An Insight into Survivin in Relevance to Hematological, Biochemical and Genetic Characteristics in Tobacco Chewers with Oral Squamous Cell Carcinoma
Abstract
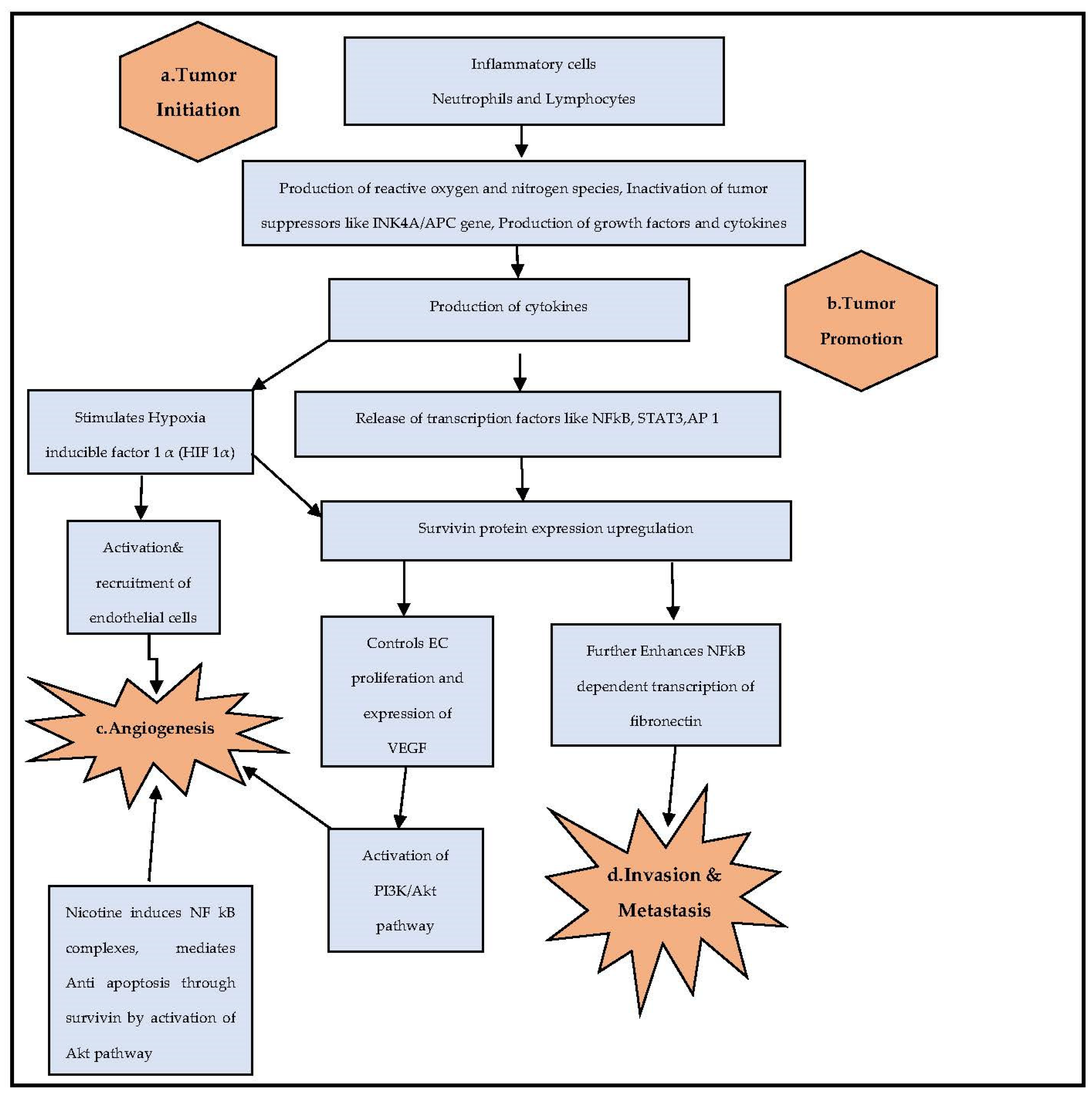
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Methods
2.2.1. Sample Collection
2.2.2. Human Survivin Assay
2.2.3. Pretreatment Hematological Parameters
2.2.4. Evidence of Tissue Survivin Protein by Immunohistochemistry
2.2.5. BIRC5/Survivin Gene Analysis
PCR Amplification of BIRC5 Gene
Analysis of PCR Ampliconson Agarose Gel Electrophoresis
BIRC5 Gene Sequencing and Sequence Analysis
2.3. Statistical Analysis
3. Results
3.1. Patient Characteristic Details
3.2. Analysis of Tissue Survivin Data
3.3. Statistical Representation of Pretreatment Hematological Parameters
3.4. Demonstration of Survivin Protein by Immunohistochemistry Technique
3.5. BIRC5 Gene Sequence Analysis
4. Discussion
5. Conclusions
6. Limitations of the Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parkin, D.M.; Pisan, P.; Ferlay, J. Global cancer statistics. Am. Cancer Soc. 1999, 49, 33–64. [Google Scholar] [CrossRef]
- Daftary, D.K.; Murti, P.R.; Bhonsle, R.B.; Gupta, P.C.; Mehta, F.S.; Pindborg, J.J. Risk factors and risk markers for oral cancer in high incidence areas of the world. In Oral Cancer Cambridge; Johnson, N.W., Ed.; Cambridge University Press: Cambridge, UK, 1991; Volume 2, pp. 29–63. [Google Scholar]
- Gupta, P.C.; Nandakumar, A. Oral cancer scene in India. Oral Dis. 1999, 5, 1–2. [Google Scholar] [CrossRef]
- Das, B.R.; Nagpal, J.K. Understanding the biology of oral cancer. Med. Sci. Monit. 2002, 8, 258–267. [Google Scholar]
- Gupta, P.C. Mouth cancer in India: A new epidemic?–Data analysis report. J. Indian Med. Assoc. 1999, 97, 370–373. [Google Scholar]
- Murthy, N.S.; Mathew, A. Cancer epidemiology, preventionand control. Curr. Sci. 2004, 86, 518–527. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Lorente, D.; Mateo, J.; Templeton, A.J.; Zafeiriou, Z.; Bianchini, D.; Ferraldeschi, R. Baseline neutrophil lymphocyte ratio (NLR) is associated with survival and response to treatment with second-line chemotherapy for advanced prostate cancer independent of baseline steroid use. Ann. Oncol. 2015, 26, 750–755. [Google Scholar] [CrossRef]
- Reed, J.C. The survivin saga goes in vivo. J. Clin. Investig. 2001, 108, 965–969. [Google Scholar] [CrossRef]
- Yoshimura, A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006, 97, 439–447. [Google Scholar] [CrossRef]
- Cooper, C.S.; Foster, C.S. Concepts of epigenetics in prostate cancer development. Br. J. Cancer 2009, 100, 240–245. [Google Scholar] [CrossRef]
- Chen, X.; Xu, H.; Yuan, P.; Fang, F.; Huss, M.; Vega, V.B. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008, 133, 1106–1117. [Google Scholar] [CrossRef]
- Karin, M. Nuclear factor-kappaB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef]
- Gupta, P.C.; Murti, P.R.; Bhonsle, R.B. Epidemiology of cancer by tobacco products and the significance of TSNA. Crit. Rev. Toxicol. 1996, 26, 183–198. [Google Scholar] [CrossRef]
- Znaor, A.; Brennan, P.; Gajalakshmi, V.; Mathew, A.; Shanta, V.; Varghese, C.; Boffetta, P. Independent and combined effects of tobacco smoking, chewing and alcohol drinking on the risk of oral, pharyngeal and esophageal cancers in Indian men. Int. J. Cancer 2003, 105, 681–686. [Google Scholar] [CrossRef]
- Balaram, P.; Sridhar, H.; Rajkumar, T.; Vaccarella, S.; Herrero, R.; Nandakumar, A.; Ravichandran, K.; Ramdas, K.; Sankaranarayanan, R.; Gajalakshmi, V.; et al. Oral cancer in southern India: The influence of smoking, drinking, paan-chewing and oral hygiene. Int. J. Cancer 2002, 98, 440–445. [Google Scholar] [CrossRef]
- Warnakulasuriya, K.A.; Johnson, N.W.; Linklater, K.M.; Bell, J. Cancer of mouth, pharynx and nasopharynx in Asian and Chinese immigrants resident in Thames regions. Oral. Oncol. 1999, 35, 471–475. [Google Scholar] [CrossRef]
- Quanri, J.; David, G.M.; Li, M.; Waun, K.H.; Lee, H.-Y. Survivin expression in normal human bronchial epithelial cells: An early and critical step in tumorigenesis induced by tobacco exposure. Carcinogenesis 2008, 29, 1614–1622. [Google Scholar]
- Dasgupta, P.; Kinkade, R.; Joshi, B.; DeCook, C.; Haura, E.; Chellappan, S. Nicotine inhibits apoptosis induced bychemotherapeutic drugs by up-regulating XIAP and survivin. Proc. Natl. Acad. Sci. USA 2006, 103, 6332–6337. [Google Scholar] [CrossRef]
- Dhirendra, N.S.; Rizwan, S.A.; Prakash, C.G. Smokeless tobacco-associated cancers: A systematic review and meta-analysis of Indian studies. Int. J. Cancer 2016, 138, 1368–1379. [Google Scholar]
- Guangyan, M.; Jiayi, W.; Zhiyan, L.; Hanxu, Z.; Shuang, Z.; Qian, X.; Yimin, C. Association between smokeless tobacco use and oral cavity cancer risk in women compared with men: A systematic review and meta-analysis. BMC Cancer 2021, 21, 960. [Google Scholar]
