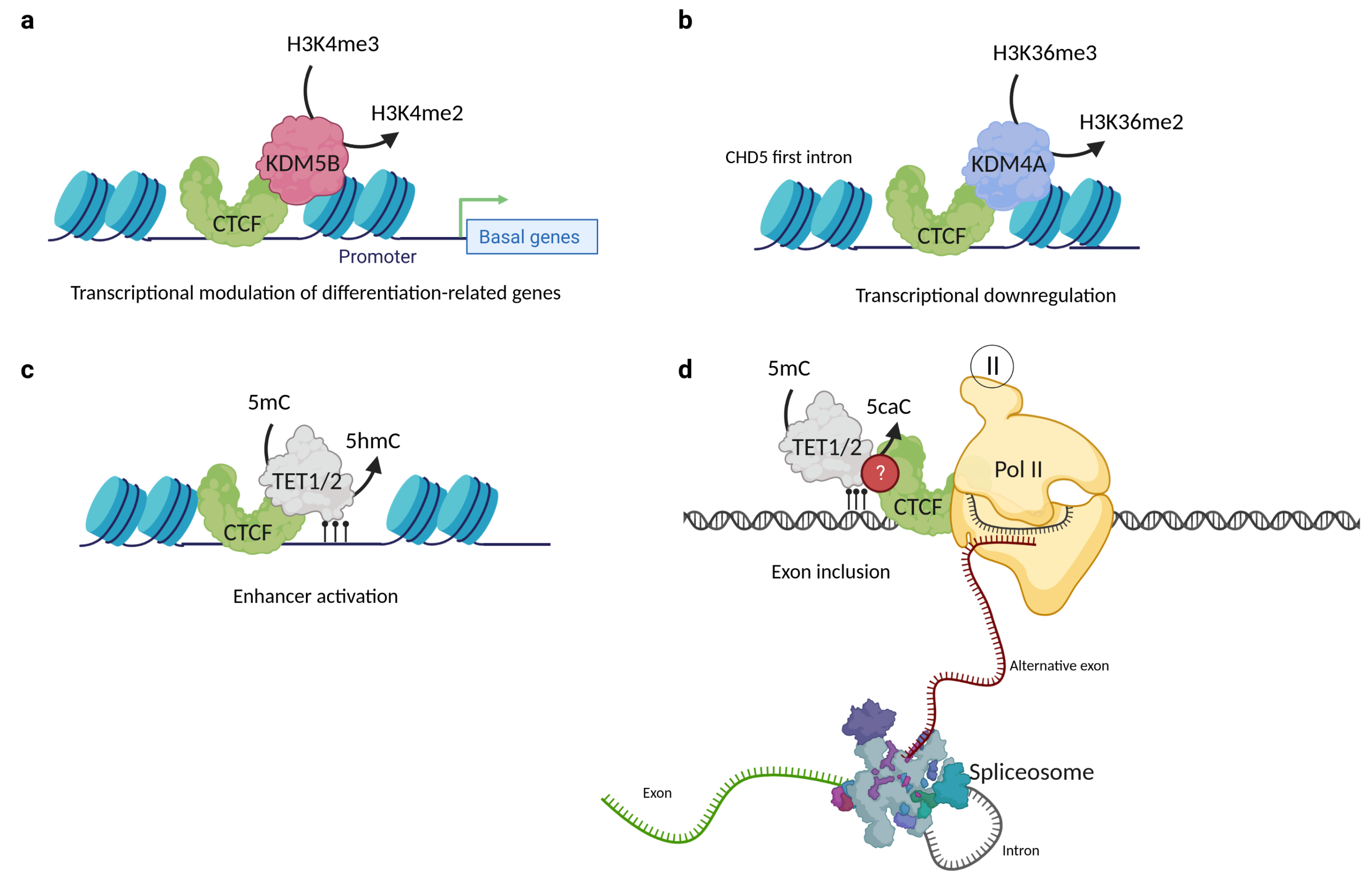
Until now, few studies have demonstrated the association between CTCF and histone demethylases; in fact, only two KDM partners have been found. The first was reported in 2014 by Yamamoto et al., who found via co-immunoprecipitation that CTCF formed a complex with the H3K4me3 and H3K4me2 specific demethylase KDM5B. Moreover, when conducting ChIP-seq assays, they discovered that KDM5B sites overlap with those of CTCF in most mammary cancer cell lines, and this overlapping phenomenon correlates with a lower H3K4me3 signal compared to those non-overlapping sites (Figure 5a). The role of the KDM5B-CTCF complex is not clear, but the authors suggest that CTCF takes part in a finely tuned regulation of basal/stem cell genes, such as ACTG2, APOE, CTGF, FN1, and TGFβ2, among others. The perturbation of these transcriptional changes could promote breast cancer progression [73,101].
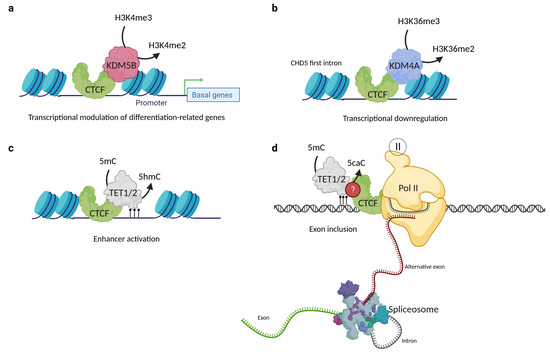
Figure 5.
CTCF interactions with histone and DNA demethylases. (a) CTCF interacts with KDM5B and regulates the transcription rate of basal/stem cell genes in luminal breast cancer lines. (b) The interaction of CTCF with KDM4A is involved in the down-regulation of CHD5 gene expression in MCF7 cells. (c) CTCF interaction with TET1 and TET2 proteins is involved in enhancer activation. (d) CTCF can also interact with 5caC, which leads to RNA pol II pausing and alternative exon inclusion of the CD45+ gene. CTCF and TET protein–protein interaction is possible but remains uncharacterized for this mechanism. Created with BioRender.com (accessed on 23 April 2023).
Another CTCF histone demethylase partner is KDM4A. The first clue that CTCF could be a KDM4A partner was reported in 2011 by Kang’s group, who performed transfection and immunofluorescence assays and observed that the demethylation frequency of KDM4A was enhanced by the presence of CTCF [102]. This study opened the window to another report in 2018, where co-immunoprecipitation was used to demonstrate that CTCF and KDM4A form a protein complex. Furthermore, it was shown by ChIP-qPCR and ChIP-Re/ChIP-qPCR that CTCF and KDM4A coexist in the first intron of CHD5, the promoter of WRAP53, and the region located at −1922 bp of the ASCL2 transcription starting site. The coexistence of CTCF and KDM4A correlates with the reduction of H3K36me3/2 histone modifications at the first intron of CHD5 and is associated with its transcriptional down-regulation (Figure 5b). Moreover, CTCF or KDM4A depletion mediated by siRNAs leads to the CHD5 reactivation expression, proposing that both proteins are involved in the negative regulation of this gene. The knockout of KDM4A by CRISPR/Cas9 restored the expression of CHD5 and H3K36me3 and H3K36me2 histone marks, without disturbing the CTCF localization [72]. Nevertheless, it is currently unknown whether this complex is related to a genome-wide repression or activation and if CTCF might also be one of the key proteins driving the specificity of KDM4A.
