The Clinical Significance and Role of CXCL1 Chemokine in Gastrointestinal Cancers
Abstract
1. Introduction
2. Head and Neck Cancer
3. Esophageal Cancer
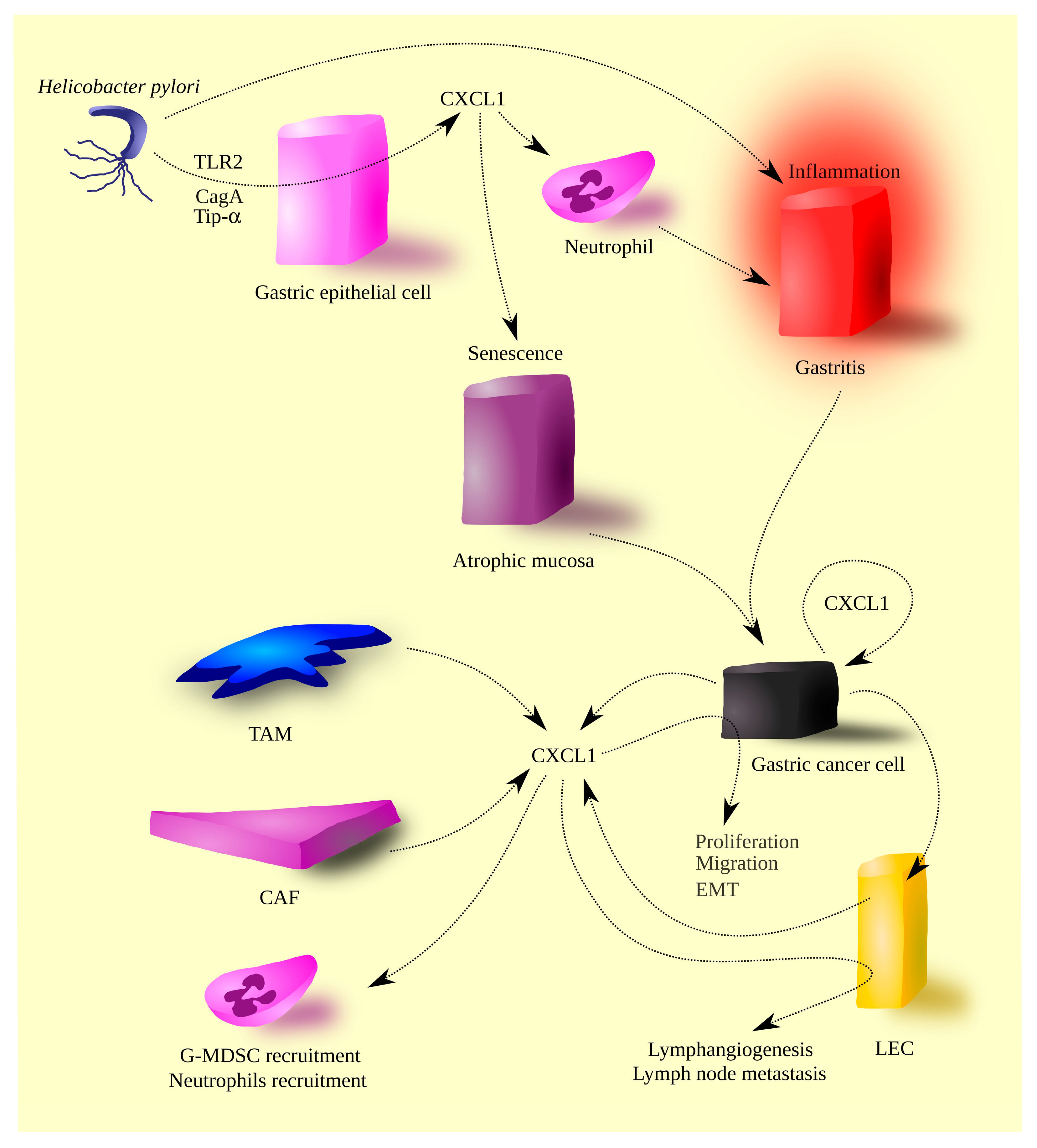
4. Gastric Cancer
5. Liver Cancer
- Chronic hepatitis B virus (HBV) infection;
- Chronic hepatitis C virus (HCV) infection;
- Consuming large amounts of alcohol, which leads to alcoholic liver disease (ALD), then to liver cirrhosis, and eventually HCC;
- Obesity, which leads to non-alcoholic fatty liver disease (NAFLD), then liver cirrhosis, and, eventually, HCC.
6. Cholangiocarcinoma
7. Pancreatic Cancer
8. Colorectal Cancer
9. CXCL1 as a Therapeutic Target in Anticancer Therapy of Gastrointestinal Tumors
10. Future Perspectives
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sabry, M.; Lowdell, M.W. Tumor-primed NK cells: Waiting for the green light. Front. Immunol. 2013, 4, 408. [Google Scholar] [CrossRef] [PubMed]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in cancer: Neutral no more. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef]
- Kumar, V.; Patel, S.; Tcyganov, E.; Gabrilovich, D.I. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol. 2016, 37, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Andersen, M.H. The role of dendritic cells in cancer. Semin. Immunopathol. 2017, 39, 307–316. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Contribution of regulatory T cells to cancer: A review. J. Cell Physiol. 2019, 234, 7983–7993. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, K.; Hana, D.; Chou, J.T.; Singh, C.; Mackiewicz, A.; Kaczmarek, M. Aspects of the Tumor Microenvironment Involved in Immune Resistance and Drug Resistance. Front. Immunol. 2021, 12, 656364. [Google Scholar] [CrossRef]
- Dinarello, C.A. Historical insights into cytokines. Eur. J. Immunol. 2007, 37 (Suppl. S1), S34–S45. [Google Scholar] [CrossRef]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- Balentien, E.; Han, J.H.; Thomas, H.G.; Wen, D.Z.; Samantha, A.K.; Zachariae, C.O.; Griffin, P.R.; Brachmann, R.; Wong, W.L.; Matsushima, K.; et al. Recombinant expression, biochemical characterization, and biological activities of the human MGSA/gro protein. Biochemistry 1990, 29, 10225–10233. [Google Scholar]
- Shattuck, R.L.; Wood, L.D.; Jaffe, G.J.; Richmond, A. MGSA/GRO transcription is differentially regulated in normal retinal pigment epithelial and melanoma cells. Mol. Cell Biol. 1994, 14, 791–802. [Google Scholar] [PubMed]
- Herjan, T.; Hong, L.; Bubenik, J.; Bulek, K.; Qian, W.; Liu, C.; Li, X.; Chen, X.; Yang, H.; Ouyang, S.; et al. IL-17-receptor-associated adaptor Act1 directly stabilizes mRNAs to mediate IL-17 inflammatory signaling. Nat. Immunol. 2018, 19, 354–365. [Google Scholar] [CrossRef]
- Loetscher, P.; Seitz, M.; Clark-Lewis, I.; Baggiolini, M.; Moser, B. Both interleukin-8 receptors independently mediate chemotaxis. Jurkat cells transfected with IL-8R1 or IL-8R2 migrate in response to IL-8, GRO alpha and NAP-2. FEBS Lett. 1994, 341, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Szabo, M.C.; Soo, K.S.; Zlotnik, A.; Schall, T.J. Chemokine class differences in binding to the Duffy antigen-erythrocyte chemokine receptor. J. Biol. Chem. 1995, 270, 25348–25351. [Google Scholar] [CrossRef]
- Fukuma, N.; Akimitsu, N.; Hamamoto, H.; Kusuhara, H.; Sugiyama, Y.; Sekimizu, K. A role of the Duffy antigen for the maintenance of plasma chemokine concentrations. Biochem. Biophys. Res. Commun. 2003, 303, 137–139. [Google Scholar] [CrossRef]
- Lee, J.S.; Frevert, C.W.; Wurfel, M.M.; Peiper, S.C.; Wong, V.A.; Ballman, K.K.; Ruzinski, J.T.; Rhim, J.S.; Martin, T.R.; Goodman, R.B. Duffy antigen facilitates movement of chemokine across the endothelium in vitro and promotes neutrophil transmigration in vitro and in vivo. J. Immunol. 2003, 170, 5244–5251. [Google Scholar] [CrossRef] [PubMed]
- Kuwano, Y.; Adler, M.; Zhang, H.; Groisman, A.; Ley, K. Gαi2 and Gαi3 differentially regulate arrest from flow and chemotaxis in mouse neutrophils. J. Immunol. 2016, 196, 3828–3833. [Google Scholar] [CrossRef]
- Raman, D.; Neel, N.F.; Sai, J.; Mernaugh, R.L.; Ham, A.J.; Richmond, A.J. Characterization of chemokine receptor CXCR2 interacting proteins using a proteomics approach to define the CXCR2 “chemosynapse”. Methods Enzymol. 2009, 460, 315–330. [Google Scholar]
- Moser, B.; Clark-Lewis, I.; Zwahlen, R.; Baggiolini, M. Neutrophil-activating properties of the melanoma growth-stimulatory activity. J. Exp. Med. 1990, 171, 1797–1802. [Google Scholar] [CrossRef]
- Richmond, A.; Lawson, D.H.; Nixon, D.W.; Chawla, R.K. Characterization of autostimulatory and transforming growth factors from human melanoma cells. Cancer Res. 1985, 45, 6390–6394. [Google Scholar]
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and neck cancer. Lancet 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Rettig, E.M.; D’Souza, G. Epidemiology of head and neck cancer. Surg. Oncol. Clin. N. Am. 2015, 24, 379–396. [Google Scholar] [CrossRef] [PubMed]
- Kolář, M.; Szabo, P.; Dvořánková, B.; Lacina, L.; Gabius, H.J.; Strnad, H.; Sáchová, J.; Vlček, C.; Plzák, J.; Chovanec, M.; et al. Upregulation of IL-6, IL-8 and CXCL-1 production in dermal fibroblasts by normal/malignant epithelial cells in vitro: Immunohistochemical and transcriptomic analyses. Biol. Cell 2012, 104, 738–751. [Google Scholar] [CrossRef]
- Ye, M.Y.; Chen, M.Y.; Chang, Y.H.; Huang, J.S.; Huang, T.T.; Wong, T.Y.; Hong, T.M.; Chen, Y.L. Growth-regulated oncogene-α from oral submucous fibrosis fibroblasts promotes malignant transformation of oral precancerous cells. J. Oral Pathol. Med. 2018, 47, 880–886. [Google Scholar] [CrossRef]
- Jing, F.; Wang, J.; Zhou, L.; Ning, Y.; Xu, S.; Zhu, Y. Bioinformatics analysis of the role of CXC ligands in the microenvironment of head and neck tumor. Aging 2021, 13, 17789–17817. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, T.; Gong, S.; Zhou, H.; Yu, L.; Liang, M.; Shi, R.; Wu, Z.; Zhang, J.; Li, S. Analysis of the Prognosis and Therapeutic Value of the CXC Chemokine Family in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2021, 10, 570736. [Google Scholar] [CrossRef]
- Shintani, S.; Ishikawa, T.; Nonaka, T.; Li, C.; Nakashiro, K.; Wong, D.T.; Hamakawa, H. Growth-regulated oncogene-1 expression is associated with angiogenesis and lymph node metastasis in human oral cancer. Oncology 2004, 66, 316–322. [Google Scholar] [CrossRef]
- Ye, H.; Yu, T.; Temam, S.; Ziober, B.L.; Wang, J.; Schwartz, J.L.; Mao, L.; Wong, D.T.; Zhou, X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genom. 2008, 9, 69. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Multani, S.; Dabholkar, J.; Saranath, D. Whole genome expression profiling in chewing-tobacco-associated oral cancers: A pilot study. Med. Oncol. 2015, 32, 60. [Google Scholar] [CrossRef]
- Wei, L.Y.; Lee, J.J.; Yeh, C.Y.; Yang, C.J.; Kok, S.H.; Ko, J.Y.; Tsai, F.C.; Chia, J.S. Reciprocal activation of cancer-associated fibroblasts and oral squamous carcinoma cells through CXCL1. Oral Oncol. 2019, 88, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Xu, X. Expression of GRO-1 and its relationship with VEGF in squamous cell carcinoma of larynx. Lin Chuang Er Bi Yan Hou Ke Za Zhi 2006, 20, 541–544. [Google Scholar] [PubMed]
- Han, L.; Liu, W.; Chen, Y.; Wu, H.; Zhang, Y.; Jiang, B. GROα expression and its prognostic implications in laryngeal squamous cell carcinoma. Neoplasma 2015, 62, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Reyimu, A.; Chen, Y.; Song, X.; Zhou, W.; Dai, J.; Jiang, F. Identification of latent biomarkers in connection with progression and prognosis in oral cancer by comprehensive bioinformatics analysis. World J. Surg. Oncol. 2021, 19, 240. [Google Scholar] [CrossRef]
- Yang, B.; Dong, K.; Guo, P.; Guo, P.; Jie, G.; Zhang, G.; Li, T. Identification of Key Biomarkers and Potential Molecular Mechanisms in Oral Squamous Cell Carcinoma by Bioinformatics Analysis. J. Comput. Biol. 2020, 27, 40–54. [Google Scholar] [CrossRef]
- Sun, W.; Qiu, Z.; Huang, W.; Cao, M. Gene expression profiles and protein-protein interaction networks during tongue carcinogenesis in the tumor microenvironment. Mol. Med. Rep. 2018, 17, 165–171. [Google Scholar] [CrossRef]
- Brøndum, L.; Eriksen, J.G.; Singers Sørensen, B.; Mortensen, L.S.; Toustrup, K.; Overgaard, J.; Alsner, J. Plasma proteins as prognostic biomarkers in radiotherapy treated head and neck cancer patients. Clin. Transl. Radiat. Oncol. 2017, 2, 46–52. [Google Scholar] [CrossRef]
- Michiels, K.; Schutyser, E.; Conings, R.; Lenaerts, J.P.; Put, W.; Nuyts, S.; Delaere, P.; Jacobs, R.; Struyf, S.; Proost, P.; et al. Carcinoma cell-derived chemokines and their presence in oral fluid. Eur. J. Oral Sci. 2009, 117, 362–368. [Google Scholar] [CrossRef]