- Sanner, T.; Grimsrud, T.K. Nicotine: Carcinogenicity and effects on response to cancer treatment—A review. Front. Oncol. 2015, 5, 196. [Google Scholar] [CrossRef]
- Fernandez, J.G.; Rodríguez, D.A.; Valenzuela, M.; Calderon, C.; Urzúa, U.; Munroe, D.; Wright, M.C. Survivin expression promotes VEGF-induced tumor angiogenesis via PI3K/Akt enhanced β-catenin/ Tcf-Lef dependent transcription. Molecular Cancer 2014, 13, 209. [Google Scholar] [CrossRef]
- Hu, H.; Shikama, Y.; Matsuoka, I.; Kimura, J. Terminally differentiated neutrophils predominantly express Survivin-2α, a dominant-negative isoform of Survivin. J. Leukoc. Biol. 2008, 83, 393–400. [Google Scholar] [CrossRef]
- Altieri, D.C. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene 2003, 22, 8581–8589. [Google Scholar] [CrossRef]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997, 3, 917–921. [Google Scholar] [CrossRef]
- Tanaka, K.; Iwamoto, S.; Gon, G.; Nohara, T.; Iwamoto, M.; Tanigawa, N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin. Cancer Res. 2000, 6, 127–134. [Google Scholar]
- Nasu, S.; Yagihashi, A.; Izawa, A.; Saito, K.; Asanuma, K.; Nakamura, M. Survivin mRNA expression in patients with breast cancer. Anticancer Res. 2002, 22, 1839–1843. [Google Scholar]
- Ito, T.; Shiraki, K.; Sugimoto, K.; Yamanaka, T.; Fujikawa, K.; Ito, M. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology 2000, 31, 1080–1085. [Google Scholar] [CrossRef]
- Cohen, C.; Lohmann, C.M.; Cotsonis, G.; Lawson, D.; Santoianni, R. Survivin expression in ovarian carcinoma: Correlation with apoptotic markers and prognosis. Mod. Pathol. 2003, 16, 574–583. [Google Scholar] [CrossRef]
- Lehner, R.; Lucia, M.S.; Jarboe, E.A.; Orlicky, D.; Shroyer, A.L.; McGregor, J.A. Immunohistochemical localization of the IAP protein survivin in bladder mucosa and transitional cell carcinoma. Appl. Immunohistochem. Mol. Morphol. 2002, 10, 134–138. [Google Scholar] [CrossRef]
- Monzo, M.; Rosell, R.; Felip, E.; Astudillo, J.; Sanchez, J.J.; Maestre, J. A novel anti-apoptosis gene: Re-expression of survivin messenger RNA as a prognosis marker in non-small-cell lung cancers. J. Clin. Oncol. 1999, 17, 2100–2104. [Google Scholar] [CrossRef]
- Yu, J.; Leung, W.K.; Ebert, M.P.; Ng, E.K.; Go, M.Y.; Wang, H.B. Increased expression of survivin in gastric cancer patients and in first degree relatives. Br. J. Cancer 2002, 87, 91–97. [Google Scholar] [CrossRef]
- Lu, C.D.; Altieri, D.C.; Tanigawa, N. Expression of a novel anti apoptosis gene, survivin, correlated with tumor cell apoptosis andp53 accumulation in gastric carcinomas. Cancer Res. 1998, 58, 1808–1812. [Google Scholar]
- Khan, Z.; Tiwari, R.P.; Mulherkar, R.; Sah, N.K.; Prasad, G.B.; Shrivastava, B.R. Detection of survivin and p53 in human oral cancer: Correlation with clinicopathologic findings. Head Neck 2009, 31, 1039–1348. [Google Scholar] [CrossRef]
- Lauxen, I.; Oliveira, M.G.; Rados, P.V.; Lingen, M.W.; Nor, J.F.; Sant’ana Filho, M. Immunoprofiling of oral squamous cell carcinomas reveals high p63 and survivin expression. Oral. Dis. 2014, 20, 76–80. [Google Scholar] [CrossRef][Green Version]
- Mori, A.; Wada, H.; Nishimura, Y.; Okamoto, T.; Takemoto, Y.; Kakishita, E. Expression of the antiapoptosis gene survivin in human leukemia. Int. J. Hematol. 2002, 75, 161–165. [Google Scholar] [CrossRef]
- Kato, J.; Kuwabara, Y.; Mitani, M.; Shinoda, N.; Sato, A.; Toyama, T.; Mitsui, A.; Nishiwaki, T.; Moriyama, S.; Kudo, J.; et al. Expression of survivin in esophageal cancer: Correlation with the prognosis and response to chemotherapy. Int. J. Cancer 2001, 95, 305–310. [Google Scholar] [CrossRef]
- Yesupatham, S.T.; Dayanand, C.D.; Azeem Mohiyuddin, S.M. Cellular Concentration of Survivin and Caspase 3 in Habitual Tobacco Chewers with and without Oral Squamous Cell Carcinoma in South Indian Rural Population—A Case Control Study. Diagnostics 2022, 12, 2249. [Google Scholar] [CrossRef]
- Dai, D.; Liang, Y.; Xie, Z. Survivin deficiency induces apoptosis and cell cycle arrest in HepG2 hepatocellular carcinoma cells. Oncol. Rep. 2012, 27, 621–627. [Google Scholar] [CrossRef][Green Version]
- Rosa, J.; Canovas, P.; Islam, A.; Altieri, D.C.; Doxsey, S.J. Survivin modulates micro-tubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell 2006, 17, 1483–1493. [Google Scholar] [CrossRef]
- Khan, Z.; Khan, A.A.; Yadav, H.; Prasad, G.B.; Bisen, P.S. Survivin, a molecular target for therapeutic interventions in Squamous cell carcinoma. Cell. Mol. Biol. Lett. 2017, 22, 8. [Google Scholar] [CrossRef]
- Srivastava, K.; Srivastava, A.; Mittal, B. Survivin promoter -31G/C (rs9904341) polymorphism and cancer susceptibility: A meta-analysis. Mol. Biol. Rep. 2012, 39, 1509–1516. [Google Scholar] [CrossRef]