To the best of our knowledge, there are no studies evaluating the association between CTCF and other histone demethylases. Nevertheless, ChIP-seq studies demonstrate some overlap between KDM5A, KDM5C, KDM1A, and CTCF, suggesting that CTCF could be involved in their regulation; however, further studies are required to determine the participation of CTCF in the modulation of these enzymes.
6. CTCF and the TET Enzymes
DNA methylation is an epigenetic process involving a methyl group transfer to the C5 position of the cytosine to form 5-methylcytosine (5mC). DNA methylation has several functions; although it is generally associated with transcriptional repression; it is also involved in other vital processes, such as genomic imprinting, X chromosome inactivation, and retrotransposon element suppression [103,104]. Similarly to histones, DNA can be demethylated; this process can be accomplished either passively, by simply not methylating the new DNA strand after replication, or actively, by a replication-independent process that involves the ten-eleven translocation (TET) enzymes [105].
The first evidence of the enzyme-mediated DNA demethylation was observed in 2007, with the identification of the Trypanosoma cruzi enzymes JBP1 and JBP2 that are responsible for gene silencing through the hydroxylation and glycosylation of a thymine methyl group (known as J Base). This discovery pointed toward the existence of “eraser” proteins that are in charge of removing DNA methylation [106]. Shortly after, in 2009, when looking for mammalian homologs of the trypanosome thymidine hydroxylases, the three human ten eleven translocation (TET) proteins, TET1, TET2, and TET3 were identified [107]. Nevertheless, the TET proteins were not at a central stage until they were found to oxidize 5mC to 5-hydroxymethyl-cytosine (5hmC) as part of the DNA demethylation mechanism [108,109]. Subsequent reports revealed that TET proteins further oxidize 5hmC to 5-formyl-cytosine (5fC) and 5-carboxyl-cytosine (5caC), both of which are removed through the Base Excision Repair (BER) pathway, thereby completing the demethylation process [108,110].
Because DNA methylation is an epigenetic marker that is essential for correct cellular function and organism development [111,112], TET proteins must be subjected to finely controlled regulatory mechanisms. These enzymes have fundamental roles in epigenetic reprogramming, embryogenesis, development, and tumorigenesis, and it is well-known that their inactivation contributes to the local DNA hypermethylation observed in cancer [113,114]. Apart from catalytic activity regulation, TET1 and TET3 are more likely recruited to their genomic target sites through the direct binding of their respective CXXC domains to the DNA [115]. In vitro binding assays and in vivo chromatin immunoprecipitation assays confirm that these domains can bind CpG-rich oligonucleotides with a slight preference for unmethylated versus methylated substrates [116,117,118]. In contrast, TET2 does not have any obvious DNA-binding domains, and it is therefore potentially recruited through the direct binding of DNA-targeting partners [119]. In fact, it has been demonstrated that the TET2 protein binds tissue-specific transcription factors such as the early B cell factor 1 (EBF1) [120] and WT1 [121,122]. The dynamic expression of DNA-binding factors and their interactions with TET2 can likely concede the tissue-specific and temporal modulation of TET activity on a limited set of genomic loci [123]. Furthermore, interaction with several binding partners is likely to alter the genomic location and stability of TET proteins [124].
Since TET enzymes form protein complexes with other epigenetic components to modify gene transcription, the interaction of these proteins with CTCF is of particular interest. It is known that synchronized fluctuations of DNA methylation, demethylation, nucleosome positioning, and CTCF chromatin binding have an important role in establishing cell-type-specific chromatin states during differentiation. Loss of CTCF in regions such as the boundaries of chromatin loops, promoters, and TADs can be associated with the spread of DNA methylation and demethylation, and can be linked to the down-regulation of adjacent genes. A hierarchical interaction between cytosine modifications, nucleosome positioning, and DNA sequences controls CTCF binding and regulates gene expression [125,126].
It has been proposed that CTCF binding to low methylated regions could mediate local DNA demethylation through TET recruitment [127]. The first evidence was an oscillating 5hmC pattern observed around the binding sites of CTCF in mouse embryonic stem cells, which suggests that accessibility and 5hmC deposition could be related to CTCF binding [128]. The genomic co-localization of CTCF, TET1, TET2, and 5hmc was probed by co-immunoprecipitation assays on 3T3-L1 and HEK293T cell lines and correlated with enhancer activation on differentiated cells through the facilitation of the hydroxymethylation of DNA [129]. This concludes that CTCF directly interacts with the TET enzymes and promotes the DNA hydroxymethylation of enhancers driving adipocyte differentiation (Figure 5c). Nevertheless, the relationship between CTCF and TET demethylases is not only relevant to cell differentiation processes, since a study in 2016 revealed that dynamic TET1 and TET2-catalyzed DNA oxidation stimulates CTCF-dependent alternative splicing in human lymphocytes. This study found that CTCF directly interacts with 5caC in vitro and that this mark was strongly associated with alternative exon inclusion [6]. Moreover, a study demonstrated that 5caC could reinforce CTCF binding to the DNA (Figure 5d). These findings suggest that the TET mediated-induction of 5caC is a potential way to regulate CTCF binding and further reinforces the idea that there is a close relationship between CTCF and the TET proteins [130]. More studies are needed to better describe the exact functions that DNA oxidation plays in transcriptional regulatory events; additional explorations will be required to define the way in which CTCF binding is associated with 5caC in vivo.
Taken together, the above information suggests that CTCF could interact, directly or indirectly, with histone and DNA demethylases; it is still unknown whether these complexes are related to repression, activation, or other transcriptional processes.
7. Long Non-Coding RNAs as Non-Protein Partners of CTCF
Long non-coding RNAs (lncRNAs) have emerged as important regulators of chromatin structure and gene expression. They act as scaffolds, guides, or decoys that recruit chromatin modifiers to specific genomic regions, mediate higher-order chromatin organization, and influence gene expression [131]. LncRNAs have been demonstrated to play a critical role in the formation and maintenance of chromatin domains, such as TADs. In this context, lncRNAs have been found to interact with chromatin-associated proteins, CTCF for instance, to modulate their function and impact on chromatin structure and gene regulation [19,132]. Recently, lncRNAs have been identified as key regulators of CTCF [20]. CTCF interacts with RNA through the RNA-binding domains in ZF1 and ZF10. Some studies have even reported a consensus sequence for RNAs that bind to CTCF, and it has been suggested that it could have around 5000 potential RNA partners in the genome [21,133].