- Wolff, H.A.; Rolke, D.; Rave-Fränk, M.; Schirmer, M.; Eicheler, W.; Doerfler, A.; Hille, A.; Hess, C.F.; Matthias, C.; Rödel, R.M.; et al. Analysis of chemokine and chemokine receptor expression in squamous cell carcinoma of the head and neck (SCCHN) cell lines. Radiat. Environ. Biophys. 2011, 50, 145–154. [Google Scholar] [CrossRef]
- Lee, C.H.; Syu, S.H.; Liu, K.J.; Chu, P.Y.; Yang, W.C.; Lin, P.; Shieh, W.Y. Interleukin-1 beta transactivates epidermal growth factor receptor via the CXCL1-CXCR2 axis in oral cancer. Oncotarget 2015, 6, 38866–38880. [Google Scholar] [CrossRef]
- Kaneko, T.; Zhang, Z.; Mantellini, M.G.; Karl, E.; Zeitlin, B.; Verhaegen, M.; Soengas, M.S.; Lingen, M.; Strieter, R.M.; Nunez, G.; et al. Bcl-2 orchestrates a cross-talk between endothelial and tumor cells that promotes tumor growth. Cancer Res. 2007, 67, 9685–9693. [Google Scholar] [CrossRef]
- Karl, E.; Warner, K.; Zeitlin, B.; Kaneko, T.; Wurtzel, L.; Jin, T.; Chang, J.; Wang, S.; Wang, C.Y.; Strieter, R.M.; et al. Bcl-2 acts in a proangiogenic signaling pathway through nuclear factor-kappaB and CXC chemokines. Cancer Res. 2005, 65, 5063–5069. [Google Scholar] [CrossRef]
- Loukinova, E.; Chen, Z.; Van Waes, C.; Dong, G. Expression of proangiogenic chemokine Gro 1 in low and high metastatic variants of Pam murine squamous cell carcinoma is differentially regulated by IL-1alpha, EGF and TGF-beta1 through NF-kappaB dependent and independent mechanisms. Int. J. Cancer 2001, 94, 637–644. [Google Scholar] [CrossRef]
- Lyons, J.G.; Patel, V.; Roue, N.C.; Fok, S.Y.; Soon, L.L.; Halliday, G.M.; Gutkind, J.S. Snail up-regulates proinflammatory mediators and inhibits differentiation in oral keratinocytes. Cancer Res. 2008, 68, 4525–4530. [Google Scholar] [CrossRef]
- Bae, J.Y.; Kim, E.K.; Yang, D.H.; Zhang, X.; Park, Y.J.; Lee, D.Y.; Che, C.M.; Kim, J. Reciprocal interaction between carcinoma-associated fibroblasts and squamous carcinoma cells through interleukin-1α induces cancer progression. Neoplasia 2014, 16, 928–938. [Google Scholar] [CrossRef]
- Kim, E.K.; Moon, S.; Kim, D.K.; Zhang, X.; Kim, J. CXCL1 induces senescence of cancer-associated fibroblasts via autocrine loops in oral squamous cell carcinoma. PLoS ONE 2018, 13, e0188847. [Google Scholar] [CrossRef]
- Khurram, S.A.; Bingle, L.; McCabe, B.M.; Farthing, P.M.; Whawell, S.A. The chemokine receptors CXCR1 and CXCR2 regulate oral cancer cell behaviour. J. Oral Pathol. Med. 2014, 43, 667–674. [Google Scholar] [CrossRef]
- Zhang, Z.; Pan, J.; Li, L.; Han, B.; Xiao, W. Altered phenotype of lymphatic endothelial cells induced by highly metastatic OTSCC cells contributed to the lymphatic metastasis of OTSCC cells. Cancer Sci. 2010, 101, 686–692. [Google Scholar] [CrossRef]
- Warner, K.A.; Miyazawa, M.; Cordeiro, M.M.; Love, W.J.; Pinsky, M.S.; Neiva, K.G.; Spalding, A.C.; Nör, J.E. Endothelial cells enhance tumor cell invasion through a crosstalk mediated by CXC chemokine signaling. Neoplasia 2008, 10, 131–139. [Google Scholar] [CrossRef]
- Brú, A.; Souto, J.C.; Alcolea, S.; Antón, R.; Remacha, A.; Camacho, M.; Soler, M.; Brú, I.; Porres, A.; Vila, L. Tumour cell lines HT-29 and FaDu produce proinflammatory cytokines and activate neutrophils in vitro: Possible applications for neutrophil-based antitumour treatment. Mediat. Inflamm. 2009, 2009, 817498. [Google Scholar] [CrossRef]
- Dufies, M.; Grytsai, O.; Ronco, C.; Camara, O.; Ambrosetti, D.; Hagege, A.; Parola, J.; Mateo, L.; Ayrault, M.; Giuliano, S.; et al. New CXCR1/CXCR2 inhibitors represent an effective treatment for kidney or head and neck cancers sensitive or refractory to reference treatments. Theranostics 2019, 9, 5332–5346. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Lagergren, J.; Fitzgerald, R.C.; Lordick, F.; Shah, M.A.; Lagergren, P.; Cunningham, D. Oesophageal cancer. Nat. Rev. Dis. Prim. 2017, 3, 17048. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Hendricks, D.T.; Wamunyokoli, F.; Parker, M.I. A growth-related oncogene/CXC chemokine receptor 2 autocrine loop contributes to cellular proliferation in esophageal cancer. Cancer Res. 2006, 66, 3071–3077. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Qu, J.; Liu, X.; Liang, J.; Li, Y.; Jiang, J.; Zhang, H.; Tian, H. Integrated bioinformatics analysis of differentially expressed genes and immune cell infiltration characteristics in Esophageal Squamous cell carcinoma. Sci. Rep. 2021, 11, 16696. [Google Scholar] [CrossRef]
- Zhang, H.; Yue, J.; Jiang, Z.; Zhou, R.; Xie, R.; Xu, Y.; Wu, S. CAF-secreted CXCL1 conferred radioresistance by regulating DNA damage response in a ROS-dependent manner in esophageal squamous cell carcinoma. Cell Death Dis. 2017, 8, e2790. [Google Scholar] [CrossRef]
- Hasib, F.M.Y. Esophageal squamous cell carcinoma: Integrated bioinformatics analysis for differential gene expression with identification of hub genes and lncRNA. Biochem. Biophys. Rep. 2022, 30, 101262. [Google Scholar] [CrossRef]
- Aversa, J.; Song, M.; Shimazu, T.; Inoue, M.; Charvat, H.; Yamaji, T.; Sawada, N.; Pfeiffer, R.M.; Karimi, P.; Dawsey, S.M.; et al. Prediagnostic circulating inflammation biomarkers and esophageal squamous cell carcinoma: A case-cohort study in Japan. Int. J. Cancer 2020, 147, 686–691. [Google Scholar] [CrossRef]
- Alvarez, H.; Opalinska, J.; Zhou, L.; Sohal, D.; Fazzari, M.J.; Yu, Y.; Montagna, C.; Montgomery, E.A.; Canto, M.; Dunbar, K.B.; et al. Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PLoS Genet. 2011, 7, e1001356. [Google Scholar] [CrossRef]
- Fang, L.; Che, Y.; Zhang, C.; Huang, J.; Lei, Y.; Lu, Z.; Sun, N.; He, J. LAMC1 upregulation via TGFβ induces inflammatory cancer-associated fibroblasts in esophageal squamous cell carcinoma via NF-κB-CXCL1-STAT3. Mol. Oncol. 2021, 15, 3125–3146. [Google Scholar] [CrossRef]
- Wang, B.; Khachigian, L.M.; Esau, L.; Birrer, M.J.; Zhao, X.; Parker, M.I.; Hendricks, D.T. A key role for early growth response-1 and nuclear factor-kappaB in mediating and maintaining GRO/CXCR2 proliferative signaling in esophageal cancer. Mol. Cancer Res. 2009, 7, 755–764. [Google Scholar] [CrossRef]
- Urakawa, N.; Utsunomiya, S.; Nishio, M.; Shigeoka, M.; Takase, N.; Arai, N.; Kakeji, Y.; Koma, Y.; Yokozaki, H. GDF15 derived from both tumor-associated macrophages and esophageal squamous cell carcinomas contributes to tumor progression via Akt and Erk pathways. Lab. Investig. 2015, 95, 491–503. [Google Scholar] [CrossRef]
- Peng, D.F.; Hu, T.L.; Soutto, M.; Belkhiri, A.; El-Rifai, W. Loss of glutathione peroxidase 7 promotes TNF-α-induced NF-κB activation in Barrett’s carcinogenesis. Carcinogenesis 2014, 35, 1620–1628. [Google Scholar] [CrossRef]
- Qin, G.; Lian, J.; Huang, L.; Zhao, Q.; Liu, S.; Zhang, Z.; Chen, X.; Yue, D.; Li, L.; Li, F.; et al. Metformin blocks myeloid-derived suppressor cell accumulation through AMPK-DACH1-CXCL1 axis. Oncoimmunology 2018, 7, e1442167. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Yan, L.; Chen, Y.; Chen, F.; Tao, T.; Hu, Z.; Wang, J.; You, J.; Wong, B.C.Y.; Chen, J.; Ye, W. Effect of Helicobacter pylori Eradication on Gastric Cancer Prevention: Updated Report from a Randomized Controlled Trial with 26.5 Years of Follow-up. Gastroenterology 2022, 163, 154–162.e3. [Google Scholar] [CrossRef]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef]
- Yin, H.; Chu, A.; Liu, S.; Yuan, Y.; Gong, Y. Identification of DEGs and transcription factors involved in H. pylori-associated inflammation and their relevance with gastric cancer. PeerJ 2020, 8, e9223. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.K.; Liao, L.M.; Huang, R.L.; Zhou, W. The relationship between gastric cancer and Helicobacter pylori cytotoxin-related gene A genotypes. Cell Mol. Biol. 2020, 66, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Cover, T.L.; Lacy, D.B.; Ohi, M.D. The Helicobacter pylori Cag Type IV Secretion System. Trends Microbiol. 2020, 28, 682–695. [Google Scholar] [CrossRef]
- de Brito, B.B.; da Silva, F.A.F.; Soares, A.S.; Pereira, V.A.; Santos, M.L.C.; Sampaio, M.M.; Neves, P.H.M.; de Melo, F.F. Pathogenesis and clinical management of Helicobacter pylori gastric infection. World J. Gastroenterol. 2019, 25, 5578–5589. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Kita, M.; Kodama, T.; Sawai, N.; Tanahashi, T.; Kashima, K.; Imanishi, J. Chemokines in the gastric mucosa in Helicobacter pylori infection. Gut 1998, 42, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Shi, P.; Yuan, Y.; Peng, J.; Ou, X.; Zhou, W.; Li, J.; Su, T.; Lin, L.; Cai, S.; et al. Inflammation-Associated Senescence Promotes Helicobacter pylori-Induced Atrophic Gastritis. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 857–880. [Google Scholar] [CrossRef]
- Tran, C.T.; Garcia, M.; Garnier, M.; Burucoa, C.; Bodet, C. Inflammatory signaling pathways induced by Helicobacter pylori in primary human gastric epithelial cells. Innate Immun. 2017, 23, 165–174. [Google Scholar] [CrossRef]