- Nassar, A.; Sexton, D.; Cotsonis, G.; Cohen, C. Survivin expression in breast carcinoma: Correlation with apoptosis and prognosis. Appl. Immunohistochem. Mol. Morphol. 2008, 16, 221–226. [Google Scholar] [CrossRef]
- Ryan, B.M.; Konecny, G.E.; Kahlert, S. Survivin expression in breast cancer predicts clinical outcome and is associated with HER2, VEGF, urokinase plasminogen activator and PAI-1. Ann. Oncol. 2006, 17, 597–604. [Google Scholar] [CrossRef]
- Khan, S.; Bennit, H.F.; Turay, D. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014, 14, 176. [Google Scholar] [CrossRef]
- Jha, K.; Shukla, M.; Pandey, M. Survivin expression and targeting in breast cancer. Surg. Oncol. 2012, 21, 125–131. [Google Scholar] [CrossRef]
- Han, C.H.; Wei, Q.; Lu, K.K.; Liu, Z.; Mills, G.B.; Wang, L.E. Polymorphisms in the survivin promoter are associated with age of onset of ovarian cancer. Int. J. Clin. Exp. Med. 2009, 2, 289–299. [Google Scholar]
- Guo, G.; Zhang, Q.; Yu, Z. Correlation between survivin genetic polymorphisms and lung cancer susceptibility. Int. J. Clin. Exp. Pathol. 2015, 8, 7426–7430. [Google Scholar]
- Upadhyay, R.; Khurana, R.; Kumar, S.; Ghoshal, U.C.; Mittal, B. Role of survivin gene promoter polymorphism (-31G > C) in susceptibility and survival of esophageal cancer in northern India. Ann. Surg. Oncol. 2011, 18, 880–887. [Google Scholar] [CrossRef]
- Xun, C.; Wei, Z.; Shangwu, C.; Dongsheng, Y. Mutation profiles of oral squamous cell carcinoma cells. Adv. Oral Maxillofac. Surg. 2021, 2, 100026. [Google Scholar]
- Sand, L.; Jalouli, M.; Jalouli, J.; Sapkota, D.; Ibrahim, S. P53 Codon 72 polymorphism in oral exfoliated cells in a sudanese population. In Vivo 2012, 26, 59–62. [Google Scholar]
- Ralhan, R.; Agarwal, S.; Nath, N.; Mathur, M.; Wasylyk, A.S. Correlation between p53 mutations and circulating antibodies in betel and tobacco-consuming North Indian population. Oral Oncol. 2001, 37, 243–250. [Google Scholar] [CrossRef]
- Mulot, C.; Stücker, I.; Clavel, J.; Beaune, P.; Loriot, M.A. Collection of human genomic DNA from buccal cells for genetics studies: Comparison between cytobrush, mouthwash, and treated card. J. Biomed. Biotechnol. 2005, 3, 291–296. [Google Scholar] [CrossRef]
- Bendl, J.; Musil, M.; Stourac, J.; Zendulka, J.; Damborsky, J.; Brezovsky, J. PredictSNP2: A Unified Platform for Accurately Evaluating SNP Effects by Exploiting the Different Characteristics of Variants in Distinct Genomic Regions. PLoS Comput. Biol. 2016, 12, e1004962. [Google Scholar] [CrossRef]
- Saurabh, B.; Balasubramaniam, G.; Prabhashankar, M.; Aanchal, J. Role of Monocyte Count and Neutrophil-to-Lymphocyte Ratio in Survival of Oral Cancer Patients. Int. Arch. Otorhinolaryngol. 2017, 21, 21–27. [Google Scholar]
- Satoshi, K.; Akihiro, H.; Hiromitsu, H.; Takatsugu, M.; Tomohiro, S.; Tomohiko, K.; Satoshi, F. Pretreatment lymphocyte-to-monocyte ratio as an independent prognostic factor for head and neck cancer. Head Neck 2016, 39, 247–253. [Google Scholar]
- Wu, Y.; Chang, K.; Ho, T. Comparative prognostic value of different preoperative complete blood count cell ratios in patients with oral cavity cancer treated with surgery and postoperative radiotherapy. Cancer Med. 2021, 10, 1975–1988. [Google Scholar] [CrossRef]
- Jane, C.; Nerurkar, A.V.; Shirsat, N.V.R.; Deshpande, B.; Amrapurkar, A.D.; Karjodkar, F.R. Increased survivin expression in high-grade oral squamous cell carcinoma: A study in Indian tobacco chewers. J. Oral. Pathol. Med. 2006, 35, 595–601. [Google Scholar] [CrossRef]
- Caro, J.J.; Salas, M.; Ward, A. Anemia as an independent prognostic factor for survival in patients with cancer: A systemic, quantitative review. Cancer 2001, 91, 2214–2221. [Google Scholar] [CrossRef]
- Chen, M.H.; Chang, P.M.; Chen, P.M.; Tzeng, C.H.; Chu, P.Y.; Chang, S.Y. Prognostic Significance of a Pretreatment Hematologic Profile in Patients with Head and Neck Cancer. J. Cancer Res. Clin. Oncol. 2009, 135, 1783–1790. [Google Scholar] [CrossRef]
- Alameddine, R.S.; Hamieh, L.; Shamseddine, A. From Sprouting Angiogenesis to Erythrocytes Generation by Cancer Stem Cells: Evolving Concepts in Tumor Microcirculation. BioMed Res. Int. 2014, 2014, 986768. [Google Scholar] [CrossRef]
- Huang, S.H.; Waldron, J.N.; Milosevic, M.; Shen, X.; Ringash, J.; Su, J. Prognostic Value of Pretreatment Circulating Neutrophils, Monocytes, and Lymphocytes in Oropharyngeal Cancer Stratified by Human Papillomavirus Status. Cancer 2015, 121, 545–555. [Google Scholar] [CrossRef]
- Hsueh, C.; Tao, L.; Zhang, M.; Cao, W.; Gong, H.; Zhou, J. The Prognostic Value of Preoperative Neutrophils, Platelets, Lymphocytes, Monocytes and Calculated Ratios in Patients with Laryngeal Squamous Cell Cancer. Oncotarget 2017, 8, 60514–60527. [Google Scholar] [CrossRef]