LncRNAs contribute to the functions of CTCF by recruiting it to specific genomic sites, modulating chromatin loops, and regulating the formation of TADs. One of the most studied cases is CTCF-mediated Xist transcriptional repression. Xist is a lncRNA involved in X chromosome inactivation. CTCF represses Xist expression by binding to its promoter; however, Jpx is a lncRNA that binds to CTCF and removes it from the Xist promoter, allowing its expression and subsequent X chromosome inactivation [19,134]. Recently, it was found that Jpx can also compete for CTCF binding sites in the DNA, altering the loop formation and the overall conformation of the chromatin [22]. The interplay between CTCF and other RNA-binding proteins is also important for the maintenance of TADs. As shown in Figure 4, CTCF interacts with several RNA-binding proteins. Among them, DDX5 is an RNA helicase involved in many steps of RNA-related processes, such as alternative splicing, miRNA biogenesis, and RNA unwinding [135]. It has been described that both DDX5 and the lncRNA steroid receptor RNA activator (SRA) interact with the CTCF-cohesin complex and stabilize it. Such an interaction is required for the insulation activity of CTCF [136].
Several other lncRNAs have been identified to interact with CTCF and modulate its function. HOTTIP, for instance, can recruit CTCF to specific genomic regions and promote TAD formation [137]. Similarly, GATA6-AS1 contributes to TAD formation by forming an RNA-DNA triplex and interacting with CTCF [138]. LncRNAs also regulate gene expression through the recruitment or detachment of CTCF [139,140,141]. PACERR recruits CTCF and p300 to promoter regions to activate gene transcription through histone acetylation [142]. LncRNAs have also been associated to increase protein stability; for instance, the lncRNA ELDR inhibits CTCF degradation by the proteasome, increasing protein levels without modifying transcript levels. Table 2 shows known interactions between CTCF and lncRNAs, along with the putative function of the complexes.

Table 2.
Long noncoding RNAs (lncRNAs) that are known to directly interact with CTCF.
Overall, lncRNAs represent an exciting new area of research in the field of chromatin biology and gene regulation. The interaction between lncRNAs and CTCF offers a new level of complexity to the already intricate network of molecular interactions that govern gene expression and chromatin architecture.
8. Conclusions and Final Remarks
CTCF is a nuclear factor that is involved in several chromatin-related processes, including transcriptional regulation, three-dimensional chromatin topology, and epigenetics. Part of its relevance lies in its versatility, as shown in this review, CTCF relies on a broad network of protein and RNA partners to achieve its different tasks. The role of the protein partners is clear upon comparison with CTCF–s and BORIS. In the first case, the lack of the N-terminal domain leads to the loss of the most studied CTCF interacting partners, the cohesin complex. The second case is more complex, since BORIS binding to the DNA has completely opposite consequences than CTCF, besides sharing a high degree of identity at their DNA binding domains. Most of this could be explained by their interactions with different protein partners through their unconserved terminal domains. There is current research going on in this regard, and without a doubt, the study of the interplay between CTCF and BORIS in cancer will help to understand CTCF’s role in chromatin organization and other epigenetic processes.
Chromatin is finely organized inside the nucleus through a complex system that has not yet been elucidated. As mentioned previously in this review, many of the epigenetic factors that help to establish and maintain chromatin structure lack DNA binding domains; thus, it has been hypothesized that their action should rely on other proteins. CTCF is capable of binding to thousands of sites in the genome, and due to the flexibility of its DNA binding domain, it is considered a multivalent protein. In this review, we demonstrate that CTCF interacts with a wide array of epigenetic factors which suggests that it could serve as a scaffold for the assembly of different protein complexes. Nevertheless, the logistics involved in partner election, the impact of each complex, and the crosstalk between different partners is an exciting point of view that is worth further study.
Since epigenetic markers such as histone and DNA methylation are highly dependent on the chromatin context, the interaction between CTCF and different components of the epigenetic complexes is interesting. So far only a few protein–protein interactions between CTCF and other epigenetic factors have been fully characterized, and in most cases, the studies have been conducted on a specific gene or promoter; thus, genomic scale experiments could be helpful to identify the overall impact and localization of the complexes.
LncRNAs add another layer of complexity to the CTCF-mediated chromatin regulation. Currently, the role of most of these complexes in biological processes remains unknown. However, several studies hint towards the existence of a broad CTCF-RNA interaction network. The role of some of these complexes has been discussed here; among them, Jpx is the most remarkable example due to its ability to detach CTCF from its binding sites. Further studies will help to understand the role of CTCF-lncRNA interactions.
Because of CTCF’s versatility, it could likely function as a scaffold for many of the epigenetic complexes required for a proper genomic organization. Without a doubt, there are still undiscovered mechanisms for CTCF; the study of this protein could aid to understand the complex mechanisms that regulate chromatin organization and gene expression.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells12101357/s1, Table S1: Protein-Protein interactions for CTCF.
Author Contributions
Conceptualization, A.D.M.-M. and E.S.-R.; investigation and data curation M.S.-A., Y.S.-P. and A.D.M.-M.; original draft preparation, A.D.M.-M. and M.S.-A.; review and editing, E.S.-R. and N.K.W.; supervision, E.S.-R. and J.B.; funding acquisition, E.S.-R. and J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Consejo Nacional de Ciencia y Tecnología (CONACyT) through the Fondo CB-SEP-CONACyT (284748, PROMED 250690) and by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grants 01ZX1910D and 01ZX2210D). Soto-Reyes, E. was supported by the Natural Science Department at UAM Cuajimalpa (DCNI-07-243-23). Del Moral-Morales, A. is a doctoral student from Programa de Maestría y Doctorado en Ciencias Bioquímicas, UNAM, and received a fellowship from CONACyT (CVU894530). Del Moral-Morales, A. was also a beneficiary of the German Academic Exchange Service (DAAD Grant No. 91833882). Baumbach, J. was partially funded by his VILLUM Young Investigator Grant nr.13154.