- Mustapha, P.; Paris, I.; Garcia, M.; Tran, C.T.; Cremniter, J.; Garnier, M.; Faure, J.P.; Barthes, T.; Boneca, I.G.; Morel, F.; et al. Chemokines and antimicrobial peptides have a cag-dependent early response to Helicobacter pylori infection in primary human gastric epithelial cells. Infect. Immun. 2014, 82, 2881–2889. [Google Scholar] [CrossRef]
- Suzuki, H.; Mori, M.; Sakaguchi, A.A.; Suzuki, M.; Miura, S.; Ishii, H. Enhanced levels of C-X-C chemokine, human GROalpha, in Helicobacter pylori-associated gastric disease. J. Gastroenterol. Hepatol. 1998, 13, 516–520. [Google Scholar] [CrossRef]
- Sierra, J.C.; Asim, M.; Verriere, T.G.; Piazuelo, M.B.; Suarez, G.; Romero-Gallo, J.; Delgado, A.G.; Wroblewski, L.E.; Barry, D.P.; Peek, R.M., Jr.; et al. Epidermal growth factor receptor inhibition downregulates Helicobacter pylori-induced epithelial inflammatory responses, DNA damage and gastric carcinogenesis. Gut 2018, 67, 1247–1260. [Google Scholar] [CrossRef]
- Neuper, T.; Frauenlob, T.; Sarajlic, M.; Posselt, G.; Wessler, S.; Horejs-Hoeck, J. TLR2, TLR4 and TLR10 Shape the Cytokine and Chemokine Release of H. pylori-Infected Human DCs. Int. J. Mol. Sci. 2020, 21, 3897. [Google Scholar] [CrossRef]
- Kuzuhara, T.; Suganuma, M.; Kurusu, M.; Fujiki, H. Helicobacter pylori-secreting protein Tipalpha is a potent inducer of chemokine gene expressions in stomach cancer cells. J. Cancer Res. Clin. Oncol. 2007, 133, 287–296. [Google Scholar] [CrossRef]
- Sakitani, K.; Hirata, Y.; Hayakawa, Y.; Serizawa, T.; Nakata, W.; Takahashi, R.; Kinoshita, H.; Sakamoto, K.; Nakagawa, H.; Akanuma, M.; et al. Role of interleukin-32 in Helicobacter pylori-induced gastric inflammation. Infect. Immun. 2012, 80, 3795–3803. [Google Scholar] [CrossRef] [PubMed]
- Sieveking, D.; Mitchell, H.M.; Day, A.S. Gastric epithelial cell CXC chemokine secretion following Helicobacter pylori infection in vitro. J. Gastroenterol. Hepatol. 2004, 19, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Lee, I. Critical pathogenic steps to high risk Helicobacter pylori gastritis and gastric carcinogenesis. World J. Gastroenterol. 2014, 20, 6412–6419. [Google Scholar] [CrossRef] [PubMed]
- Eck, M.; Schmausser, B.; Scheller, K.; Toksoy, A.; Kraus, M.; Menzel, T.; Müller-Hermelink, H.K.; Gillitzer, R. CXC chemokines Gro(alpha)/IL-8 and IP-10/MIG in Helicobacter pylori gastritis. Clin. Exp. Immunol. 2000, 122, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Kihira, K.; Kawata, H.; Tokumaru, K.; Kumakura, Y.; Ishino, Y.; Kawakami, S.; Inoue, K.; Kojima, T.; Satoh, Y.; et al. p53 expression in the gastric mucosa before and after eradication of Helicobacter pylori. Helicobacter 2001, 6, 31–36. [Google Scholar] [CrossRef]
- Li, J.H.; Shi, X.Z.; Lv, S.; Liu, M.; Xu, G.W. Effect of Helicobacter pylori infection on p53 expression of gastric mucosa and adenocarcinoma with microsatellite instability. World J. Gastroenterol. 2005, 11, 4363–4366. [Google Scholar] [CrossRef]
- Shu, X.; Yang, Z.; Li, Z.H.; Chen, L.; Zhou, X.D.; Xie, Y.; Lu, N.H. Helicobacter pylori Infection Activates the Akt-Mdm2-p53 Signaling Pathway in Gastric Epithelial Cells. Dig. Dis. Sci. 2015, 60, 876–886. [Google Scholar] [CrossRef]
- Triantafyllou, K.; Papadopoulos, V.; Emanouil, T.; Gkolfakis, P.; Damaskou, V.; Tziatzios, G.; Panayiotides, I.G.; Vafiadis, I.; Ladas, S.D. Eradication of Helicobacter pylori Infection Restores ki67, p53, and Cyclin D1 Immunoreactivity in the Human Gastric Epithelium. Clin. Med. Insights Gastroenterol. 2016, 9, 73–78. [Google Scholar] [CrossRef]
- Toller, I.M.; Neelsen, K.J.; Steger, M.; Hartung, M.L.; Hottiger, M.O.; Stucki, M.; Kalali, B.; Gerhard, M.; Sartori, A.A.; Lopes, M.; et al. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA 2011, 108, 14944–14949. [Google Scholar] [CrossRef]
- André, A.R.; Ferreira, M.V.; Mota, R.M.; Ferrasi, A.C.; Pardini, M.I.; Rabenhorst, S.H. Gastric adenocarcinoma and Helicobacter pylori: Correlation with p53 mutation and p27 immunoexpression. Cancer Epidemiol. 2010, 34, 618–625. [Google Scholar] [CrossRef]
- Imai, S.; Ooki, T.; Murata-Kamiya, N.; Komura, D.; Tahmina, K.; Wu, W.; Takahashi-Kanemitsu, A.; Knight, C.T.; Kunita, A.; Suzuki, N.; et al. Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell Host Microbe 2021, 29, 941–958.e10. [Google Scholar] [CrossRef]
- Bussière, F.I.; Michel, V.; Mémet, S.; Avé, P.; Vivas, J.R.; Huerre, M.; Touati, E.H. pylori-induced promoter hypermethylation downregulates USF1 and USF2 transcription factor gene expression. Cell Microbiol. 2010, 12, 1124–1133. [Google Scholar] [CrossRef]
- Costa, L.; Corre, S.; Michel, V.; Le Luel, K.; Fernandes, J.; Ziveri, J.; Jouvion, G.; Danckaert, A.; Mouchet, N.; Da Silva Barreira, D.; et al. USF1 defect drives p53 degradation during Helicobacter pylori infection and accelerates gastric carcinogenesis. Gut 2020, 69, 1582–1591. [Google Scholar] [CrossRef]
- Wei, J.; Nagy, T.A.; Vilgelm, A.; Zaika, E.; Ogden, S.R.; Romero-Gallo, J.; Piazuelo, M.B.; Correa, P.; Washington, M.K.; El-Rifai, W.; et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology 2010, 139, 1333–1343. [Google Scholar] [CrossRef]
- Abu-Lubad, M.A.; Helaly, G.F.; Haddadin, W.J.; Jarajreh, D.A.K.; Aqel, A.A.; Al-Zeer, M.A. Loss of p53 Expression in Gastric Epithelial Cells of Helicobacter pylori-Infected Jordanian Patients. Int. J. Microbiol. 2022, 2022, 7779770. [Google Scholar] [CrossRef]
- Junnila, S.; Kokkola, A.; Mizuguchi, T.; Hirata, K.; Karjalainen-Lindsberg, M.L.; Puolakkainen, P.; Monni, O. Gene expression analysis identifies over-expression of CXCL1, SPARC, SPP1, and SULF1 in gastric cancer. Genes Chromosom. Cancer 2010, 49, 28–39. [Google Scholar] [CrossRef]
- Cheng, W.L.; Wang, C.S.; Huang, Y.H.; Tsai, M.M.; Liang, Y.; Lin, K.H. Overexpression of CXCL1 and its receptor CXCR2 promote tumor invasion in gastric cancer. Ann. Oncol. 2011, 22, 2267–2276. [Google Scholar] [CrossRef]
- Wei, Z.W.; Xia, G.K.; Wu, Y.; Chen, W.; Xiang, Z.; Schwarz, R.E.; Brekken, R.A.; Awasthi, N.; He, Y.L.; Zhang, C.H. CXCL1 promotes tumor growth through VEGF pathway activation and is associated with inferior survival in gastric cancer. Cancer Lett. 2015, 359, 335–343. [Google Scholar] [CrossRef]
- Chen, X.; Jin, R.; Chen, R.; Huang, Z. Complementary action of CXCL1 and CXCL8 in pathogenesis of gastric carcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 1036–1045. [Google Scholar]
- Eck, M.; Schmausser, B.; Scheller, K.; Brändlein, S.; Müller-Hermelink, H.K. Pleiotropic effects of CXC chemokines in gastric carcinoma: Differences in CXCL8 and CXCL1 expression between diffuse and intestinal types of gastric carcinoma. Clin. Exp. Immunol. 2003, 134, 508–515. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kuroda, K.; Sera, T.; Sugimoto, A.; Kushiyama, S.; Nishimura, S.; Togano, S.; Okuno, T.; Yoshii, M.; Tamura, T.; et al. The Clinicopathological Significance of the CXCR2 Ligands, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, and CXCL8 in Gastric Cancer. Anticancer Res. 2019, 39, 6645–6652. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.J.; Noh, S.; Jeung, H.C.; Jung, M.; Kim, T.S.; Noh, S.H.; Roh, J.K.; Chung, H.C.; Rha, S.Y. Chemokine growth-regulated oncogene 1 as a putative biomarker for gastric cancer progression. Cancer Sci. 2010, 101, 2200–2206. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, D.; Zhang, W.; Tang, B.; Li, Q.Q.; Li, L. Evaluation of proteomics-identified CCL18 and CXCL1 as circulating tumor markers for differential diagnosis between ovarian carcinomas and benign pelvic masses. Int. J. Biol. Mrk. 2011, 26, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, N.; Sakao, Y.; Hayashi, S.; Hadden, W.A., 3rd; Harmon, C.L.; Miller, E.J. alpha-Chemokine growth factors for adenocarcinomas; a synthetic peptide inhibitor for alpha-chemokines inhibits the growth of adenocarcinoma cell lines. J. Cancer Res. Clin. Oncol. 2000, 126, 19–26. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, C.; Xu, J.; Wu, H.; Peng, J.; Cai, S.; He, Y. CXCL1 gene silencing inhibits HGC803 cell migration and invasion and acts as an independent prognostic factor for poor survival in gastric cancer. Mol. Med. Rep. 2016, 14, 4673–4679. [Google Scholar] [CrossRef]
- Shrestha, S.; Yang, C.D.; Hong, H.C.; Chou, C.H.; Tai, C.S.; Chiew, M.Y.; Chen, W.L.; Weng, S.L.; Chen, C.C.; Chang, Y.A.; et al. Integrated MicroRNA-mRNA Analysis Reveals miR-204 Inhibits Cell Proliferation in Gastric Cancer by Targeting CKS1B, CXCL1 and GPRC5A. Int. J. Mol. Sci. 2017, 19, 87. [Google Scholar] [CrossRef]
- Zhou, Z.; Xia, G.; Xiang, Z.; Liu, M.; Wei, Z.; Yan, J.; Chen, W.; Zhu, J.; Awasthi, N.; Sun, X.; et al. A C-X-C Chemokine Receptor Type 2-Dominated Cross-talk between Tumor Cells and Macrophages Drives Gastric Cancer Metastasis. Clin. Cancer Res. 2019, 25, 3317–3328. [Google Scholar] [CrossRef]