- Yuzhen, L.; Xiang, S.; Wenchao, L.; Lijun, M.; Zheng, Y.; Xiaohong, L.; Liuqun, Q.; Wuning, M. Evaluation of the clinical value of hematological parameters in patients with urothelial carcinoma of the bladder. Medicine 2018, 97, 14. [Google Scholar]
- Nakashima, H.; Matsuoka, Y.; Yoshida, R.; Nagata, M.; Hirosue, A.; Kawahara, K. Pre-treatment neutrophil to lymphocyte ratio predicts the chemoradiotherapy outcome and survival in patients with oral squamous cell carcinoma: A retrospective study. BMC Cancer 2016, 16, 41. [Google Scholar] [CrossRef]
- Phulari, R.G.; Rathore, R.S.; Shah, A.K.; Agnani, S.S. Neutrophil: Lymphocyte ratio and oral squamous cell carcinoma: A preliminary study. J. Oral. Maxillofac. Pathol. 2019, 23, 78–81. [Google Scholar]
- Lee, A.; Whyte, M.K.; Haslett, C. Inhibition of apoptosis andprolongation of neutrophil functional longevity by inflammatory mediators. J. Leukoc. Biol. 1993, 54, 283–288. [Google Scholar] [CrossRef]
- Brach, M.A.; DeVos, S.; Gruss, H.J.; Herrmann, F. Prolongationof survival of human polymorphonuclear neutrophils by granulocyte-macrophage colony-stimulating factor is caused by inhibition of programmedcell death. Blood 1992, 80, 2920–2924. [Google Scholar] [CrossRef]
- Altznauer, F.; Martinelli, S.; Yousefi, S.; Thurig, C.; Schmid, I.; Conway, E.M.; Schoni, M.H.; Vogt, P.; Mueller, C.; Fey, M.F.; et al. Inflammation-associated cell cycle-independent block of apoptosis by survivin in terminally differentiated neutrophils. J. Exp. Med. 2004, 199, 1343–1354. [Google Scholar] [CrossRef]
- Temraz, S.; Mukherji, D.; Farhat, Z.A.; Nasr, R.; Charafeddine, M.; Shahait, M.; Wehbe, M.R.; Ghaida, R.A.; Gheida, I.A.; Shamseddine, A. Preoperative lymphocyte-to-monocyte ratio predicts clinical outcome in patients undergoing radical cystectomy for transitional cell carcinoma of the bladder: A retrospective analysis. BMC Urol. 2014, 76, 14–19. [Google Scholar] [CrossRef]
- Stotz, M.; Szkandera, J.; Stojakovic, T.; Seidel, J.; Samonigg, H.; Kornprat, P.; Schaberl-Moser, R.; Seggewies, F.; Hoefler, G.; Gerger, A.; et al. The lymphocyte to monocyte ratio in peripheral blood represents a novel prognostic marker in patients with pancreatic cancer. Clin. Chem. Lab. Med. 2015, 53, 499–506. [Google Scholar] [CrossRef]
- Mantovani, A.; Bottazzi, B.; Colotta, F.; Sozzani, S.; Ruco, L. The origin and function of tumor-associated macrophages. Immunol. Today 1992, 13, 265–270. [Google Scholar] [CrossRef]
- Condeelis, J.; Pollard, J.W. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006, 124, 263–266. [Google Scholar] [CrossRef]
- Pollard, J.W. Tumour-educated macrophages promote tumour progressionand metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef]
- Dirkx, A.E.; Oude Egbrink, M.G.; Wagstaff, J.; Griffioen, A.W. Monocyte/macrophage infiltration in tumors: Modulators of angiogenesis. J. Leukoc. Biol. 2006, 80, 1183–1196. [Google Scholar] [CrossRef]
- Lewis, C.E.; Pollard, J.W. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006, 66, 605–612. [Google Scholar] [CrossRef]
- Tsai, Y.D.; Wang, C.P.; Chen, C.Y.; Lin, L.W.; Hwang, T.Z.; Lu, L.F.; Hsu, H.F.; Chung, F.M.; Lee, Y.J.; Houng, J.Y. Pretreatment circulating monocyte count associated with poor prognosis in patients with oral cavity cancer. Head Neck 2014, 36, 947–953. [Google Scholar] [CrossRef]
- Duzlu, M.; Karamert, R.; Tutar, H.; Şahin, M.; Turkcan, A.; Yılmaz, M. Diagnostic role of neutrophil-lymphocyte ratio in oral cavity cancers. Niger. J. Clin. Pract. 2018, 21, 49–53. [Google Scholar]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Rabinowich, H.; Cohen, R.; Bruderman, I.; Steiner, Z.; Klajman, A. Functional analysis of mononuclear cells infiltrating into tumors:lysis of autologous human tumor cells by cultured infiltrating lymphocytes. Cancer Res. 1987, 47, 173–177. [Google Scholar]
- Cho, H.; Hur, H.W.; Kim, S.W. Pre-treatment neutrophil to lymphocyte ratio is elevated in epithelial ovarian cancer and predicts survival after treatment. Cancer Immunol. Immunother. 2009, 58, 2346. [Google Scholar] [CrossRef]
- Yamanaka, T.; Matsumoto, S.; Teramukai, S.; Ishiwata, R.; Nagai, Y.; Fukushima, M. The baseline ratio of neutrophils to lymphocytes is associated with patient prognosis in advanced gastric cancer. Oncology 2007, 73, 215–220. [Google Scholar] [CrossRef]
- Deng, Q.; He, B.; Liu, X. Prognostic value of pre-operative inflammatoryresponse biomarkers in gastric cancer patients and the construction of a predictive model. J. Transl. Med. 2015, 13, 66. [Google Scholar] [CrossRef]