Acknowledgments
We greatly acknowledge the support and collaboration with the Center for Data and Computing in Natural Sciences (CDCS).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Morrison, O.; Thakur, J. Molecular Complexes at Euchromatin, Heterochromatin and Centromeric Chromatin. Int. J. Mol. Sci. 2021, 22, 6922. [Google Scholar] [CrossRef] [PubMed]
- Holwerda, S.J.B.; de Laat, W. CTCF: The protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120369. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.C.; West, A.G.; Felsenfeld, G. The Protein CTCF Is Required for the Enhancer Blocking Activity of Vertebrate Insulators. Cell 1999, 98, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Velasco, M.; Kumar, M.; Lai, M.C.; Bhat, P.; Solis-Pinson, A.B.; Reyes, A.; Kleinsorg, S.; Noh, K.M.; Gibson, T.J.; Zaugg, J.B. CTCF-Mediated Chromatin Loops between Promoter and Gene Body Regulate Alternative Splicing across Individuals. Cell Syst. 2017, 5, 628–637.e6. [Google Scholar] [CrossRef] [PubMed]
- Marina, R.J.; Sturgill, D.; Bailly, M.A.; Thenoz, M.; Varma, G.; Prigge, M.F.; Nanan, K.K.; Shukla, S.; Haque, N.; Oberdoerffer, S. TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. EMBO J. 2016, 35, 335–355. [Google Scholar] [CrossRef]
- Vostrov, A.A.; Taheny, M.J.; Quitschke, W.W. A region to the N-terminal side of the CTCF zinc finger domain is essential for activating transcription from the amyloid precursor protein promoter. J. Biol. Chem. 2002, 277, 1619–1627. [Google Scholar] [CrossRef]
- Pugacheva, E.M.; Kubo, N.; Loukinov, D.; Tajmul, M.; Kang, S.; Kovalchuk, A.L.; Strunnikov, A.V.; Zentner, G.E.; Ren, B.; Lobanenkov, V.V. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl. Acad. Sci. USA 2020, 117, 2020–2031. [Google Scholar] [CrossRef]
- Felsenfeld, G.; Groudine, M. Controlling the double helix. Nature 2003, 421, 448–453. [Google Scholar] [CrossRef]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Medvedeva, Y.A.; Lennartsson, A.; Ehsani, R.; Kulakovskiy, I.V.; Vorontsov, I.E.; Panahandeh, P.; Khimulya, G.; Kasukawa, T.; FANTOM Consortium; Drabløs, F. EpiFactors: A comprehensive database of human epigenetic factors and complexes. Database 2015, 2015, bav067. [Google Scholar] [CrossRef] [PubMed]
- Marakulina, D.; Vorontsov, I.E.; Kulakovskiy, I.V.; Lennartsson, A.; Drabløs, F.; Medvedeva, Y.A. EpiFactors 2022: Expansion and enhancement of a curated database of human epigenetic factors and complexes. Nucleic Acids Res. 2023, 51, D564–D570. [Google Scholar] [CrossRef] [PubMed]
- Hore, T.A.; Deakin, J.E.; Marshall Graves, J.A. The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes. PLoS Genet. 2008, 4, e1000169. [Google Scholar] [CrossRef] [PubMed]
- Vatolin, S.; Abdullaev, Z.; Pack, S.D.; Flanagan, P.T.; Custer, M.; Loukinov, D.I.; Pugacheva, E.; Hong, J.A.; Morse, H., 3rd; Schrump, D.S.; et al. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 2005, 65, 7751–7762. [Google Scholar] [CrossRef]
- Nguyen, P.; Cui, H.; Bisht, K.S.; Sun, L.; Patel, K.; Lee, R.S.; Kugoh, H.; Oshimura, M.; Feinberg, A.P.; Gius, D. CTCFL/BORIS is a methylation-independent DNA-binding protein that preferentially binds to the paternal H19 differentially methylated region. Cancer Res. 2008, 68, 5546–5551. [Google Scholar] [CrossRef]
- Hong, J.A.; Kang, Y.; Abdullaev, Z.; Flanagan, P.T.; Pack, S.D.; Fischette, M.R.; Adnani, M.T.; Loukinov, D.I.; Vatolin, S.; Risinger, J.I.; et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005, 65, 7763–7774. [Google Scholar] [CrossRef]
- Guh, C.Y.; Hsieh, Y.H.; Chu, H.P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020, 27, 44. [Google Scholar] [CrossRef]
- Saldaña-Meyer, R.; Rodriguez-Hernaez, J.; Escobar, T.; Nishana, M.; Jácome-López, K.; Nora, E.P.; Bruneau, B.G.; Tsirigos, A.; Furlan-Magaril, M.; Skok, J.; et al. RNA Interactions Are Essential for CTCF-Mediated Genome Organization. Mol. Cell 2019, 76, 412–422.e5. [Google Scholar] [CrossRef]
- Hansen, A.S.; Hsieh, T.H.S.; Cattoglio, C.; Pustova, I.; Saldaña-Meyer, R.; Reinberg, D.; Darzacq, X.; Tjian, R. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol. Cell 2019, 76, 395–411.e13. [Google Scholar] [CrossRef]
- Gavrilov, A.A.; Sultanov, R.I.; Magnitov, M.D.; Galitsyna, A.A.; Dashinimaev, E.B.; Lieberman Aiden, E.; Razin, S.V. RedChIP identifies noncoding RNAs associated with genomic sites occupied by Polycomb and CTCF proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2116222119. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.J.; Aguilar, R.; Kesner, B.; Lee, H.G.; Kriz, A.J.; Chu, H.P.; Lee, J.T. Jpx RNA regulates CTCF anchor site selection and formation of chromosome loops. Cell 2021, 184, 6157–6173.e24. [Google Scholar] [CrossRef] [PubMed]
- Lobanenkov, V.V.; Nicolas, R.H.; Adler, V.V.; Paterson, H.; Klenova, E.M.; Polotskaja, A.V.; Goodwin, G.H. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5’-flanking sequence of the chicken c-myc gene. Oncogene 1990, 5, 1743–1753. [Google Scholar] [PubMed]
- Heger, P.; Marin, B.; Bartkuhn, M.; Schierenberg, E.; Wiehe, T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc. Natl. Acad. Sci. USA 2012, 109, 17507–17512. [Google Scholar] [CrossRef] [PubMed]
- Bonchuk, A.; Kamalyan, S.; Mariasina, S.; Boyko, K.; Popov, V.; Maksimenko, O.; Georgiev, P. N-terminal domain of the architectural protein CTCF has similar structural organization and ability to self-association in bilaterian organisms. Sci. Rep. 2020, 10, 2677. [Google Scholar] [CrossRef]
- Martinez, S.R.; Miranda, J.J.L. CTCF terminal segments are unstructured. Protein Sci. 2010, 19, 1110. [Google Scholar] [CrossRef]
- Maksimenko, O.G.; Fursenko, D.V.; Belova, E.V.; Georgiev, P.G. CTCF As an Example of DNA-Binding Transcription Factors Containing Clusters of C2H2-Type Zinc Fingers. Acta Nat. 2021, 13, 31. [Google Scholar] [CrossRef]
- Ohlsson, R.; Renkawitz, R.; Lobanenkov, V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001, 17, 520–527. [Google Scholar] [CrossRef]