- Naito, Y.; Yamamoto, Y.; Sakamoto, N.; Shimomura, I.; Kogure, A.; Kumazaki, M.; Yokoi, A.; Yashiro, M.; Kiyono, T.; Yanagihara, K.; et al. Cancer extracellular vesicles contribute to stromal heterogeneity by inducing chemokines in cancer-associated fibroblasts. Oncogene 2019, 38, 5566–5579. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Li, G.; Wu, H.; Sun, K.; Chen, J.; Feng, Y.; Chen, C.; Cai, S.; Xu, J.; et al. CXCL1 from tumor-associated lymphatic endothelial cells drives gastric cancer cell into lymphatic system via activating integrin β1/FAK/AKT signaling. Cancer Lett. 2017, 385, 28–38. [Google Scholar] [CrossRef]
- Xiang, Z.; Zhou, Z.J.; Xia, G.K.; Zhang, X.H.; Wei, Z.W.; Zhu, J.T.; Yu, J.; Chen, W.; He, Y.; Schwarz, R.E.; et al. A positive crosstalk between CXCR4 and CXCR2 promotes gastric cancer metastasis. Oncogene 2017, 36, 5122–5133. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, C.; He, Y.; Wu, H.; Wang, Z.; Song, W.; Li, W.; He, W.; Cai, S.; Zhan, W. Lymphatic endothelial cell-secreted CXCL1 stimulates lymphangiogenesis and metastasis of gastric cancer. Int. J. Cancer 2012, 130, 787–797. [Google Scholar] [CrossRef]
- Kasashima, H.; Yashiro, M.; Nakamae, H.; Masuda, G.; Kinoshita, H.; Morisaki, T.; Fukuoka, T.; Hasegawa, T.; Nakane, T.; Hino, M.; et al. Clinicopathologic significance of the CXCL1-CXCR2 axis in the tumor microenvironment of gastric carcinoma. PLoS ONE 2017, 12, e0178635. [Google Scholar] [CrossRef]
- Zhou, X.; Fang, D.; Liu, H.; Ou, X.; Zhang, C.; Zhao, Z.; Zhao, S.; Peng, J.; Cai, S.; He, Y.; et al. PMN-MDSCs accumulation induced by CXCL1 promotes CD8+ T cells exhaustion in gastric cancer. Cancer Lett. 2022, 532, 215598. [Google Scholar] [CrossRef]
- Kasashima, H.; Yashiro, M.; Nakamae, H.; Kitayama, K.; Masuda, G.; Kinoshita, H.; Fukuoka, T.; Hasegawa, T.; Nakane, T.; Hino, M.; et al. CXCL1-Chemokine (C-X-C Motif) Receptor 2 Signaling Stimulates the Recruitment of Bone Marrow-Derived Mesenchymal Cells into Diffuse-Type Gastric Cancer Stroma. Am. J. Pathol. 2016, 186, 3028–3039. [Google Scholar] [CrossRef]
- Xiang, Z.; Jiang, D.P.; Xia, G.G.; Wei, Z.W.; Chen, W.; He, Y.; Zhang, C.H. CXCL1 expression is correlated with Snail expression and affects the prognosis of patients with gastric cancer. Oncol. Lett. 2015, 10, 2458–2464. [Google Scholar] [CrossRef]
- Chen, X.; Chen, R.; Jin, R.; Huang, Z. The role of CXCL chemokine family in the development and progression of gastric cancer. Int. J. Clin. Exp. Pathol. 2020, 13, 484–492. [Google Scholar]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Nischalke, H.D.; Berger, C.; Luda, C.; Müller, T.; Berg, T.; Coenen, M.; Krämer, B.; Körner, C.; Trebicka, J.; Grünhage, F.; et al. The CXCL1 rs4074 A allele is associated with enhanced CXCL1 responses to TLR2 ligands and predisposes to cirrhosis in HCV genotype 1-infected Caucasian patients. J. Hepatol. 2012, 56, 758–764. [Google Scholar] [CrossRef]
- Nischalke, H.D.; Berger, C.; Lutz, P.; Langhans, B.; Wolter, F.; Eisenhardt, M.; Krämer, B.; Kokordelis, P.; Glässner, A.; Müller, T.; et al. Influence of the CXCL1 rs4074 A allele on alcohol induced cirrhosis and HCC in patients of European descent. PLoS ONE 2013, 8, e80848. [Google Scholar] [CrossRef]
- Wilson, C.L.; Jurk, D.; Fullard, N.; Banks, P.; Page, A.; Luli, S.; Elsharkawy, A.M.; Gieling, R.G.; Chakraborty, J.B.; Fox, C.; et al. NFκB1 is a suppressor of neutrophil-driven hepatocellular carcinoma. Nat. Commun. 2015, 6, 6818. [Google Scholar] [CrossRef]
- Yoo, Y.D.; Ueda, H.; Park, K.; Flanders, K.C.; Lee, Y.I.; Jay, G.; Kim, S.J. Regulation of transforming growth factor-beta 1 expression by the hepatitis B virus (HBV) X transactivator. Role in HBV pathogenesis. J. Clin. Investig. 1996, 97, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Ju, D.; Zhang, D.W.; Li, H.; Kong, L.M.; Guo, Y.; Li, C.; Wang, X.L.; Chen, Z.N.; Bian, H. Activation of TGF-β1-CD147 positive feedback loop in hepatic stellate cells promotes liver fibrosis. Sci. Rep. 2015, 5, 16552. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.P.; Ju, D.; Li, H.; Yuan, L.; Cui, J.; Luo, D.; Chen, Z.N.; Bian, H. CD147 Promotes CXCL1 Expression and Modulates Liver Fibrogenesis. Int. J. Mol. Sci. 2018, 19, 1145. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xi, Y.; Li, Y.; Zuo, Z.; Zeng, C.; Fan, J.; Zhang, D.; Tao, H.; Guo, Y. Ethanol promoting the upregulation of C-X-C Motif Chemokine Ligand 1 (CXCL1) and C-X-C Motif Chemokine Ligand 6 (CXCL6) in models of early alcoholic liver disease. Bioengineered 2022, 13, 4688–4701. [Google Scholar] [CrossRef]
- Maltby, J.; Wright, S.; Bird, G.; Sheron, N. Chemokine levels in human liver homogenates: Associations between GRO alpha and histopathological evidence of alcoholic hepatitis. Hepatology 1996, 24, 1156–1160. [Google Scholar] [CrossRef]
- Roh, Y.S.; Zhang, B.; Loomba, R.; Seki, E. TLR2 and TLR9 contribute to alcohol-mediated liver injury through induction of CXCL1 and neutrophil infiltration. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G30–G41. [Google Scholar] [CrossRef]
- Stefanovic, L.; Brenner, D.A.; Stefanovic, B. Direct hepatotoxic effect of KC chemokine in the liver without infiltration of neutrophils. Exp. Biol. Med. 2005, 230, 573–586. [Google Scholar] [CrossRef]
- Casini, A.; Ceni, E.; Salzano, R.; Biondi, P.; Parola, M.; Galli, A.; Foschi, M.; Caligiuri, A.; Pinzani, M.; Surrenti, C. Neutrophil-derived superoxide anion induces lipid peroxidation and stimulates collagen synthesis in human hepatic stellate cells: Role of nitric oxide. Hepatology 1997, 25, 361–367. [Google Scholar] [CrossRef]
- Asselah, T.; Bièche, I.; Laurendeau, I.; Paradis, V.; Vidaud, D.; Degott, C.; Martinot, M.; Bedossa, P.; Valla, D.; Vidaud, M.; et al. Liver gene expression signature of mild fibrosis in patients with chronic hepatitis C. Gastroenterology 2005, 129, 2064–2075. [Google Scholar] [CrossRef]
- Johansson, S.; Talloen, W.; Tuefferd, M.; Darling, J.M.; Scholliers, A.; Fanning, G.; Fried, M.W.; Aerssens, J. Plasma levels of growth-related oncogene (CXCL1-3) associated with fibrosis and platelet counts in HCV-infected patients. Aliment. Pharmacol. Ther. 2015, 42, 1111–1121. [Google Scholar] [CrossRef]
- Cao, Z.; Fu, B.; Deng, B.; Zeng, Y.; Wan, X.; Qu, L. Overexpression of Chemokine (C-X-C) ligand 1 (CXCL1) associated with tumor progression and poor prognosis in hepatocellular carcinoma. Cancer Cell Int. 2014, 14, 86. [Google Scholar] [CrossRef]
- Han, K.Q.; Han, H.; He, X.Q.; Wang, L.; Guo, X.D.; Zhang, X.M.; Chen, J.; Zhu, Q.G.; Nian, H.; Zhai, X.F.; et al. Chemokine CXCL1 may serve as a potential molecular target for hepatocellular carcinoma. Cancer Med. 2016, 5, 2861–2871. [Google Scholar] [CrossRef]
- Wu, F.X.; Wang, Q.; Zhang, Z.M.; Huang, S.; Yuan, W.P.; Liu, J.Y.; Ban, K.C.; Zhao, Y.N. Identifying serological biomarkers of hepatocellular carcinoma using surface-enhanced laser desorption/ionization-time-of-flight mass spectroscopy. Cancer Lett. 2009, 279, 163–170. [Google Scholar] [CrossRef]
- Slany, A.; Haudek-Prinz, V.; Zwickl, H.; Stättner, S.; Grasl-Kraupp, B.; Gerner, C. Myofibroblasts are important contributors to human hepatocellular carcinoma: Evidence for tumor promotion by proteome profiling. Electrophoresis 2013, 34, 3315–3325. [Google Scholar] [CrossRef]
- Ye, L.Y.; Chen, W.; Bai, X.L.; Xu, X.Y.; Zhang, Q.; Xia, X.F.; Sun, X.; Li, G.G.; Hu, Q.D.; Fu, Q.H.; et al. Hypoxia-Induced Epithelial-to-Mesenchymal Transition in Hepatocellular Carcinoma Induces an Immunosuppressive Tumor Microenvironment to Promote Metastasis. Cancer Res. 2016, 76, 818–830. [Google Scholar] [CrossRef]
- Li, Y.M.; Liu, Z.Y.; Wang, J.C.; Yu, J.M.; Li, Z.C.; Yang, H.J.; Tang, J.; Chen, Z.N. Receptor-Interacting Protein Kinase 3 Deficiency Recruits Myeloid-Derived Suppressor Cells to Hepatocellular Carcinoma through the Chemokine (C-X-C Motif) Ligand 1-Chemokine (C-X-C Motif) Receptor 2 Axis. Hepatology 2019, 70, 1564–1581. [Google Scholar] [CrossRef]
- Tang, K.H.; Ma, S.; Lee, T.K.; Chan, Y.P.; Kwan, P.S.; Tong, C.M.; Ng, I.O.; Man, K.; To, K.F.; Lai, P.B.; et al. CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology 2012, 55, 807–820. [Google Scholar] [CrossRef]
- Han, K.Q.; He, X.Q.; Ma, M.Y.; Guo, X.D.; Zhang, X.M.; Chen, J.; Han, H.; Zhang, W.W.; Zhu, Q.G.; Zhao, W.Z. Targeted silencing of CXCL1 by siRNA inhibits tumor growth and apoptosis in hepatocellular carcinoma. Int. J. Oncol. 2015, 47, 2131–2140. [Google Scholar] [CrossRef]
- Ueda, S.; Basaki, Y.; Yoshie, M.; Ogawa, K.; Sakisaka, S.; Kuwano, M.; Ono, M. PTEN/Akt signaling through epidermal growth factor receptor is prerequisite for angiogenesis by hepatocellular carcinoma cells that is susceptible to inhibition by gefitinib. Cancer Res. 2006, 66, 5346–5353. [Google Scholar] [CrossRef]
- Liu, L.; Sun, H.; Wu, S.; Tan, H.; Sun, Y.; Liu, X.; Si, S.; Xu, L.; Huang, J.; Zhou, W.; et al. IL-17A promotes CXCR2-dependent angiogenesis in a mouse model of liver cancer. Mol. Med. Rep. 2019, 20, 1065–1074. [Google Scholar] [CrossRef]