- Han, L.H.; Jia, Y.B.; Song, Q.X.; Wang, J.B.; Wang, N.N.; Cheng, Y.F. Prognostic significance of preoperative lymphocyte-monocyte ratio in patients withresectable esophageal squamous cell carcinoma. Asian Pac. J. Cancer Prev. 2015, 16, 2245–2250. [Google Scholar] [CrossRef]
- Neal, C.P.; Cairns, V.; Jones, M.J. Prognostic performance of inflammation-based prognostic indices in patients with resectable colorectalliver metastases. Med. Oncol. 2015, 32, 144–147. [Google Scholar] [CrossRef]
- Yamagishi, T.; Fujimoto, N.; Nishi, H. Prognostic significance of the lymphocyte-to-monocyte ratio in patients with malignant pleural mesothelioma. Lung Cancer 2015, 90, 111–117. [Google Scholar] [CrossRef]
- Ying, H.Q.; Deng, Q.W.; He, B.S. The prognostic value of preoperativeNLR, d-NLR, PLR and LMR for predicting clinical outcome in surgical colorectal cancer patients. Med. Oncol. 2014, 31, 305–309. [Google Scholar] [CrossRef]
- Zhang, G.M.; Zhu, Y.; Luo, L. Preoperative lymphocyte-monocyte andplatelet-lymphocyte ratios as predictors of overall survival in patients withbladder cancer undergoing radical cystectomy. Tumour. Biol. 2015, 36, 8537–8543. [Google Scholar] [CrossRef]
- Borst, J.; Ahrends, T.; Babala, N.; Melief, C.J.; Kastenmuller, W. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018, 18, 635–647. [Google Scholar] [CrossRef]
- Kumagai, K.; Hamada, Y.; Gotoh, A.; Kobayashi, H.; Kawaguchi, K.; Horie, A. Evidence for the changes of antitumor immune response during lymph node metastasis in head and neck squamous cell carcinoma. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 2010, 110, 341–350. [Google Scholar] [CrossRef]
- Bin-Alee, F.; Arayataweegool, A.; Buranapraditkun, S.; Mahattanasakul, P.; Tangjaturonrasme, N.; Mutirangura, A.; Kitkumthorn, N. Evaluation of lymphocyte apoptosis in patients with oral cancer. Appl. Oral. Sci. 2020, 28, E20200124. [Google Scholar] [CrossRef]
- Lu, B.; Gonzalez, A.; Massion, P.P. Nuclear survivin as a biomarker for non-small-cell lung cancer. Br. J. Cancer 2004, 91, 537–540. [Google Scholar] [CrossRef][Green Version]
- Grabowski, P.; Kuhnel, T.; Muhr-Wilkenshoff, F. Prognostic value of nuclear survivin expression in oesophageal squamous cell carcinoma. Br. J. Cancer. 2003, 88, 115–159. [Google Scholar] [CrossRef]
- Tonini, G.; Vicenzi, B.; Santini, D.; Scarpa, D.; Vasaturo, T.; Malacrino, C.; Coppola, R. Nuclear and cytoplasmic expression of survivin in 67 surgically resected pancreatic cancer patients. Br. J. Cancer 2005, 92, 2225–2232. [Google Scholar] [CrossRef]
- Poomsawat, S.; Punyasingh, J.; Vejchapipat, P. Overexpression of survivin and caspase 3 in oral carcinogenesis. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 65–71. [Google Scholar] [CrossRef]
- Wheatley, S.P.; Altieri, D.C. Survivin at a glance. J. Cell Sci. 2019, 132, jcs223826. [Google Scholar] [CrossRef]
- Boidot, R.; Vegran, F.; Lizard-Nacol, S. Transcriptional regulation of the survivin gene. Mol. Biol. Rep. 2014, 41, 233–240. [Google Scholar] [CrossRef]
- Aspe, J.R.; Wall, N.R. Survivin-T34A: Molecular mechanism and therapeutic potential. Onco. Targets. Ther. 2010, 3, 247–254. [Google Scholar]
- Li, M.; Gao, F.; Yu, X.; Zhao, Q.; Zhou, L.; Liu, W.; Li, W. Promotion of ubiquitination-dependent survivin destruction contributes to xanthohumol-mediated tumor suppression and overcomes radioresistance in human oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 88. [Google Scholar] [CrossRef]
- Yazdani, N.; Sayahpour, F.A.; Haghpanah, V. Survivin gene polymorphism association with papillary thyroid carcinoma. Pathol. Res. Pract. 2012, 208, 100–103. [Google Scholar] [CrossRef]
- Chen, J.; Cui, X.; Zhou, H. Functional promoter -31G/C variant of survivin gene predict prostate cancer susceptibility among Chinese: A case control study. BMC Cancer 2013, 13, 356. [Google Scholar] [CrossRef]
- Yang, X.; Xiong, G.; Chen, X. Survivin expression in esophageal cancer: Correlation with p53 mutations and promoter polymorphism. Dis Esophagus. 2009, 22, 223–230. [Google Scholar] [CrossRef]
- Li, X.B.; Li, S.N.; Yang, Z.H.; Cao, L.; Duan, F.L.; Sun, X.W. Polymorphisms of survivin and its protein expression are associated with colorectal cancer susceptibility in Chinese population. DNA Cell Biol. 2013, 32, 236–242. [Google Scholar] [CrossRef]
- Dunajova, L.; Cash, E.; Markus, R.; Rochette, S.; Townley, A.R.; Wheatley, S.P. The N-terminus of survivin is a mitochondrial-targeting sequence and Src regulator. J. Cell Sci. 2016, 129, 2707–2712. [Google Scholar] [CrossRef]
- Dohi, T.; Beltrami, E.; Wall, N.R.; Plescia, J.; Altieri, D.C. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J. Clin. Investig. 2004, 114, 1117–1127. [Google Scholar] [CrossRef]
- Xia, F.; Canovas, P.M.; Guadagno, T.M.; Altieri, D.C. A survivin-ran complex regulates spindle formation in tumor cells. Mol. Cell Biol. 2008, 28, 5299–5311. [Google Scholar] [CrossRef]