- Nakahashi, H.; Kieffer Kwon, K.R.; Resch, W.; Vian, L.; Dose, M.; Stavreva, D.; Hakim, O.; Pruett, N.; Nelson, S.; Yamane, A.; et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013, 3, 1678–1689. [Google Scholar] [CrossRef]
- Yin, M.; Wang, J.; Wang, M.; Li, X.; Zhang, M.; Wu, Q.; Wang, Y. Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res. 2017, 27, 1365–1377. [Google Scholar] [CrossRef]
- Hashimoto, H.; Wang, D.; Horton, J.R.; Zhang, X.; Corces, V.G.; Cheng, X. Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol. Cell 2017, 66, 711–720.e3. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Ma, R.; Zhang, J.; Liu, Z.; Wu, B.; Peng, J.; Zhai, Y.; Gong, Q.; Shi, Y.; Wu, J.; et al. Dynamic Nature of CTCF Tandem 11 Zinc Fingers in Multivalent Recognition of DNA As Revealed by NMR Spectroscopy. J. Phys. Chem. Lett. 2018, 9, 4020–4028. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.; Schwalie, P.C.; Wilson, M.D.; Ballester, B.; Gonçalves, A.; Kutter, C.; Brown, G.D.; Marshall, A.; Flicek, P.; Odom, D.T. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 2012, 148, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Abdullaev, Z.K.; Smith, A.D.; Ching, K.A.; Loukinov, D.I.; Green, R.D.; Zhang, M.Q.; Lobanenkov, V.V.; Ren, B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007, 128, 1231–1245. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.K.; Iyer, V.R. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J. Biol. Chem. 2012, 287, 30906–30913. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Yue, F.; McCleary, D.F.; Ye, Z.; Edsall, L.; Kuan, S.; Wagner, U.; Dixon, J.; Lee, L.; Lobanenkov, V.V.; et al. A map of the cis-regulatory sequences in the mouse genome. Nature 2012, 488, 116–120. [Google Scholar] [CrossRef]
- Fiorito, E.; Sharma, Y.; Gilfillan, S.; Wang, S.; Singh, S.K.; Satheesh, S.V.; Katika, M.R.; Urbanucci, A.; Thiede, B.; Mills, I.G.; et al. CTCF modulates Estrogen Receptor function through specific chromatin and nuclear matrix interactions. Nucleic Acids Res. 2016, 44, 10588–10602. [Google Scholar] [CrossRef]
- Kaczmarczyk, L.S.; Levi, N.; Segal, T.; Salmon-Divon, M.; Gerlitz, G. CTCF supports preferentially short lamina-associated domains. Chromosome Res. 2022, 30, 123–136. [Google Scholar] [CrossRef]
- Kentepozidou, E.; Aitken, S.J.; Feig, C.; Stefflova, K.; Ibarra-Soria, X.; Odom, D.T.; Roller, M.; Flicek, P. Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains. Genome Biol. 2020, 21, 5. [Google Scholar] [CrossRef]
- Ong, C.T.; Corces, V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014, 15, 234–246. [Google Scholar] [CrossRef]
- Ren, G.; Jin, W.; Cui, K.; Rodrigez, J.; Hu, G.; Zhang, Z.; Larson, D.R.; Zhao, K. CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression. Mol. Cell 2017, 67, 1049–1058.e6. [Google Scholar] [CrossRef] [PubMed]
- Schwalie, P.C.; Ward, M.C.; Cain, C.E.; Faure, A.J.; Gilad, Y.; Odom, D.T.; Flicek, P. Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biol. 2013, 14, R148. [Google Scholar] [CrossRef] [PubMed]
- Chernukhin, I.V.; Shamsuddin, S.; Robinson, A.F.; Carne, A.F.; Paul, A.; El-Kady, A.I.; Lobanenkov, V.V.; Klenova, E.M. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 2000, 275, 29915–29921. [Google Scholar] [CrossRef] [PubMed]
- Defossez, P.A.; Kelly, K.F.; Filion, G.J.; Pérez-Torrado, R.; Magdinier, F.; Menoni, H.; Nordgaard, C.L.; Daniel, J.M.; Gilson, E. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J. Biol. Chem. 2005, 280, 43017–43023. [Google Scholar] [CrossRef]
- Wei, L.; Liu, Q.; Huang, Y.; Liu, Z.; Zhao, R.; Li, B.; Zhang, J.; Sun, C.; Gao, B.; Ding, X.; et al. Knockdown of CTCF reduces the binding of EZH2 and affects the methylation of the SOCS3 promoter in hepatocellular carcinoma. Int. J. Biochem. Cell Biol. 2020, 120, 105685. [Google Scholar] [CrossRef] [PubMed]
- Gabriele, M.; Brandão, H.B.; Grosse-Holz, S.; Jha, A.; Dailey, G.M.; Cattoglio, C.; Hsieh, T.H.S.; Mirny, L.; Zechner, C.; Hansen, A.S. Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging. Science 2022, 376, 496–501. [Google Scholar] [CrossRef]
- Li, Y.; Haarhuis, J.H.I.; Sedeño Cacciatore, Á.; Oldenkamp, R.; van Ruiten, M.S.; Willems, L.; Teunissen, H.; Muir, K.W.; de Wit, E.; Rowland, B.D.; et al. The structural basis for cohesin–CTCF-anchored loops. Nature 2020, 578, 472–476. [Google Scholar] [CrossRef]
- Wutz, G.; Várnai, C.; Nagasaka, K.; Cisneros, D.A.; Stocsits, R.R.; Tang, W.; Schoenfelder, S.; Jessberger, G.; Muhar, M.; Hossain, M.J.; et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017, 36, 3573–3599. [Google Scholar] [CrossRef]
- Li, J.; Huang, K.; Hu, G.; Babarinde, I.A.; Li, Y.; Dong, X.; Chen, Y.S.; Shang, L.; Guo, W.; Wang, J.; et al. An alternative CTCF isoform antagonizes canonical CTCF occupancy and changes chromatin architecture to promote apoptosis. Nat. Commun. 2019, 10, 1535. [Google Scholar] [CrossRef]
- Rivero-Hinojosa, S.; Pugacheva, E.M.; Kang, S.; Méndez-Catalá, C.F.; Kovalchuk, A.L.; Strunnikov, A.V.; Loukinov, D.; Lee, J.T.; Lobanenkov, V.V. The combined action of CTCF and its testis-specific paralog BORIS is essential for spermatogenesis. Nat. Commun. 2021, 12, 3846. [Google Scholar] [CrossRef]
- Loukinov, D.I.; Pugacheva, E.; Vatolin, S.; Pack, S.D.; Moon, H.; Chernukhin, I.; Mannan, P.; Larsson, E.; Kanduri, C.; Vostrov, A.A.; et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl. Acad. Sci. USA 2002, 99, 6806–6811. [Google Scholar] [CrossRef] [PubMed]
- Janssen, S.M.; Moscona, R.; Elchebly, M.; Papadakis, A.I.; Redpath, M.; Wang, H.; Rubin, E.; van Kempen, L.C.; Spatz, A. BORIS/CTCFL promotes a switch from a proliferative towards an invasive phenotype in melanoma cells. Cell Death Discov. 2020, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Pugacheva, E.M.; Rivero-Hinojosa, S.; Espinoza, C.A.; Méndez-Catalá, C.F.; Kang, S.; Suzuki, T.; Kosaka-Suzuki, N.; Robinson, S.; Nagarajan, V.; Ye, Z.; et al. Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome Biol. 2015, 16, 161. [Google Scholar] [CrossRef]