- Dahlquist, K.J.V.; Voth, L.C.; Fee, A.J.; Stoeckman, A.K. An Autocrine Role for CXCL1 in Progression of Hepatocellular Carcinoma. Anticancer Res. 2020, 40, 6075–6081. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Wu, J.; Zhou, W.; Zhang, M.; Zhao, K.; Liu, J.; Tian, D.; Liao, J. SLC7A2 deficiency promotes hepatocellular carcinoma progression by enhancing recruitment of myeloid-derived suppressors cells. Cell Death Dis. 2021, 12, 570. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, Z.; Gao, J.; Gao, P.J.; Ni, Y.B.; Zhu, J.Y. Elevated CXCL1 increases hepatocellular carcinoma aggressiveness and is inhibited by miRNA-200a. Oncotarget 2016, 7, 65052–65066. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Pan, Q.; Yang, W.; Koo, S.C.; Tian, C.; Li, L.; Lu, M.; Brown, A.; Ju, B.; Easton, J.; et al. A developmentally prometastatic niche to hepatoblastoma in neonatal liver mediated by the Cxcl1/Cxcr2 axis. Hepatology 2022, 76, 1275–1290. [Google Scholar] [CrossRef]
- Vansaun, M.N.; Mendonsa, A.M.; Lee Gorden, D. Hepatocellular proliferation correlates with inflammatory cell and cytokine changes in a murine model of nonalchoholic fatty liver disease. PLoS ONE 2013, 8, e73054. [Google Scholar] [CrossRef]
- Wolf, B.; Krieg, K.; Falk, C.; Breuhahn, K.; Keppeler, H.; Biedermann, T.; Schmid, E.; Warmann, S.; Fuchs, J.; Vetter, S.; et al. Inducing Differentiation of Premalignant Hepatic Cells as a Novel Therapeutic Strategy in Hepatocarcinoma. Cancer Res. 2016, 76, 5550–5561. [Google Scholar] [CrossRef]
- Li, L.; Xu, L.; Yan, J.; Zhen, Z.J.; Ji, Y.; Liu, C.Q.; Lau, W.Y.; Zheng, L.; Xu, J. CXCR2-CXCL1 axis is correlated with neutrophil infiltration and predicts a poor prognosis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 129. [Google Scholar] [CrossRef]
- Zhao, W.; Xu, Y.; Xu, J.; Wu, D.; Zhao, B.; Yin, Z.; Wang, X. Subsets of myeloid-derived suppressor cells in hepatocellular carcinoma express chemokines and chemokine receptors differentially. Int. Immunopharmacol. 2015, 26, 314–321. [Google Scholar] [CrossRef]
- Zhou, S.L.; Zhou, Z.J.; Hu, Z.Q.; Huang, X.W.; Wang, Z.; Chen, E.B.; Fan, J.; Cao, Y.; Dai, Z.; Zhou, J. Tumor-Associated Neutrophils Recruit Macrophages and T-Regulatory Cells to Promote Progression of Hepatocellular Carcinoma and Resistance to Sorafenib. Gastroenterology 2016, 150, 1646–1658.e17. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef]
- Shen, Q.; Hu, G.; Wu, J.; Lv, L. A new clinical prognostic nomogram for liver cancer based on immune score. PLoS ONE 2020, 15, e0236622. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C.; Chen, X.; Li, Y.; Li, A.; Liu, D.; Li, F.; Luo, T. Functions of CXC chemokines as biomarkers and potential therapeutic targets in the hepatocellular carcinoma microenvironment. Transl. Cancer Res. 2021, 10, 2169–2187. [Google Scholar] [CrossRef]
- Krasinskas, A.M. Cholangiocarcinoma. Surg. Pathol. Clin. 2018, 11, 403–429. [Google Scholar] [CrossRef]
- Shaib, Y.; El-Serag, H.B. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004, 24, 115–125. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Sugimoto, A.; Maruo, K.; Tsujio, G.; Sera, T.; Kushiyama, S.; Nishimura, S.; Kuroda, K.; Togano, S.; Eguchi, S.; et al. CXCR2 signaling might have a tumor-suppressive role in patients with cholangiocarcinoma. PLoS ONE 2022, 17, e0266027. [Google Scholar] [CrossRef]
- Haga, H.; Yan, I.K.; Takahashi, K.; Wood, J.; Zubair, A.; Patel, T. Tumour cell-derived extracellular vesicles interact with mesenchymal stem cells to modulate the microenvironment and enhance cholangiocarcinoma growth. J. Extracell. Vesicles 2015, 4, 24900. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, C.; Duan, Y.; Heinrich, B.; Rosato, U.; Diggs, L.P.; Ma, L.; Roy, S.; Fu, Q.; Brown, Z.J.; et al. Gut Microbiome Directs Hepatocytes to Recruit MDSCs and Promote Cholangiocarcinoma. Cancer Discov. 2021, 11, 1248–1267. [Google Scholar] [CrossRef]
- Sueoka, H.; Hirano, T.; Uda, Y.; Iimuro, Y.; Yamanaka, J.; Fujimoto, J. Blockage of CXCR2 suppresses tumor growth of intrahepatic cholangiocellular carcinoma. Surgery 2014, 155, 640–649. [Google Scholar] [CrossRef]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Nagawkar, S.S.; Abu-Funni, S.; Simon, E.; Bick, T.; Prinz, E.; Sabo, E.; Ben-Izhak, O.; Hershkovitz, D. Intratumor Heterogeneity of KRAS Mutation Status in Pancreatic Ductal Adenocarcinoma Is Associated with Smaller Lesions. Pancreas 2016, 45, 876–881. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, C.; Nagee, K.; Mohammadi, A.; Monteiro, C. Characterization of KRAS Mutation in Acinar and Langerhans Islet Cells of Patients with Pancreatic Ductal Adenocarcinoma. Pancreas 2016, 45, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.H.; Mejlænder-Andersen, E.; Manueldas, S.; El Jellas, K.; Steine, S.J.; Tjensvoll, K.; Sætran, H.A.; Knappskog, S.; Hoem, D.; Nordgård, O.; et al. Mutation analysis by deep sequencing of pancreatic juice from patients with pancreatic ductal adenocarcinoma. BMC Cancer 2019, 19, 11. [Google Scholar] [CrossRef]
- Lian, S.; Zhai, X.; Wang, X.; Zhu, H.; Zhang, S.; Wang, W.; Wang, Z.; Huang, J. Elevated expression of growth-regulated oncogene-alpha in tumor and stromal cells predicts unfavorable prognosis in pancreatic cancer. Medicine 2016, 95, e4328. [Google Scholar] [CrossRef] [PubMed]
- Nywening, T.M.; Belt, B.A.; Cullinan, D.R.; Panni, R.Z.; Han, B.J.; Sanford, D.E.; Jacobs, R.C.; Ye, J.; Patel, A.A.; Gillanders, W.E.; et al. Targeting both tumour-associated CXCR2+ neutrophils and CCR2+ macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut 2018, 67, 1112–1123. [Google Scholar] [CrossRef]
- Huang, J.; Chen, Z.; Ding, C.; Lin, S.; Wan, D.; Ren, K. Prognostic Biomarkers and Immunotherapeutic Targets among CXC Chemokines in Pancreatic Adenocarcinoma. Front. Oncol. 2021, 11, 711402. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Molczyk, C.; Purohit, A.; Ehrhorn, E.; Goel, P.; Prajapati, D.R.; Atri, P.; Kaur, S.; Grandgenett, P.M.; Hollingsworth, M.A.; et al. Differential expression profile of CXC-receptor-2 ligands as potential biomarkers in pancreatic ductal adenocarcinoma. Am. J. Cancer Res. 2022, 12, 68–90. [Google Scholar]
- Steele, C.W.; Karim, S.A.; Leach, J.D.G.; Bailey, P.; Upstill-Goddard, R.; Rishi, L.; Foth, M.; Bryson, S.; McDaid, K.; Wilson, Z.; et al. CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer Cell 2016, 29, 832–845. [Google Scholar] [CrossRef]
- Takamori, H.; Oades, Z.G.; Hoch, O.C.; Burger, M.; Schraufstatter, I.U. Autocrine growth effect of IL-8 and GROalpha on a human pancreatic cancer cell line, Capan-1. Pancreas 2000, 21, 52–56. [Google Scholar] [CrossRef]
- Somerville, T.D.; Biffi, G.; Daßler-Plenker, J.; Hur, S.K.; He, X.Y.; Vance, K.E.; Miyabayashi, K.; Xu, Y.; Maia-Silva, D.; Klingbeil, O.; et al. Squamous trans-differentiation of pancreatic cancer cells promotes stromal inflammation. eLife 2020, 9, e53381. [Google Scholar] [CrossRef]
- Matsuo, Y.; Campbell, P.M.; Brekken, R.A.; Sung, B.; Ouellette, M.M.; Fleming, J.B.; Aggarwal, B.B.; Der, C.J.; Guha, S. K-Ras promotes angiogenesis mediated by immortalized human pancreatic epithelial cells through mitogen-activated protein kinase signaling pathways. Mol. Cancer Res. 2009, 7, 799–808. [Google Scholar] [CrossRef]
- Purohit, A.; Varney, M.; Rachagani, S.; Ouellette, M.M.; Batra, S.K.; Singh, R.K. CXCR2 signaling regulates KRAS(G¹²D)-induced autocrine growth of pancreatic cancer. Oncotarget 2016, 7, 7280–7296. [Google Scholar] [CrossRef]
- Woodstock, D.L.; Sammons, M.A.; Fischer, M. p63 and p53: Collaborative Partners or Dueling Rivals? Front. Cell Dev. Biol. 2021, 9, 701986. [Google Scholar] [CrossRef]
- Wu, H.H.; Hwang-Verslues, W.W.; Lee, W.H.; Huang, C.K.; Wei, P.C.; Chen, C.L.; Shew, J.Y.; Lee, E.Y.; Jeng, Y.M.; Tien, Y.W.; et al. Targeting IL-17B-IL-17RB signaling with an anti-IL-17RB antibody blocks pancreatic cancer metastasis by silencing multiple chemokines. J. Exp. Med. 2015, 212, 333–349. [Google Scholar] [CrossRef]
- Kemp, S.B.; Carpenter, E.S.; Steele, N.G.; Donahue, K.L.; Nwosu, Z.C.; Pacheco, A.; Velez-Delgado, A.; Menjivar, R.E.; Lima, F.; The, S.; et al. Apolipoprotein E Promotes Immune Suppression in Pancreatic Cancer through NF-κB-Mediated Production of CXCL1. Cancer Res. 2021, 81, 4305–4318. [Google Scholar] [CrossRef]
- Hrabák, P.; Kalousová, M.; Krechler, T.; Zima, T. Pancreatic stellate cells—Rising stars in pancreatic pathologies. Physiol. Res. 2021, 70, S597–S616. [Google Scholar] [CrossRef]
- Takikawa, T.; Masamune, A.; Yoshida, N.; Hamada, S.; Kogure, T.; Shimosegawa, T. Exosomes Derived from Pancreatic Stellate Cells: MicroRNA Signature and Effects on Pancreatic Cancer Cells. Pancreas 2017, 46, 19–27. [Google Scholar] [CrossRef]
- Hoffman, M.T.; Kemp, S.B.; Salas-Escabillas, D.J.; Zhang, Y.; Steele, N.G.; The, S.; Long, D.; Benitz, S.; Yan, W.; Margolskee, R.F.; et al. The Gustatory Sensory G-Protein GNAT3 Suppresses Pancreatic Cancer Progression in Mice. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 349–369. [Google Scholar] [CrossRef]