- Song, Z.; Liu, S.; He, H.; Hoti, N.; Wang, Y.; Feng, S.; Wu, M. A single amino acid change (Asp53-Ala53) converts survivin from anti-apoptotic to pro-apoptotic. Mol. Biol. Cell 2004, 15, 1287–1296. [Google Scholar] [CrossRef]
- Verdecia, M.A.; Bowman, M.E.; Lu, K.P.; Hunter, T.; Noel, J.P. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 2000, 7, 602–608. [Google Scholar] [CrossRef]
- Pavlyukov, M.S.; Antipova, N.V.; Balashova, M.V.; Vinogradova, T.V.; Kopantzev, E.P.; Shakhparonov, M.I. Survivin monomer plays an essential role in apoptosis regulation. J. Biol. Chem. 2011, 286, 23296–23307. [Google Scholar] [CrossRef]
- Jeyaprakash, A.A.; Basquin, C.; Jayachandran, U.; Conti, E. Structural basis for the recognition of phosphorylated histone H3 by the Survivin subunit of the chromosomal passenger complex. Structure 2011, 19, 1625–1634. [Google Scholar] [CrossRef]
- Stauber, R.H.; Rabenhorst, U.; Rekik, A.; Engels, K.; Bier, C.; Knauer, S.K. Nucleocytoplasmic shuttling and the biological activity of mouse survivin are regulated by an active nuclear export signal. Traffic 2007, 7, 1461–1472. [Google Scholar] [CrossRef]
- Colnaghi, R.; Connell, C.M.; Barrett, R.M.A.; Wheatley, S.P. Separating the anti-apoptotic and mitotic roles of survivin. J. Biol. Chem. 2006, 281, 33450–33456. [Google Scholar] [CrossRef]
- Temme, A.; Diestelkoetter-Bachert, P.; Schmitz, M.; Morgenroth, A.; Weigle, B.; Rieger, M.A.; Kiessling, A.; Rieber, E.P. Increased p21ras activity in human fibroblasts transduced with survivin enhances cell proliferation. Biochem. Biophys. Res. Commun. 2005, 327, 765–773. [Google Scholar] [CrossRef]
- Al-Khalaf, H.H.; Aboussekhra, A. Survivin expression increases during aging and enhances the resistance of aged human fibroblasts to genotoxic stress. Age 2013, 35, 549–562. [Google Scholar] [CrossRef]
- Lens, S.M.A.; Wolthuis, R.M.; Klompmaker, R.; Kauw, J.; Agami, R.; Brummelkamp, T.; Kops, G.; Medema, R.H. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBOJ 2003, 22, 2934–2947. [Google Scholar] [CrossRef]
- Carvalho, A.; Carmena, M.; Sambade, C.; Earnshaw, W.C.; Wheatley, S.P. Survivin is required for stable checkpoint activation in response to loss of spindle tension in HeLa cells. J. Cell Sci. 2003, 116, 2987–2998. [Google Scholar] [CrossRef]
- Rajagopalan, S.; Balasubramanian, M.K. Schizosaccharomyces pombe Bir1p, a nuclear protein that localizes to kinetochores and the spindle midzone, is essential for chromosome condensation and spindle elongation during mitosis. Genetics 2002, 160, 445–456. [Google Scholar] [CrossRef]



| Locus | Primer | Sequence | Product Size |
|---|---|---|---|
| Promoter | Forward | GCCTCTCAAAGTGTTGGGATTA | 343 bp |
| Reverse | GGGCCAGTTCTTGAATGTAGAG | ||
| Exon1 | Forward | CCGCCTCTACTCCCAGAA | 226 bp |
| Reverse | CGCAGCCCTCCAAGAAG | ||
| Exon2 | Forward | CTCCCTGCTTTGTCCCCAT | 341 bp |
| Reverse | GAGGTATCCGTTCACAACAGC | ||
| Exon3 | Forward | GCAGTTCTGGTAACGGTGATAG | 230 bp |
| Reverse | CCGTATTAGCCAAGATGGTCTC | ||
| Exon4 | Forward | ATGTCCACAGGGAGAGAGAA | 536 bp |
| Reverse | GAGAATCACTTGAACCCGAGAG | ||
| Exon5 | Forward (Internal Primer) | GAAGCGTCTGGCAGATAC | 1070 bp |
| Reverse | AGTCTAGGCGGTTGCACTT |
| Analytes | Groups | Mean ± SD | 95% Confidence Interval for Mean | p Value | |
|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||
| Survivin (pg/mL) | Group 1 | 1670.9 ± 796.21 | 1466.94 | 1874.75 | <0.001 |
| Group 2 | 1096.02 ± 346.17 | 1008.11 | 1183.93 | ||
| Group 3 | 397.5 ± 96.1 | 373.29 | 421.69 | ||
| SL. No. | Blood Parameters | Mean ± SD (N = 63) |
|---|---|---|
| 1 | Hemoglobin (gm%) | 11.67 ± 1.68 |
| 2 | RBC (mil/cu.mm) | 4.47 ± 0.57 |
| 3 | Packed Cell Volume (%) | 36.14 ± 4.60 |
| 4 | Mean Corpuscular Volume (fl) | 80.85 ± 6.22 |
| 5 | Mean Corpuscular Hemoglobin Concentration (%) | 32.23 ± 1.51 |
| 6 | Red Cell Distribution Width (%) | 15.81 ± 3.04 |
| 7 | White Blood Cell count (thousands/cu.mm) | 7.67 ± 2.75 |
| 8 | Absolute Neutrophil Count × 109/L | 4.38 ± 2.18 |
| 9 | Absolute Lymphocyte Count × 109/L | 1.96 ± 1.02 |
| 10 | Absolute Monocyte Count × 109/L | 0.90 ± 1.76 |
| 11 | Platelet Count × 109/L | 341.1 ± 113.63 |
| 12 | Neutrophil/Lymphocyte ratio | 2.86 ± 2.34 |
| 13 | Platelet/Lymphocyte Ratio | 226.86 ± 144.98 |
| 14 | Lymphocyte/Monocyte Ratio | 3.12 ± 1.50 |
| SL. No. | Survivin pg/mL | Univariate Analysis | |
|---|---|---|---|
| Pearson Correlation | Sig. (2-Tailed) | ||
| 1 | Hemoglobin | −0.014 | 0.91 |
| 2 | RBC | 0.023 | 0.86 |
| 3 | PCV | 0.075 | 0.56 |
| 4 | MCV | 0.077 | 0.55 |
| 5 | MCHC | −0.221 | 0.08 |
| 6 | RDW | 0.193 | 0.13 |
| 7 | WBC | −0.143 | 0.26 |
| 8 | ANC | −0.094 | 0.46 |
| 9 | ALC | −0.221 | 0.08 |
| 10 | AMC | −0.089 | 0.49 |
| 11 | PLT | 0.037 | 0.78 |