- Sleutels, F.; Soochit, W.; Bartkuhn, M.; Heath, H.; Dienstbach, S.; Bergmaier, P.; Franke, V.; Rosa-Garrido, M.; van de Nobelen, S.; Caesar, L.; et al. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics Chromatin 2012, 5, 8. [Google Scholar] [CrossRef]
- Bergmaier, P.; Weth, O.; Dienstbach, S.; Boettger, T.; Galjart, N.; Mernberger, M.; Bartkuhn, M.; Renkawitz, R. Choice of binding sites for CTCFL compared to CTCF is driven by chromatin and by sequence preference. Nucleic Acids Res. 2018, 46, 7097–7107. [Google Scholar] [CrossRef]
- Nishana, M.; Ha, C.; Rodriguez-Hernaez, J.; Ranjbaran, A.; Chio, E.; Nora, E.P.; Badri, S.B.; Kloetgen, A.; Bruneau, B.G.; Tsirigos, A.; et al. Defining the relative and combined contribution of CTCF and CTCFL to genomic regulation. Genome Biol. 2020, 21, 108. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.; Bar-Sela, G.; Sun, L.; Bisht, K.S.; Cui, H.; Kohn, E.; Feinberg, A.P.; Gius, D. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol. Cell. Biol. 2008, 28, 6720–6729. [Google Scholar] [CrossRef]
- Gaykalova, D.; Vatapalli, R.; Glazer, C.A.; Bhan, S.; Shao, C.; Sidransky, D.; Ha, P.K.; Califano, J.A. Dose-dependent activation of putative oncogene SBSN by BORIS. PLoS ONE 2012, 7, e40389. [Google Scholar] [CrossRef]
- Rosa-Garrido, M.; Ceballos, L.; Alonso-Lecue, P.; Abraira, C.; Delgado, M.D.; Gandarillas, A. A cell cycle role for the epigenetic factor CTCF-L/BORIS. PLoS ONE 2012, 7, e39371. [Google Scholar] [CrossRef]
- Kemp, C.J.; Moore, J.M.; Moser, R.; Bernard, B.; Teater, M.; Smith, L.E.; Rabaia, N.A.; Gurley, K.E.; Guinney, J.; Busch, S.E.; et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014, 7, 1020–1029. [Google Scholar] [CrossRef]
- Renaud, S.; Loukinov, D.; Alberti, L.; Vostrov, A.; Kwon, Y.W.; Bosman, F.T.; Lobanenkov, V.; Benhattar, J. BORIS/CTCFL-mediated transcriptional regulation of the hTERT telomerase gene in testicular and ovarian tumor cells. Nucleic Acids Res. 2011, 39, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Ulaner, G.A.; Vu, T.H.; Li, T.; Hu, J.F.; Yao, X.M.; Yang, Y.; Gorlick, R.; Meyers, P.; Healey, J.; Ladanyi, M.; et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum. Mol. Genet. 2003, 12, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Oshimura, M.; Nakao, M. CTCF-Dependent Chromatin Insulator Is Linked to Epigenetic Remodeling. Mol. Cell 2006, 23, 733–742. [Google Scholar] [CrossRef] [PubMed]
- van de Nobelen, S.; Rosa-Garrido, M.; Leers, J.; Heath, H.; Soochit, W.; Joosen, L.; Jonkers, I.; Demmers, J.; van der Reijden, M.; Torrano, V.; et al. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin 2010, 3, 19. [Google Scholar] [CrossRef]
- Jelinic, P.; Stehle, J.C.; Shaw, P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006, 4, e355. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Eichelser, C.; Steinbach, B.; Tadewaldt, J.; Pantel, K.; Lobanenkov, V.; Loukinov, D. Differential regulation of MAGE-A1 promoter activity by BORIS and Sp1, both interacting with the TATA binding protein. BMC Cancer 2014, 14, 796. [Google Scholar] [CrossRef]
- Kang, Y.; Hong, J.A.; Chen, G.A.; Nguyen, D.M.; Schrump, D.S. Dynamic transcriptional regulatory complexes including BORIS, CTCF and Sp1 modulate NY-ESO-1 expression in lung cancer cells. Oncogene 2007, 26, 4394–4403. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, M.; Li, S.; Xu, H.; Ren, J.; Tu, L.; Zuo, B.; Yao, W.; Liang, G. BTApep-TAT peptide inhibits ADP-ribosylation of BORIS to induce DNA damage in cancer. Mol. Cancer 2022, 21, 158. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Kotlyar, M.; Pastrello, C.; Ahmed, Z.; Chee, J.; Varyova, Z.; Jurisica, I. IID 2021: Towards context-specific protein interaction analyses by increased coverage, enhanced annotation and enrichment analysis. Nucleic Acids Res. 2022, 50, D640–D647. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, J.; Cao, C. CTCF and Its Partners: Shaper of 3D Genome during Development. Genes 2022, 13, 1383. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Calderas, L.; González-Barrios, R.; Patiño, C.C.; Alcaraz, N.; Salgado-Albarrán, M.; de León, D.C.; Hernández, C.C.; Sánchez-Pérez, Y.; Maldonado-Martínez, H.A.; De la Rosa-Velazquez, I.A.; et al. CTCF-KDM4A complex correlates with histone modifications that negatively regulate gene expression in cancer cell lines. Oncotarget 2018, 9, 17028–17042. [Google Scholar] [CrossRef]
- Yamamoto, S.; Wu, Z.; Russnes, H.G.; Takagi, S.; Peluffo, G.; Vaske, C.; Zhao, X.; Moen Vollan, H.K.; Maruyama, R.; Ekram, M.B.; et al. JARID1B Is a Luminal Lineage-Driving Oncogene in Breast Cancer. Cancer Cell 2014, 25, 762–777. [Google Scholar] [CrossRef]
- Allen, M.D.; Religa, T.L.; Freund, S.M.V.; Bycroft, M. Solution structure of the BRK domains from CHD7. J. Mol. Biol. 2007, 371, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yang, B.; Xie, X.; Wei, L.; Liu, W.; Yang, W.; Ge, Y.; Zhu, Q.; Zhang, J.; Jiang, L.; et al. Vigilin interacts with CCCTC-binding factor (CTCF) and is involved in CTCF-dependent regulation of the imprinted genes Igf2 and H19. FEBS J. 2014, 281, 2713–2725. [Google Scholar] [CrossRef]
- Yusufzai, T.M.; Tagami, H.; Nakatani, Y.; Felsenfeld, G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 2004, 13, 291–298. [Google Scholar] [CrossRef]
- Witcher, M.; Emerson, B.M. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell 2009, 34, 271–284. [Google Scholar] [CrossRef]
- Marino, M.M.; Rega, C.; Russo, R.; Valletta, M.; Gentile, M.T.; Esposito, S.; Baglivo, I.; De Feis, I.; Angelini, C.; Xiao, T.; et al. Interactome mapping defines BRG1, a component of the SWI/SNF chromatin remodeling complex, as a new partner of the transcriptional regulator CTCF. J. Biol. Chem. 2019, 294, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, M.E.; Zhang, L.F.; Xu, N.; Shi, Y.; Lee, J.T. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell 2007, 25, 43–56. [Google Scholar] [CrossRef]