- Erez, N.; Truitt, M.; Olson, P.; Arron, S.T.; Hanahan, D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell 2010, 17, 135–147. [Google Scholar] [CrossRef]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef]
- Shao, C.; Tu, C.; Cheng, X.; Xu, Z.; Wang, X.; Shen, J.; Chai, K.; Chen, W. Inflammatory and Senescent Phenotype of Pancreatic Stellate Cells Induced by Sqstm1 Downregulation Facilitates Pancreatic Cancer Progression. Int. J. Biol. Sci. 2019, 15, 1020–1029. [Google Scholar] [CrossRef]
- Acosta, J.C.; O’Loghlen, A.; Banito, A.; Guijarro, M.V.; Augert, A.; Raguz, S.; Fumagalli, M.; Da Costa, M.; Brown, C.; Popov, N.; et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 2008, 133, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Liu, Z.; Xu, B.; Hu, H.; Wei, Z.; Liu, Q.; Zhang, X.; Ding, X.; Wang, Y.; Zhao, M.; et al. Chemokine receptor CXCR2 is transactivated by p53 and induces p38-mediated cellular senescence in response to DNA damage. Aging Cell 2013, 12, 1110–1121. [Google Scholar] [CrossRef] [PubMed]
- Awaji, M.; Saxena, S.; Wu, L.; Prajapati, D.R.; Purohit, A.; Varney, M.L.; Kumar, S.; Rachagani, S.; Ly, Q.P.; Jain, M.; et al. CXCR2 signaling promotes secretory cancer-associated fibroblasts in pancreatic ductal adenocarcinoma. FASEB J. 2020, 34, 9405–9418. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, Z.; Li, N.; Li, Y.; Chang, A.; Zhao, T.; Wang, X.; Wang, H.; Gao, S.; Yang, S.; et al. Interleukin 35 Expression Correlates with Microvessel Density in Pancreatic Ductal Adenocarcinoma, Recruits Monocytes, and Promotes Growth and Angiogenesis of Xenograft Tumors in Mice. Gastroenterology 2018, 154, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Bellomo, G.; Rainer, C.; Quaranta, V.; Astuti, Y.; Raymant, M.; Boyd, E.; Stafferton, R.; Campbell, F.; Ghaneh, P.; Halloran, C.M.; et al. Chemotherapy-induced infiltration of neutrophils promotes pancreatic cancer metastasis via Gas6/AXL signalling axis. Gut 2022, 71, 2284–2299. [Google Scholar] [CrossRef]
- Lesina, M.; Wörmann, S.M.; Morton, J.; Diakopoulos, K.N.; Korneeva, O.; Wimmer, M.; Einwächter, H.; Sperveslage, J.; Demir, I.E.; Kehl, T.; et al. RelA regulates CXCL1/CXCR2-dependent oncogene-induced senescence in murine Kras-driven pancreatic carcinogenesis. J. Clin. Investig. 2016, 126, 2919–2932. [Google Scholar] [CrossRef]
- Maeda, S.; Kuboki, S.; Nojima, H.; Shimizu, H.; Yoshitomi, H.; Furukawa, K.; Miyazaki, M.; Ohtsuka, M. Duffy antigen receptor for chemokines (DARC) expressing in cancer cells inhibits tumor progression by suppressing CXCR2 signaling in human pancreatic ductal adenocarcinoma. Cytokine 2017, 95, 12–21. [Google Scholar] [CrossRef]
- Neote, K.; Mak, J.Y.; Kolakowski, L.F., Jr.; Schall, T.J. Functional and biochemical analysis of the cloned Duffy antigen: Identity with the red blood cell chemokine receptor. Blood 1994, 84, 44–52. [Google Scholar] [CrossRef]
- Sano, M.; Ijichi, H.; Takahashi, R.; Miyabayashi, K.; Fujiwara, H.; Yamada, T.; Kato, H.; Nakatsuka, T.; Tanaka, Y.; Tateishi, K.; et al. Blocking CXCLs-CXCR2 axis in tumor-stromal interactions contributes to survival in a mouse model of pancreatic ductal adenocarcinoma through reduced cell invasion/migration and a shift of immune-inflammatory microenvironment. Oncogenesis 2019, 8, 8. [Google Scholar] [CrossRef]
- Purohit, A.; Saxena, S.; Varney, M.; Prajapati, D.R.; Kozel, J.A.; Lazenby, A.; Singh, R.K. Host Cxcr2-Dependent Regulation of Pancreatic Cancer Growth, Angiogenesis, and Metastasis. Am. J. Pathol. 2021, 191, 759–771. [Google Scholar] [CrossRef]
- Miyake, M.; Hori, S.; Morizawa, Y.; Tatsumi, Y.; Nakai, Y.; Anai, S.; Torimoto, K.; Aoki, K.; Tanaka, N.; Shimada, K.; et al. CXCL1-Mediated Interaction of Cancer Cells with Tumor-Associated Macrophages and Cancer-Associated Fibroblasts Promotes Tumor Progression in Human Bladder Cancer. Neoplasia 2016, 18, 636–646. [Google Scholar] [CrossRef]
- Seifert, L.; Werba, G.; Tiwari, S.; Giao Ly, N.N.; Alothman, S.; Alqunaibit, D.; Avanzi, A.; Barilla, R.; Daley, D.; Greco, S.H.; et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016, 532, 245–249. [Google Scholar] [CrossRef]
- Di Mitri, D.; Mirenda, M.; Vasilevska, J.; Calcinotto, A.; Delaleu, N.; Revandkar, A.; Gil, V.; Boysen, G.; Losa, M.; Mosole, S.; et al. Re-education of Tumor-Associated Macrophages by CXCR2 Blockade Drives Senescence and Tumor Inhibition in Advanced Prostate Cancer. Cell Rep. 2019, 28, 2156–2168.e5. [Google Scholar] [CrossRef]
- Hu, J.; Zhao, Q.; Kong, L.Y.; Wang, J.; Yan, J.; Xia, X.; Jia, Z.; Heimberger, A.B.; Li, S. Regulation of tumor immune suppression and cancer cell survival by CXCL1/2 elevation in glioblastoma multiforme. Sci. Adv. 2021, 7, eabc2511. [Google Scholar] [CrossRef]
- Ijichi, H.; Chytil, A.; Gorska, A.E.; Aakre, M.E.; Bierie, B.; Tada, M.; Mohri, D.; Miyabayashi, K.; Asaoka, Y.; Maeda, S.; et al. Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J. Clin. Investig. 2011, 121, 4106–4117. [Google Scholar] [CrossRef]
- Aikawa, T.; Gunn, J.; Spong, S.M.; Klaus, S.J.; Korc, M. Connective tissue growth factor-specific antibody attenuates tumor growth, metastasis, and angiogenesis in an orthotopic mouse model of pancreatic cancer. Mol. Cancer Ther. 2006, 5, 1108–1116. [Google Scholar] [CrossRef]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Kwong, T.N.Y.; Wang, X.; Nakatsu, G.; Chow, T.C.; Tipoe, T.; Dai, R.Z.W.; Tsoi, K.K.K.; Wong, M.C.S.; Tse, G.; Chan, M.T.V.; et al. Association between Bacteremia from Specific Microbes and Subsequent Diagnosis of Colorectal Cancer. Gastroenterology 2018, 155, 383–390.e8. [Google Scholar] [CrossRef]
- Cuenca, R.E.; Azizkhan, R.G.; Haskill, S. Characterization of GRO alpha, beta and gamma expression in human colonic tumours: Potential significance of cytokine involvement. Surg. Oncol. 1992, 1, 323–329. [Google Scholar] [CrossRef]
- Baier, P.K.; Eggstein, S.; Wolff-Vorbeck, G.; Baumgartner, U.; Hopt, U.T. Chemokines in human colorectal carcinoma. Anticancer Res. 2005, 25, 3581–3584. [Google Scholar]
- Wen, Y.; Giardina, S.F.; Hamming, D.; Greenman, J.; Zachariah, E.; Bacolod, M.D.; Liu, H.; Shia, J.; Amenta, P.S.; Barany, F.; et al. GROalpha is highly expressed in adenocarcinoma of the colon and down-regulates fibulin-1. Clin. Cancer Res. 2006, 12, 5951–5959. [Google Scholar] [CrossRef] [PubMed]
- Rubie, C.; Frick, V.O.; Wagner, M.; Schuld, J.; Gräber, S.; Brittner, B.; Bohle, R.M.; Schilling, M.K. ELR+ CXC chemokine expression in benign and malignant colorectal conditions. BMC Cancer 2008, 8, 178. [Google Scholar] [CrossRef] [PubMed]
- Erreni, M.; Bianchi, P.; Laghi, L.; Mirolo, M.; Fabbri, M.; Locati, M.; Mantovani, A.; Allavena, P. Expression of chemokines and chemokine receptors in human colon cancer. Methods Enzymol. 2009, 460, 105–121. [Google Scholar] [PubMed]
- Oladipo, O.; Conlon, S.; O’Grady, A.; Purcell, C.; Wilson, C.; Maxwell, P.J.; Johnston, P.G.; Stevenson, M.; Kay, E.W.; Wilson, R.H.; et al. The expression and prognostic impact of CXC-chemokines in stage II and III colorectal cancer epithelial and stromal tissue. Br. J. Cancer 2011, 104, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Zhang, Y.; Guo, Y.C.; Yang, Z.H.; Xu, Y.C. A Prognostic Model Based on the Immune-related Genes in Colon Adenocarcinoma. Int. J. Med. Sci. 2020, 17, 1879–1896. [Google Scholar] [CrossRef]
- Yang, J.; Gao, S.; Qiu, M.; Kan, S. Integrated Analysis of Gene Expression and Metabolite Data Reveals Candidate Molecular Markers in Colorectal Carcinoma. Cancer Biother. Radiopharm. 2020, 37, 907–916. [Google Scholar] [CrossRef]
- Yu, L.; Yang, X.; Xu, C.; Sun, J.; Fang, Z.; Pan, H.; Han, W. Comprehensive analysis of the expression and prognostic value of CXC chemokines in colorectal cancer. Int. Immunopharmacol. 2020, 89, 107077. [Google Scholar] [CrossRef]
- Chen, B.; Song, L.; Nie, X.; Lin, F.; Yu, Z.; Kong, W.; Qi, X.; Wang, W. CXCL1 Regulated by miR-302e Is Involved in Cell Viability and Motility of Colorectal Cancer via Inhibiting JAK-STAT Signaling Pathway. Front. Oncol. 2021, 10, 577229. [Google Scholar] [CrossRef]
- Gong, Y.Z.; Ma, H.; Ruan, G.T.; Zhu, L.C.; Liao, X.W.; Wang, S.; Yan, L.; Huang, W.; Huang, K.T.; Xie, H.; et al. Diagnosis and prognostic value of C-X-C motif chemokine ligand 1 in colon adenocarcinoma based on the Cancer Genome Atlas and Guangxi cohort. J. Cancer 2021, 12, 5506–5518. [Google Scholar] [CrossRef]
- Yang, M.Q.; Bai, L.L.; Wang, Z.; Lei, L.; Zheng, Y.W.; Li, Z.H.; Huang, W.J.; Liu, C.C.; Xu, H.T. DEK is highly expressed in breast cancer and is associated with malignant phenotype and progression. Oncol. Lett. 2021, 21, 440. [Google Scholar] [CrossRef]