| 12 | NLR | 0.166 | 0.19 |
| 13 | PLR | 0.321 * | 0.01 * |
| 14 | LMR | −0.196 | 0.12 |
| Variables | T. Staging | N | Mean | SD | t Value | p Value | Node Involvement | N | Mean | SD | t Value | p Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hb | T1–T2 | 13 | 12 | 1.50 | 0.762 | 0.449 | N0 | 14 | 11.79 | 1.63 | 0.296 | 0.769 |
| T3–T4 | 50 | 11.6 | 1.7 | N1–N3 | 49 | 11.64 | 1.70 | |||||
| RBC | T1–T2 | 13 | 4.53 | 0.42 | 0.455 | 0.651 | N0 | 14 | 4.34 | 0.67 | −0.952 | 0.345 |
| T3–T4 | 50 | 4.45 | 0.61 | N1–N3 | 49 | 4.51 | 0.54 | |||||
| PCV | T1–T2 | 13 | 36.76 | 3.76 | 0.542 | 0.590 | N0 | 14 | 35.84 | 4.41 | −0.281 | 0.779 |
| T3–T4 | 50 | 35.98 | 4.81 | N1–N3 | 49 | 36.23 | 4.69 | |||||
| MCV | T1–T2 | 13 | 81.05 | 7.39 | 0.129 | 0.898 | N0 | 14 | 82.25 | 4.14 | 0.957 | 0.342 |
| T3–T4 | 50 | 80.79 | 5.96 | N1–N3 | 49 | 80.45 | 6.68 | |||||
| MCHC | T1–T2 | 13 | 32.82 | 1.92 | 1.610 | 0.113 | N0 | 14 | 32.43 | 2.01 | 0.554 | 0.581 |
| (T3–T4 | 50 | 32.08 | 1.37 | N1–N3 | 49 | 32.17 | 1.36 | |||||
| RDW | T1–T2 | 13 | 13.86 | 2.26 | −2.723 | 0.008 * | N0 | 14 | 14.13 | 1.43 | −2.436 | 0.018 * |
| T3–T4 | 50 | 16.31 | 3.03 | N1–N3 | 49 | 16.29 | 3.21 | |||||
| WBC | T1–T2 | 13 | 9.35 | 3.50 | 2.588 | 0.012 | N0 | 14 | 8.10 | 2.33 | 0.656 | 0.514 |
| T3–T4 | 50 | 7.23 | 2.37 | N1–N3 | 49 | 7.55 | 2.87 | |||||
| ABN | T1–T2 | 13 | 5.20 | 1.82 | 1.538 | 0.129 | N0 | 14 | 4.79 | 1.85 | 0.786 | 0.435 |
| T3–T4 | 50 | 4.17 | 2.23 | N1–N3 | 49 | 4.27 | 2.27 | |||||
| ABL | T1–T2 | 13 | 2.68 | 1.62 | 3.063 | 0.003 | N0 | 14 | 2.00 | 0.70 | 0.190 | 0.850 |
| T3–T4 | 50 | 1.77 | 0.70 | N1–N3 | 49 | 1.94 | 1.09 | |||||
| ABM | T1–T2 | 13 | 2.20 | 3.68 | 1.608 | 0.134 | N0 | 14 | 0.64 | 0.22 | −0.610 | 0.544 |
| T3–T4 | 50 | 0.56 | 0.17 | N1–N3 | 49 | 0.97 | 1.99 | |||||
| PLT | T1–T2 | 13 | 413.23 | 151.28 | 2.063 | 0.057 | N0 | 14 | 315.77 | 153.06 | −0.945 | 0.348 |
| T3–T4 | 50 | 322.36 | 94.83 | N1–N3 | 49 | 348.35 | 100.45 | |||||
| NLR | T1–T2 | 13 | 2.67 | 1.57 | −0.328 | 0.744 | N0 | 14 | 2.69 | 1.60 | −0.301 | 0.764 |
| T3–T4 | 50 | 2.91 | 2.52 | N1–N3 | 49 | 2.91 | 2.53 | |||||
| PLR | T1–T2 | 13 | 235.70 | 174.41 | 0.245 | 0.807 | N0 | 14 | 175.80 | 101.72 | −1.883 | 0.069 |
| T3–T4 | 50 | 224.56 | 138.28 | N1–N3 | 49 | 241.45 | 152.88 | |||||
| LMR | T1–T2 | 13 | 2.64 | 1.60 | −1.313 | 0.194 | N0 | 14 | 3.30 | 1.07 | 0.497 | 0.621 |
| T3–T4 | 50 | 3.25 | 1.45 | N1–N3 | 49 | 3.07 | 1.60 |
| Dependent Variable: Survivin | Hematology Parameters | Unstandardized Coefficients | Standardized Coefficients | t Value | p Value | |
| B | Std. Error | Beta | ||||
|---|---|---|---|---|---|---|
| (Constant) | 12,981.379 | 12,985.118 | 1.000 | 0.324 | ||
| Hb | −175.851 | 221.050 | −0.677 | −0.796 | 0.431 | |
| RBC | 681.616 | 645.181 | 0.897 | 1.056 | 0.297 | |
| PCV | 17.702 | 100.027 | 0.187 | 0.177 | 0.860 | |
| MCV | −88.496 | 132.679 | −1.266 | −0.667 | 0.509 | |
| MCHC | −471.015 | 329.721 | −1.637 | −1.429 | 0.161 | |
| RDW | 43.961 | 26.720 | 0.307 | 1.645 | 0.108 | |
| WBC | −205.654 | 547.736 | −1.300 | −0.375 | 0.709 | |
| ANC | 312.950 | 545.156 | 1.570 | 0.574 | 0.569 | |
| ALC | 1160.188 | 722.812 | 2.711 | 1.605 | 0.117 | |
| AMC | −3955.307 | 1909.181 | −16.024 | −2.072 | 0.045 * | |
| PLT | 1.075 | 1.364 | 0.281 | 0.788 | 0.435 | |
| NLR | 226.494 | 133.105 | 1.221 | 1.702 | 0.097 | |
| PLR | −5.289 | 2.704 | −1.765 | −1.956 | 0.058 | |
| LMR | 608.382 | 260.746 | 2.089 | 2.333 | 0.025 * | |
| Parameters | Cut-off Values | N | Mean ± Std. Deviation | t Value | p Value |
|---|---|---|---|---|---|
| WBC × 109/L | ≥7.9 | 26 | 2039.54 ± 394.83 | 1.080 | 0.284 |
| <7.9 | 37 | 1919.57 ± 459.11 | |||
| Absolute NeutrophilCount × 109/L | ≥4.9 | 21 | 2067.33 ± 411.62 | 1.276 | 0.207 |
| <4.9 | 42 | 1919.95 ± 442.07 | |||
| Absolute LymphocyteCount × 109/L | >1.980 | 20 | 2020.30 ± 404.10 | −0.635 | 0.528 |
| ≤1.980 | 43 | 1945.26 ± 450.54 | |||
| Absolute MonocyteCount × 109/L | ≥0.50 | 38 | 2116.92 ± 380.76 | 3.645 | 0.001 * |
| <0.50 | 25 | 1744.36 ± 420.71 | |||
| NLR | ≥2.39 | 21 | 2241.19 ± 203.90 | 3.896 | 0.001 * |
| <2.39 | 42 | 1833.02 ± 456.42 | |||
| LMR | >3.22 | 33 | 1717.55 ± 434.75 | 6.044 | 0.001 * |
| ≤3.22 | 30 | 2245.77 ± 209.46 | |||
| PLR | >110.6 | 47 | 2007.19 ± 395.99 | −1.197 | 0.236 |
| ≤110.6 | 16 | 1857.13 ± 530.60 |
| Promoter | Group 1 (n = 20) | Group 2 (n = 10) | Group 3 (n = 10) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Nucleotide Variants | Frequency | % | Tissue Survivin (Average pg/mL) | Frequency | % | Tissue Survivin (Average pg/mL) | Frequency | % | Tissue Survivin (Average pg/mL) |
| G → T | 7 | 35 | 2228.58 | 3 | 30 | 1104.12 | 3 | 30 | 443.65 |
| G → C | 3 | 15 | 2194.29 | - | - | - | 1 | 10 | 437.12 |