- Wiles, E.T.; Selker, E.U. H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 2017, 43, 31–37. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Niu, B.; Hu, J.F.; Ge, S.; Wang, H.; Li, T.; Ling, J.; Steelman, B.N.; Qian, G.; Hoffman, A.R. Interruption of intrachromosomal looping by CCCTC binding factor decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. J. Cell Biol. 2011, 193, 475–487. [Google Scholar] [CrossRef]
- Li, T.; Hu, J.F.; Qiu, X.; Ling, J.; Chen, H.; Wang, S.; Hou, A.; Vu, T.H.; Hoffman, A.R. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 2008, 28, 6473–6482. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Jia, L.; Huang, X.; Tan, J.; Wang, M.; Niu, J.; Hou, Y.; Sun, J.; Zeng, P.; Wang, J.; et al. CTCF organizes inter-A compartment interactions through RYBP-dependent phase separation. Cell Res. 2022, 32, 744–760. [Google Scholar] [CrossRef]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Guerra-Calderas, L.; González-Barrios, R.; Herrera, L.A.; Cantú de León, D.; Soto-Reyes, E. The role of the histone demethylase KDM4A in cancer. Cancer Genet. 2015, 208, 215–224. [Google Scholar] [CrossRef]
- Sterling, J.; Menezes, S.V.; Abbassi, R.H.; Munoz, L. Histone lysine demethylases and their functions in cancer. Int. J. Cancer 2020, 148, 2375–2388. [Google Scholar] [CrossRef]
- Anand, R.; Marmorstein, R. Structure and Mechanism of Lysine-specific Demethylase Enzymes. J. Biol. Chem. 2007, 282, 35425–35429. [Google Scholar] [CrossRef]
- Yang, M.; Gocke, C.B.; Luo, X.; Borek, D.; Tomchick, D.R.; Machius, M.; Otwinowski, Z.; Yu, H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell 2006, 23, 377–387. [Google Scholar] [CrossRef]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Couture, J.F.; Collazo, E.; Ortiz-Tello, P.A.; Brunzelle, J.S.; Trievel, R.C. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007, 14, 689–695. [Google Scholar] [CrossRef]
- Ng, S.S.; Kavanagh, K.L.; McDonough, M.A.; Butler, D.; Pilka, E.S.; Lienard, B.M.R.; Bray, J.E.; Savitsky, P.; Gileadi, O.; von Delft, F.; et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 2007, 448, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Nottke, A.C.; Shi, Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 2008, 20, 316. [Google Scholar] [CrossRef]
- Shiau, C.; Trnka, M.J.; Bozicevic, A.; Ortiz Torres, I.; Al-Sady, B.; Burlingame, A.L.; Narlikar, G.J.; Fujimori, D.G. Reconstitution of nucleosome demethylation and catalytic properties of a Jumonji histone demethylase. Chem. Biol. 2013, 20, 494–499. [Google Scholar] [CrossRef]
- Ruthenburg, A.J.; Allis, C.D.; Wysocka, J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell 2007, 25, 15–30. [Google Scholar] [CrossRef]
- Lan, F.; Zaratiegui, M.; Villén, J.; Vaughn, M.W.; Verdel, A.; Huarte, M.; Shi, Y.; Gygi, S.P.; Moazed, D.; Martienssen, R.A.; et al. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol. Cell 2007, 26, 89–101. [Google Scholar] [CrossRef]
- Metzger, E.; Wissmann, M.; Yin, N.; Müller, J.M.; Schneider, R.; Peters, A.H.F.M.; Günther, T.; Buettner, R.; Schüle, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bassets, I.; Kwon, Y.S.; Telese, F.; Prefontaine, G.G.; Hutt, K.R.; Cheng, C.S.; Ju, B.G.; Ohgi, K.A.; Wang, J.; Escoubet-Lozach, L.; et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 2007, 128, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Scully, K.; Zhu, X.; Cai, L.; Zhang, J.; Prefontaine, G.G.; Krones, A.; Ohgi, K.A.; Zhu, P.; Garcia-Bassets, I.; et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007, 446, 882–887. [Google Scholar] [CrossRef]
- Kristensen, L.H.; Nielsen, A.L.; Helgstrand, C.; Lees, M.; Cloos, P.; Kastrup, J.S.; Helin, K.; Olsen, L.; Gajhede, M. Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J. 2012, 279, 1905–1914. [Google Scholar] [CrossRef]
- Jeong, Y.S.; Park, J.S.; Ko, Y.; Kang, Y.K. JHDM3A module as an effector molecule in guide-directed modification of target chromatin. J. Biol. Chem. 2011, 286, 4461–4470. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2012, 38, 23–38. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Genest, P.A.; ter Riet, B.; Sweeney, K.; DiPaolo, C.; Kieft, R.; Christodoulou, E.; Perrakis, A.; Simmons, J.M.; Hausinger, R.P.; et al. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007, 35, 2107–2115. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef]
- He, Y.F.; Li, B.Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992, 69, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Guo, B.; Wu, H.; Tan, L.; Lu, Q. TET Family of Dioxygenases: Crucial Roles and Underlying Mechanisms. Cytogenet. Genome Res. 2015, 146, 171–180. [Google Scholar] [CrossRef]
- Williams, K.; Christensen, J.; Helin, K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011, 13, 28–35. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Clark, E.; Mulcahey, M.; Huang, S.; Shi, Y.G. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010, 20, 1390–1393. [Google Scholar] [CrossRef]
- Wu, H.; D’Alessio, A.C.; Ito, S.; Xia, K.; Wang, Z.; Cui, K.; Zhao, K.; Sun, Y.E.; Zhang, Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 2011, 473, 389–393. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, F.; Tan, L.; Kong, L.; Xiong, L.; Deng, J.; Barbera, A.J.; Zheng, L.; Zhang, H.; Huang, S.; et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol. Cell 2011, 42, 451–464. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, C.; Kato, A.; Tempel, W.; Abreu, J.G.; Bian, C.; Hu, Y.; Hu, D.; Zhao, B.; Cerovina, T.; et al. Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development. Cell 2012, 151, 1200–1213. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.; An, J.; Bandukwala, H.S.; Chavez, L.; Aijö, T.; Pastor, W.A.; Segal, M.F.; Li, H.; Koh, K.P.; Lähdesmäki, H.; et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 2013, 497, 122–126. [Google Scholar] [CrossRef]