- Sipos, F.; Germann, T.M.; Wichmann, B.; Galamb, O.; Spisák, S.; Krenács, T.; Tulassay, Z.; Molnár, B.; Műzes, G. MMP3 and CXCL1 are potent stromal protein markers of dysplasia-carcinoma transition in sporadic colorectal cancer. Eur. J. Cancer Prev. 2014, 23, 336–343. [Google Scholar] [CrossRef]
- Ogawa, R.; Yamamoto, T.; Hirai, H.; Hanada, K.; Kiyasu, Y.; Nishikawa, G.; Mizuno, R.; Inamoto, S.; Itatani, Y.; Sakai, Y.; et al. Loss of SMAD4 Promotes Colorectal Cancer Progression by Recruiting Tumor-Associated Neutrophils via the CXCL1/8-CXCR2 Axis. Clin. Cancer Res. 2019, 25, 2887–2899. [Google Scholar] [CrossRef]
- Liu, K.; Lai, M.; Wang, S.; Zheng, K.; Xie, S.; Wang, X. Construction of a CXC Chemokine-Based Prediction Model for the Prognosis of Colon Cancer. Biomed Res. Int. 2020, 2020, 6107865. [Google Scholar] [CrossRef]
- Chiu, S.T.; Hsieh, F.J.; Chen, S.W.; Chen, C.L.; Shu, H.F.; Li, H. Clinicopathologic correlation of up-regulated genes identified using cDNA microarray and real-time reverse transcription-PCR in human colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2005, 14, 437–443. [Google Scholar] [CrossRef]
- Wang, D.; Wang, H.; Brown, J.; Daikoku, T.; Ning, W.; Shi, Q.; Richmond, A.; Strieter, R.; Dey, S.K.; DuBois, R.N. CXCL1 induced by prostaglandin E2 promotes angiogenesis in colorectal cancer. J. Exp. Med. 2006, 203, 941–951. [Google Scholar] [CrossRef]
- Katoh, H.; Wang, D.; Daikoku, T.; Sun, H.; Dey, S.K.; Dubois, R.N. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 2013, 24, 631–644. [Google Scholar] [CrossRef]
- Khan, S.; Cameron, S.; Blaschke, M.; Moriconi, F.; Naz, N.; Amanzada, A.; Ramadori, G.; Malik, I.A. Differential gene expression of chemokines in KRAS and BRAF mutated colorectal cell lines: Role of cytokines. World J. Gastroenterol. 2014, 20, 2979–2994. [Google Scholar] [CrossRef]
- le Rolle, A.F.; Chiu, T.K.; Fara, M.; Shia, J.; Zeng, Z.; Weiser, M.R.; Paty, P.B.; Chiu, V.K. The prognostic significance of CXCL1 hypersecretion by human colorectal cancer epithelia and myofibroblasts. J. Transl. Med. 2015, 13, 199. [Google Scholar] [CrossRef]
- Hsu, Y.L.; Chen, Y.J.; Chang, W.A.; Jian, S.F.; Fan, H.L.; Wang, J.Y.; Kuo, P.L. Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1. Int. J. Mol. Sci. 2018, 19, 2427. [Google Scholar] [CrossRef]
- Wang, D.; Sun, H.; Wei, J.; Cen, B.; DuBois, R.N. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Res. 2017, 77, 3655–3665. [Google Scholar] [CrossRef]
- Triner, D.; Xue, X.; Schwartz, A.J.; Jung, I.; Colacino, J.A.; Shah, Y.M. Epithelial Hypoxia-Inducible Factor 2α Facilitates the Progression of Colon Tumors through Recruiting Neutrophils. Mol. Cell Biol. 2017, 37, e00481-16. [Google Scholar] [CrossRef] [PubMed]
- Tosti, N.; Cremonesi, E.; Governa, V.; Basso, C.; Kancherla, V.; Coto-Llerena, M.; Amicarella, F.; Weixler, B.; Däster, S.; Sconocchia, G.; et al. Infiltration by IL22-Producing T Cells Promotes Neutrophil Recruitment and Predicts Favorable Clinical Outcome in Human Colorectal Cancer. Cancer Immunol. Res. 2020, 8, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, B.; Li, R.; Chen, J.; Xu, G.; Zhu, Y.; Li, J.; Liang, Q.; Hua, Q.; Wang, L.; et al. KIAA1199 drives immune suppression to promote colorectal cancer liver metastasis by modulating neutrophil infiltration. Hepatology 2022, 76, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Neufeld, H.; Torlakovic, E.; Xiao, W. Uev1A-Ubc13 promotes colorectal cancer metastasis through regulating CXCL1 expression via NF-κB activation. Oncotarget 2018, 9, 15952–15967. [Google Scholar] [CrossRef] [PubMed]
- Farquharson, A.J.; Steele, R.J.; Carey, F.A.; Drew, J.E. Novel multiplex method to assess insulin, leptin and adiponectin regulation of inflammatory cytokines associated with colon cancer. Mol. Biol. Rep. 2012, 39, 5727–5736. [Google Scholar] [CrossRef]
- Cai, L.; Xu, S.; Piao, C.; Qiu, S.; Li, H.; Du, J. Adiponectin induces CXCL1 secretion from cancer cells and promotes tumor angiogenesis by inducing stromal fibroblast senescence. Mol. Carcinog. 2016, 55, 1796–1806. [Google Scholar] [CrossRef]
- Urosevic, J.; Blasco, M.T.; Llorente, A.; Bellmunt, A.; Berenguer-Llergo, A.; Guiu, M.; Cañellas, A.; Fernandez, E.; Burkov, I.; Clapés, M.; et al. ERK1/2 Signaling Induces Upregulation of ANGPT2 and CXCR4 to Mediate Liver Metastasis in Colon Cancer. Cancer Res. 2020, 80, 4668–4680. [Google Scholar] [CrossRef]
- Zhuang, W.; Niu, T.; Li, Z. MicroRNA miR-145-5p regulates cell proliferation and cell migration in colon cancer by inhibiting chemokine (C-X-C motif) ligand 1 and integrin α2. Bioengineered 2021, 12, 9909–9917. [Google Scholar] [CrossRef]
- Lv, Q.Y.; Zou, H.Z.; Xu, Y.Y.; Shao, Z.Y.; Wu, R.Q.; Li, K.J.; Deng, X.; Gu, D.N.; Jiang, H.X.; Su, M.; et al. Expression levels of chemokine (C-X-C motif) ligands CXCL1 and CXCL3 as prognostic biomarkers in rectal adenocarcinoma: Evidence from Gene Expression Omnibus (GEO) analyses. Bioengineered 2021, 12, 3711–3725. [Google Scholar] [CrossRef]
- Zhao, Z.W.; Fan, X.X.; Yang, L.L.; Song, J.J.; Fang, S.J.; Tu, J.F.; Chen, M.J.; Zheng, L.Y.; Wu, F.Z.; Zhang, D.K.; et al. The identification of a common different gene expression signature in patients with colorectal cancer. Math. Biosci. Eng. 2019, 16, 2942–2958. [Google Scholar] [CrossRef]
- Li, A.; Varney, M.L.; Singh, R.K. Constitutive expression of growth regulated oncogene (gro) in human colon carcinoma cells with different metastatic potential and its role in regulating their metastatic phenotype. Clin. Exp. Metastasis 2004, 21, 571–579. [Google Scholar] [CrossRef]
- Bandapalli, O.R.; Ehrmann, F.; Ehemann, V.; Gaida, M.; Macher-Goeppinger, S.; Wente, M.; Schirmacher, P.; Brand, K. Down-regulation of CXCL1 inhibits tumor growth in colorectal liver metastasis. Cytokine 2012, 57, 46–53. [Google Scholar] [CrossRef]
- Zhuo, C.; Wu, X.; Li, J.; Hu, D.; Jian, J.; Chen, C.; Zheng, X.; Yang, C. Chemokine (C-X-C motif) ligand 1 is associated with tumor progression and poor prognosis in patients with colorectal cancer. Biosci. Rep. 2018, 38, BSR20180580. [Google Scholar] [CrossRef]
- Guil-Luna, S.; Mena, R.; Navarrete-Sirvent, C.; López-Sánchez, L.M.; Khouadri, K.; Toledano-Fonseca, M.; Mantrana, A.; Guler, I.; Villar, C.; Díaz, C.; et al. Association of Tumor Budding with Immune Evasion Pathways in Primary Colorectal Cancer and Patient-Derived Xenografts. Front. Med. 2020, 7, 264. [Google Scholar] [CrossRef]
- Mitrovic, B.; Schaeffer, D.F.; Riddell, R.H.; Kirsch, R. Tumor budding in colorectal carcinoma: Time to take notice. Mod. Pathol. 2012, 25, 1315–1325. [Google Scholar] [CrossRef]
- Divella, R.; Daniele, A.; Abbate, I.; Bellizzi, A.; Savino, E.; Simone, G.; Giannone, G.; Giuliani, F.; Fazio, V.; Gadaleta-Caldarola, G.; et al. The presence of clustered circulating tumor cells (CTCs) and circulating cytokines define an aggressive phenotype in metastatic colorectal cancer. Cancer Causes Control 2014, 25, 1531–1541. [Google Scholar] [CrossRef]
- Tannenbaum, C.S.; Rayman, P.A.; Pavicic, P.G.; Kim, J.S.; Wei, W.; Polefko, A.; Wallace, W.; Rini, B.I.; Morris-Stiff, G.; Allende, D.S.; et al. Mediators of Inflammation-Driven Expansion, Trafficking, and Function of Tumor-Infiltrating MDSCs. Cancer Immunol. Res. 2019, 7, 1687–1699. [Google Scholar] [CrossRef]
- Ogata, H.; Sekikawa, A.; Yamagishi, H.; Ichikawa, K.; Tomita, S.; Imura, J.; Ito, Y.; Fujita, M.; Tsubaki, M.; Kato, H.; et al. GROα promotes invasion of colorectal cancer cells. Oncol. Rep. 2010, 24, 1479–1486. [Google Scholar]
- Yang, X.; Wei, Y.; Sheng, F.; Xu, Y.; Liu, J.; Gao, L.; Yang, J.; Sun, X.; Huang, J.; Guo, Q. Comprehensive analysis of the prognosis and immune infiltration for CXC chemokines in colorectal cancer. Aging 2021, 13, 17548–17567. [Google Scholar] [CrossRef]
- Divella, R.; Daniele, A.; DE Luca, R.; Simone, M.; Naglieri, E.; Savino, E.; Abbate, I.; Gadaleta, C.D.; Ranieri, G. Circulating Levels of VEGF and CXCL1 Are Predictive of Metastatic Organotropismin in Patients with Colorectal Cancer. Anticancer Res. 2017, 37, 4867–4871. [Google Scholar]
- Gong, B.; Kao, Y.; Zhang, C.; Sun, F.; Gong, Z.; Chen, J. Identification of Hub Genes Related to Carcinogenesis and Prognosis in Colorectal Cancer Based on Integrated Bioinformatics. Mediat. Inflamm. 2020, 2020, 5934821. [Google Scholar] [CrossRef] [PubMed]
- White, J.R.; Lee, J.M.; Young, P.R.; Hertzberg, R.P.; Jurewicz, A.J.; Chaikin, M.A.; Widdowson, K.; Foley, J.J.; Martin, L.D.; Griswold, D.E.; et al. Identification of a potent, selective non-peptide CXCR2 antagonist that inhibits interleukin-8-induced neutrophil migration. J. Biol. Chem. 1998, 273, 10095–10098. [Google Scholar] [CrossRef] [PubMed]
- Romanini, J.; Mielcke, T.R.; Leal, P.C.; Figueiredo, C.P.; Calixto, J.B.; Morrone, F.B.; Batista, E.L., Jr.; Campos, M.M. The role of CXCR2 chemokine receptors in the oral squamous cell carcinoma. Investig. New Drugs 2012, 30, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Goda, A.E.; Koyama, M.; Sowa, Y.; Elokely, K.M.; Yoshida, T.; Kim, B.Y.; Sakai, T. Molecular mechanisms of the antitumor activity of SB225002: A novel microtubule inhibitor. Biochem. Pharmacol. 2013, 85, 1741–1752. [Google Scholar] [CrossRef]