| A → T | 3 | 15 | 2139,39 | 1 | 10 | 1197.3 | 2 | 20 | 428.48 |
| C → T | 2 | 10 | 2204.96 | 5 | 50 | 1104.12 | 3 | 30 | 427.24 |
| C → G | 1 | 5 | 2008.01 | 2 | 20 | 1053.68 | 2 | 20 | 428.48 |
| A → G | - | - | - | 1 | 10 | 1197.3 | 1 | 10 | 424.75 |
| G → A | - | - | - | 3 | 30 | 1053.68 | - | - | - |
| T → G | 1 | 5 | 2082.88 | - | - | - | - | - | - |
| EXON 1 | Group 1 (n = 30) | Group 2 (n = 20) | Group 3 (n = 20) | ||||||
| Nucleotide Variants | Frequency | % | Tissue Survivin (Average pg/mL) | Frequency | % | Tissue Survivin (Average pg/mL) | Frequency | % | Tissue Survivin (average pg/mL) |
| G → A | 4 | 13.33 | 1400.68 | - | - | - | 2 | 10 | 116.35 |
| C → A | 10 | 33.33 | 1649.3 | 1 | 5 | 1197.3 | 1 | 5 | 180.97 |
| A → C | 3 | 10 | 1412.43 | - | - | - | - | - | - |
| G → C | 26 | 86.67 | 1632.77 | 10 | 50 | 1045.56 | 9 | 45 | 398.22 |
| G → T | 8 | 26.67 | 2015.04 | - | - | - | - | - | - |
| C → G | 2 | 6.67 | 2110.49 | - | - | - | - | - | - |
| T → G | 2 | 6.67 | 2427.92 | - | - | - | - | - | - |
| C → T | 1 | 3.33 | 1565.42 | - | - | - | - | - | - |
| EXON 2 | Group 1 (n = 30) | Group 2 (n = 20) | Group 3 (n = 20) | ||||||
| Nucleotide Variants | Frequency | % | Tissue Survivin (Average pg/mL) | Frequency | % | Tissue Survivin (Average pg/mL) | Frequency | % | Tissue Survivin (Average pg/mL) |
| A → C | - | - | - | 1 | 5 | 303.97 | 1 | 5 | 432.24 |
| G → C | 1 | 3.33 | 1074.94 | - | - | 3 | 15 | 432.24 | |
| C → T | - | - | - | - | - | 1 | 5 | 428.65 | |
| G → A | 1 | 3.33 | 1074.94 | - | - | 2 | 10 | 428.65 | |
| EXON 3 | Group 1 (n = 30) | Group 2 (n = 20) | Group 3 (n = 20) | ||||||
| Nucleotide Variants | Frequency | % | Tissue Survivin (Average pg/mL) | Frequency | % | Tissue Survivin (Average pg/mL) | Frequency | % | Tissue Survivin (Average pg/mL) |
| C → T | - | - | - | 2 | 10 | 720.05 | - | - | - |
| G → C | 1 | 3.33 | 2485.07 | - | - | - | - | - | |
| EXON 4 | Group 1 (n = 30) | Group 2 (n = 20) | Group 3 (n = 20) | ||||||
| Nucleotide Variants | Frequency | % | Tissue survivin (Average pg/mL) | Frequency | % | Tissue survivin (Average pg/mL) | Frequency | % | Tissue survivin (Average pg/mL) |
| G → C | - | - | - | 3 | 15 | 1266.55 | 1 | 5 | 422.81 |
| C → T | - | - | - | - | - | 1 | 5 | 422.81 | |
| T → C | 1 | 3.33 | 1850.2 | 2 | 10 | 1310 | - | - | |
| A → T | 2 | 6.67 | 1691.5 | 1 | 5 | 1415 | 2 | 10 | 406.12 |
| C → A | 1 | 3.33 | 2370.77 | - | - | - | - | - | - |
| A → G | 2 | 6.67 | 2272.69 | - | - | - | - | - | - |
| G → T | 3 | 10 | 1711.07 | - | - | - | - | - | - |
| T → G | 1 | 3.33 | 1850.2 | - | - | - | - | - | - |
| A → C | 2 | 6.67 | 1807.81 | - | - | - | - | - | - |
| G → A | 1 | 3.33 | 1765.42 | - | - | - | - | - | - |
| T → A | - | - | - | 2 | 10 | 1157.7 | 1 | 5 | 227.01 |
| EXON 5 | Group 1 (n = 30) | Group 2 (n = 20) | Group 3 (n = 20) | ||||||
| Nucleotide Variants | Frequency | % | Tissue survivin (Average pg/mL) | Frequency | % | Tissue survivin (Average pg/mL) | Frequency | % | Tissue survivin (Average pg/mL) |
| A → C | - | - | - | - | - | - | 1 | 5 | 422.81 |
| C → A | 1 | 3.33 | 1850.2 | - | - | - | - | - | - |
| T → A | 4 | 13.33 | 1856.94 | 2 | 10 | 1021.89 | 2 | 10 | 421.03 |
| G → A | 2 | 6.67 | 1542.09 | 1 | 5 | 628.77 | 1 | 5 | 391.69 |
| C → T | 1 | 3.33 | 1244.67 | 1 | 5 | 712.23 | 2 | 10 | 327.18 |
| G → T | 2 | 6.67 | 1542.09 | - | - | - | - | - | - |
| T → G | - | - | - | 1 | 5 | 1415 | - | - | - |
| G → C | 1 | 3.33 | 1850.2 | - | - | - | - | - | - |
| A → T | 1 | 3.33 | 1244.67 | - | - | - | 1 | 5 | 441.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yesupatham, S.T.; Dayanand, C.D.; Azeem Mohiyuddin, S.M.; Harendra Kumar, M.L. An Insight into Survivin in Relevance to Hematological, Biochemical and Genetic Characteristics in Tobacco Chewers with Oral Squamous Cell Carcinoma. Cells 2023, 12, 1444. https://doi.org/10.3390/cells12101444
Yesupatham ST, Dayanand CD, Azeem Mohiyuddin SM, Harendra Kumar ML. An Insight into Survivin in Relevance to Hematological, Biochemical and Genetic Characteristics in Tobacco Chewers with Oral Squamous Cell Carcinoma. Cells. 2023; 12(10):1444. https://doi.org/10.3390/cells12101444
Chicago/Turabian StyleYesupatham, Susanna Theophilus, C. D. Dayanand, S. M. Azeem Mohiyuddin, and M. L. Harendra Kumar. 2023. "An Insight into Survivin in Relevance to Hematological, Biochemical and Genetic Characteristics in Tobacco Chewers with Oral Squamous Cell Carcinoma" Cells 12, no. 10: 1444. https://doi.org/10.3390/cells12101444
APA StyleYesupatham, S. T., Dayanand, C. D., Azeem Mohiyuddin, S. M., & Harendra Kumar, M. L. (2023). An Insight into Survivin in Relevance to Hematological, Biochemical and Genetic Characteristics in Tobacco Chewers with Oral Squamous Cell Carcinoma. Cells, 12(10), 1444. https://doi.org/10.3390/cells12101444