- Guilhamon, P.; Eskandarpour, M.; Halai, D.; Wilson, G.A.; Feber, A.; Teschendorff, A.E.; Gomez, V.; Hergovich, A.; Tirabosco, R.; Fernanda Amary, M.; et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat. Commun. 2013, 4, 2166. [Google Scholar] [CrossRef]
- Rampal, R.; Alkalin, A.; Madzo, J.; Vasanthakumar, A.; Pronier, E.; Patel, J.; Li, Y.; Ahn, J.; Abdel-Wahab, O.; Shih, A.; et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014, 9, 1841–1855. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, M.; Chen, X.; Chen, L.; Xu, Y.; Lv, L.; Wang, P.; Yang, H.; Ma, S.; Lin, H.; et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol. Cell 2015, 57, 662–673. [Google Scholar] [CrossRef]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y. Regulation of TET protein stability by calpains. Cell Rep. 2014, 6, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Teif, V.B.; Beshnova, D.A.; Vainshtein, Y.; Marth, C.; Mallm, J.P.; Höfer, T.; Rippe, K. Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development. Genome Res. 2014, 24, 1285–1295. [Google Scholar] [CrossRef]
- Stadler, M.B.; Murr, R.; Burger, L.; Ivanek, R.; Lienert, F.; Schöler, A.; van Nimwegen, E.; Wirbelauer, C.; Oakeley, E.J.; Gaidatzis, D.; et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011, 480, 490–495. [Google Scholar] [CrossRef]
- Feldmann, A.; Ivanek, R.; Murr, R.; Gaidatzis, D.; Burger, L.; Schübeler, D. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 2013, 9, e1003994. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Terragni, J.; Borgaro, J.G.; Liu, Y.; Yu, L.; Guan, S.; Wang, H.; Sun, D.; Cheng, X.; Zhu, Z.; et al. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep. 2013, 3, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Dubois-Chevalier, J.; Oger, F.; Dehondt, H.; Firmin, F.F.; Gheeraert, C.; Staels, B.; Lefebvre, P.; Eeckhoute, J. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res. 2014, 42, 10943–10959. [Google Scholar] [CrossRef]
- Nanan, K.K.; Sturgill, D.M.; Prigge, M.F.; Thenoz, M.; Dillman, A.A.; Mandler, M.D.; Oberdoerffer, S. TET-Catalyzed 5-Carboxylcytosine Promotes CTCF Binding to Suboptimal Sequences Genome-wide. iScience 2019, 19, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef]
- Kuang, S.; Wang, L. Identification and analysis of consensus RNA motifs binding to the genome regulator CTCF. NAR Genom. Bioinform. 2020, 2, lqaa031. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Del Rosario, B.C.; Szanto, A.; Ogawa, Y.; Jeon, Y.; Lee, J.T. Jpx RNA activates Xist by evicting CTCF. Cell 2013, 153, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Sun, S.; Yan, M.; Cui, J.; Yang, Y.; Li, W.; Huang, X.; Dou, L.; Chen, B.; Tang, W.; et al. DDX5 and DDX17—Multifaceted proteins in the regulation of tumorigenesis and tumor progression. Front. Oncol. 2022, 12, 943032. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Brick, K.; Evrard, Y.; Xiao, T.; Camerini-Otero, R.D.; Felsenfeld, G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010, 24, 2543–2555. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, J.; Deng, J.; Chen, Y. Long non-coding RNA HOTTIP induces inflammation in asthma by promoting EFNA3 transcription by CCCTC-binding factor. Am. J. Transl. Res. 2022, 14, 8903–8917. [Google Scholar]
- Soibam, B. Association between Triplex-Forming Sites of Cardiac Long Noncoding RNA and Chromatin Organization. Noncoding RNA 2022, 8, 41. [Google Scholar] [CrossRef]
- Karner, H.; Webb, C.H.; Carmona, S.; Liu, Y.; Lin, B.; Erhard, M.; Chan, D.; Baldi, P.; Spitale, R.C.; Sun, S. Functional Conservation of LncRNA JPX Despite Sequence and Structural Divergence. J. Mol. Biol. 2020, 432, 283–300. [Google Scholar] [CrossRef]
- Ren, C.; Han, H.; Pan, J.; Chang, Q.; Wang, W.; Guo, X.; Bian, J. DLGAP1-AS2 promotes human colorectal cancer progression through trans-activation of Myc. Mamm. Genome 2022, 33, 672–683. [Google Scholar] [CrossRef]
- Chen, Q.; Jiang, S.; Liu, H.; Gao, Y.; Yang, X.; Ren, Z.; Gao, Y.; Xiao, L.; Hu, H.; Yu, Y.; et al. Association of lncRNA SH3PXD2A-AS1 with preeclampsia and its function in invasion and migration of placental trophoblast cells. Cell Death Dis. 2020, 11, 583. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Zhu, Y.; Cao, Y.; Wang, L.; Li, F.; Zhang, Y.; Li, Y.; Zhang, Z.; Luo, J.; et al. The CTCF/LncRNA-PACERR complex recruits E1A binding protein p300 to induce pro-tumour macrophages in pancreatic ductal adenocarcinoma via directly regulating PTGS2 expression. Clin. Transl. Med. 2022, 12, e654. [Google Scholar] [CrossRef] [PubMed]
- Sur, S.; Steele, R.; Ko, B.C.B.; Zhang, J.; Ray, R.B. Long noncoding RNA ELDR promotes cell cycle progression in normal oral keratinocytes through induction of a CTCF-FOXM1-AURKA signaling axis. J. Biol. Chem. 2022, 298, 101895. [Google Scholar] [CrossRef]
- Ou, M.; Li, X.; Zhao, S.; Cui, S.; Tu, J. Long non-coding RNA CDKN2B-AS1 contributes to atherosclerotic plaque formation by forming RNA-DNA triplex in the CDKN2B promoter. EBioMedicine 2020, 55, 102694. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.X.; He, R.Z.; Zhang, Z.; Yang, L.; Mo, Y.Y. LINC00346 promotes pancreatic cancer progression through the CTCF-mediated Myc transcription. Oncogene 2019, 38, 6770–6780. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liu, Q.; He, J.; Huang, Y.; Jiang, L.; Xie, X.; Liu, J.; Chen, L.; Wei, L.; Qin, Y. Vigilin interacts with CTCF and is involved in the maintenance of imprinting of IGF2 through a novel RNA-mediated mechanism. Int. J. Biol. Macromol. 2018, 108, 515–522. [Google Scholar] [CrossRef]
- Yang, F.; Deng, X.; Ma, W.; Berletch, J.B.; Rabaia, N.; Wei, G.; Moore, J.M.; Filippova, G.N.; Xu, J.; Liu, Y.; et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015, 16, 52. [Google Scholar] [CrossRef]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).