- Goda, A.E.; Sakai, T. Molecular insights into the microtubules depolymerizing activity of the IL-8 receptor B antagonist SB225002. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 3726–3734. [Google Scholar]
- Ahuja, S.K.; Murphy, P.M. The CXC chemokines growth-regulated oncogene (GRO) alpha, GRObeta, GROgamma, neutrophil-activating peptide-2, and epithelial cell-derived neutrophil-activating peptide-78 are potent agonists for the type B, but not the type A, human interleukin-8 receptor. J. Biol. Chem. 1996, 271, 20545–20550. [Google Scholar] [CrossRef]
- Dwyer, M.P.; Yu, Y.; Chao, J.; Aki, C.; Chao, J.; Biju, P.; Girijavallabhan, V.; Rindgen, D.; Bond, R.; Mayer-Ezel, R.; et al. Discovery of 2-hydroxy-N,N-dimethyl-3-{2-[[(R)-1-(5-methylfuran-2-yl)propyl]amino]-3,4-dioxocyclobut-1-enylamino}benzamide (SCH 527123): A potent, orally bioavailable CXCR2/CXCR1 receptor antagonist. J. Med. Chem. 2006, 49, 7603–7606. [Google Scholar] [CrossRef]
- Gonsiorek, W.; Fan, X.; Hesk, D.; Fossetta, J.; Qiu, H.; Jakway, J.; Billah, M.; Dwyer, M.; Chao, J.; Deno, G.; et al. Pharmacological characterization of Sch527123, a potent allosteric CXCR1/CXCR2 antagonist. J. Pharm. Exp. Ther. 2007, 322, 477–485. [Google Scholar] [CrossRef]
- Ning, Y.; Labonte, M.J.; Zhang, W.; Bohanes, P.O.; Gerger, A.; Yang, D.; Benhaim, L.; Paez, D.; Rosenberg, D.O.; Nagulapalli Venkata, K.C.; et al. The CXCR2 antagonist, SCH-527123, shows antitumor activity and sensitizes cells to oxaliplatin in preclinical colon cancer models. Mol. Cancer Ther. 2012, 11, 1353–1364. [Google Scholar] [CrossRef]
| Type of Tumor | Effect on the Survival at High CXCL1 Expression | Number of Patients in the Study | Notes | References |
|---|---|---|---|---|
| Head and neck cancer: squamous cell carcinoma | Worse prognosis | 491 | OS, DFS Data from the Cancer Genome Atlas (TCGA) dataset | [31,52] |
| Head and neck cancer: squamous cell carcinoma | Worse prognosis | 494 | OS, DFS/PFS Data from TCGA dataset | [51,52] |
| Head and neck cancer: squamous cell carcinoma | Worse prognosis | 499 | OS, statistically insignificant difference in RFS Data from Kaplan–Meier plotter | [26,27] |
| Head and neck cancer: larynx squamous cell carcinoma | Worse prognosis | 135 | OS | [33] |
| Type of Tumor | Effect on the Survival at High CXCL1 Expression | Number of Patients in the Study | Notes | References |
|---|---|---|---|---|
| Esophageal cancer: esophageal squamous cell carcinoma | Worse prognosis | 141 | OS, | [56] |
| Esophageal cancer: esophageal squamous cell carcinoma | Better prognosis | 877 | OS Data from the Kaplan–Meier Plotter database (http://kmplot.com access date: 16 April 2022) | [57] |
| Esophageal carcinoma | No significant impact on prognosis | 92 | RFS and OS Data from the gene expression profiling interactive analysis (GEPIA) database | [65] |
| Type of Tumor | Effect on the Survival at High CXCL1 Expression | Number of Patients in the Study | Notes | References |
|---|---|---|---|---|
| Gastric cancer | Worse prognosis | 98 | OS | [98] |
| Gastric cancer | Worse prognosis | 56 | Cumulative survival, CXCL1 in the tumor, as well as plasma CXCL1 level | [97] |
| Gastric cancer | Better prognosis | 34 | OS | [96] |
| Gastric cancer | Worse prognosis | 572 | OS, Data from the public database | [108] |
| Gastric cancer | Worse prognosis | 155 | OS | [107] |
| Gastric cancer | Worse prognosis | 127 | OS | [115] |
| Gastric cancer | Worse prognosis | 590 | OS | [101] |
| Gastric cancer | Worse prognosis | 100 | OS | [105] |
| Gastric cancer | Worse prognosis | 263 | OS, Only for stage I patients | [112] |
| Gastric cancer | Worse prognosis | 105 | OS | [109] |
| Gastric cancer | Worse prognosis | 72 | OS | [99] |
| Type of Tumor | Effect on the Survival at High CXCL1 Expression | Number of Patients in the Study | Notes | References |
|---|---|---|---|---|
| Liver cancer | Worse prognosis | 346 | OS, Data from TCGA dataset | [52,151] |
| Liver cancer: HCC | Worse prognosis | 182 | OS Data from the Kaplan–Meier plotter database | [152] |
| Liver cancer: HCC | Worse prognosis | 48 | OS | [131] |
| Liver cancer: HCC | Worse prognosis | 119 | OS, DFS | [143] |
| Liver cancer: HCC | Worse prognosis | 259 | OS, RFS Patients with a high CXCL1 expression in a tumor together with a high CXCR2 expression | [147] |
| Type of Tumor | Effect on the Survival at High CXCL1 Expression | Number of Patients in the Study | Notes | References |
|---|---|---|---|---|
| Cholangiocarcinoma | Better prognosis | 165 | OS | [155] |
| Cholangiocarcinoma | Worse prognosis | 18 | RFS No effect at OS, Data from the GEPIA database | [65] |
| Type of Tumor | Effect on the Survival at High CXCL1 Expression | Number of Patients in the Study | Notes | References |
|---|---|---|---|---|
| Pancreatic cancer | Worse prognosis | 160 | OS | [163] |
| Pancreatic cancer: pancreatic adenocarcinoma | No significant impact on prognosis | 178 | OS, DFS | [165] |
| Pancreatic adenocarcinoma | No significant impact on prognosis | 90 | OS, RFS Data from the GEPIA database | [65] |
| Type of Tumor | Effect on the Survival at High CXCL1 Expression | Number of Patients in the Study | Notes | References |
|---|---|---|---|---|
| Colorectal cancer | Worse prognosis | 62 | OS | [238] |
| Colorectal cancer | Worse prognosis | 91 | RFS, analysis only in stage III patients. In other stages, CXCL1 expression is not related to prognosis | [204] |
| Colorectal cancer | No significant impact on prognosis | 163 | RFS, OS analysis only in stage II patients | [204] |
| Colorectal cancer | No significant impact on prognosis | 270 | OS, Data from the GEPIA database | [65,206] |
| Colorectal cancer | No significant impact on prognosis | 362 | RFS, OS Data from the GEPIA database | [65,239] |
| Colorectal cancer | Worse prognosis | 276 | OS, DFS | [233] |
| Colorectal cancer | No significant impact on prognosis | 125 | OS, RFS | [212] |
| Colorectal cancer | Worse prognosis | 45 | OS, only stage IV patients | [218] |
| Colorectal cancer | No significant impact on prognosis | 70 | OS, Only stage II and III patients | [218] |
| Colorectal cancer: colon cancer | Better prognosis | 438 | OS, data from TCGA | [52,209,213] |
| Colorectal cancer: colon adenocarcinoma | No significant impact on prognosis | 171 | OS, RFS | [209] |
| Colorectal cancer: rectal adenocarcinoma | Worse prognosis | 304 | OS Data from the TCGA database | [52,229] |
| Disease for Which the Drug Is Being Tested | Drug Name | Clinical Trial Phase | ClinicalTrials.gov Identifier |
|---|---|---|---|
| Metastatic Castration-Resistant Prostate Cancer | AZD5069 | I and II | NCT03177187 |
| Myelodysplastic Syndromes | SX-682 | I | NCT04245397 |
| Pancreatic Ductal Adenocarcinoma | SX-682 | I | NCT04477343 |
| Melanoma Stage III and Stage IV | SX-682 | I | NCT03161431 |
| Inflammatory Response | RIST4721 | I | NCT04105959 |
| Chronic Obstructive Pulmonary Disease (COPD) | Danirixin (GSK1325756) | I | NCT01453478 |
| COPD | Danirixin (GSK1325756) | I | NCT03136380 |
| COPD | Danirixin (GSK1325756) | II | NCT03250689 |
| COPD | Danirixin (GSK1325756) | II | NCT02130193 |
| COPD | Navarixin (SCH 527123, MK-7123) | II | NCT01006616 |
| COPD | QBM076 | II | NCT01972776 |
| Influenza | Danirixin (GSK1325756) | II | NCT02469298 |
| Influenza | Danirixin (GSK1325756) | II | NCT02927431 |
| Respiratory Syncytial Virus (RSV) Infections | Danirixin (GSK1325756) | I | NCT02201303 |
| COPD | SB-656933 | I | NCT00504439 |
| Cystic Fibrosis | SB-656933 | II | NCT00903201 |
| Type 1 Diabetes | Ladarixin | II | NCT05035368 |
| Type 1 Diabetes | Ladarixin | III | NCT04628481 |
| Bullous Pemphigoid | DF2156A | II | NCT01571895 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korbecki, J.; Bosiacki, M.; Barczak, K.; Łagocka, R.; Chlubek, D.; Baranowska-Bosiacka, I. The Clinical Significance and Role of CXCL1 Chemokine in Gastrointestinal Cancers. Cells 2023, 12, 1406. https://doi.org/10.3390/cells12101406
Korbecki J, Bosiacki M, Barczak K, Łagocka R, Chlubek D, Baranowska-Bosiacka I. The Clinical Significance and Role of CXCL1 Chemokine in Gastrointestinal Cancers. Cells. 2023; 12(10):1406. https://doi.org/10.3390/cells12101406
Chicago/Turabian StyleKorbecki, Jan, Mateusz Bosiacki, Katarzyna Barczak, Ryta Łagocka, Dariusz Chlubek, and Irena Baranowska-Bosiacka. 2023. "The Clinical Significance and Role of CXCL1 Chemokine in Gastrointestinal Cancers" Cells 12, no. 10: 1406. https://doi.org/10.3390/cells12101406
APA StyleKorbecki, J., Bosiacki, M., Barczak, K., Łagocka, R., Chlubek, D., & Baranowska-Bosiacka, I. (2023). The Clinical Significance and Role of CXCL1 Chemokine in Gastrointestinal Cancers. Cells, 12(10), 1406. https://doi.org/10.3390/cells12101406






