Cellular Functions of High-Temperature Requirement Factor A4 in Placenta
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects and Samples
2.2. Cell Culture
2.3. Construction of Expression Vectors
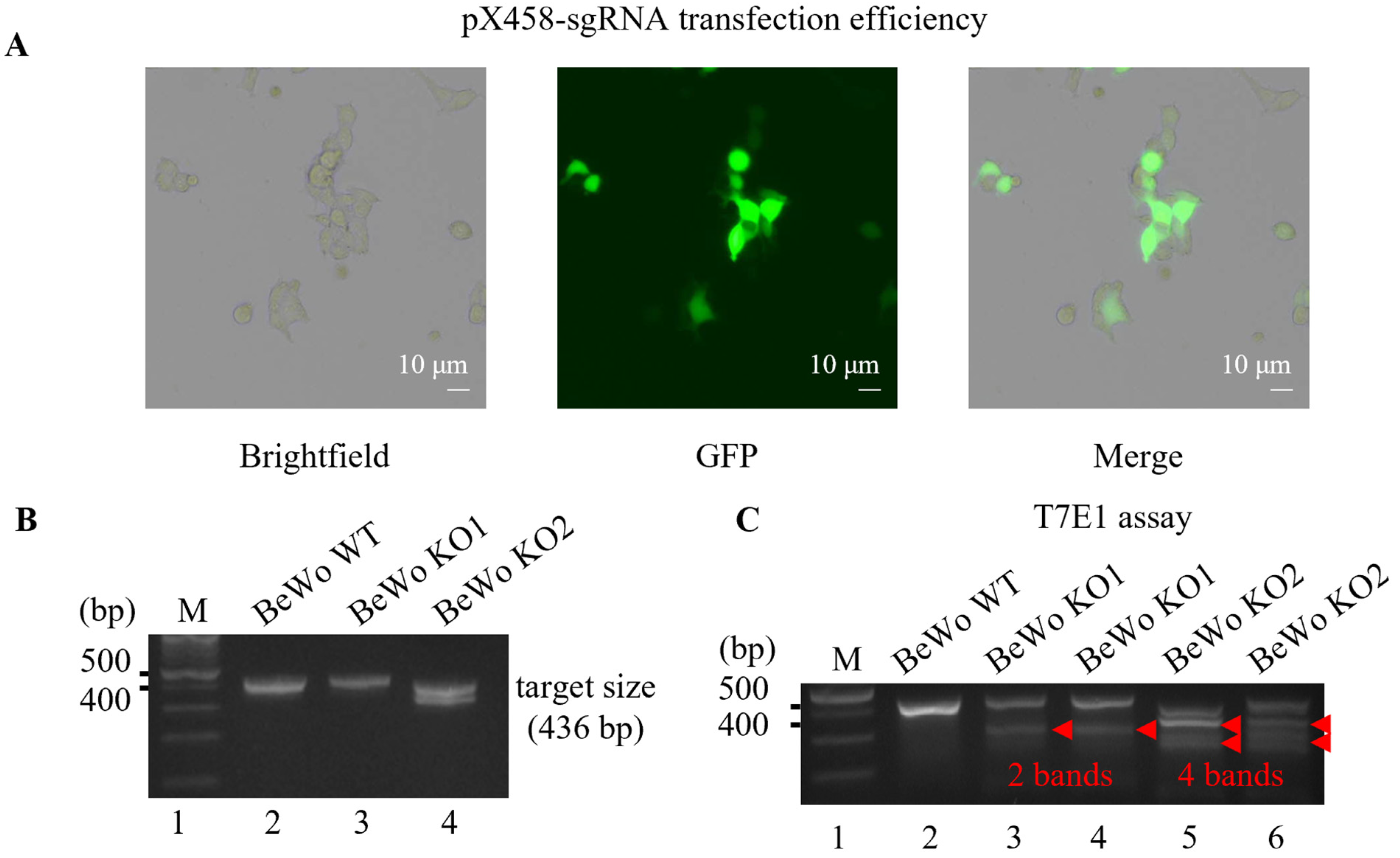
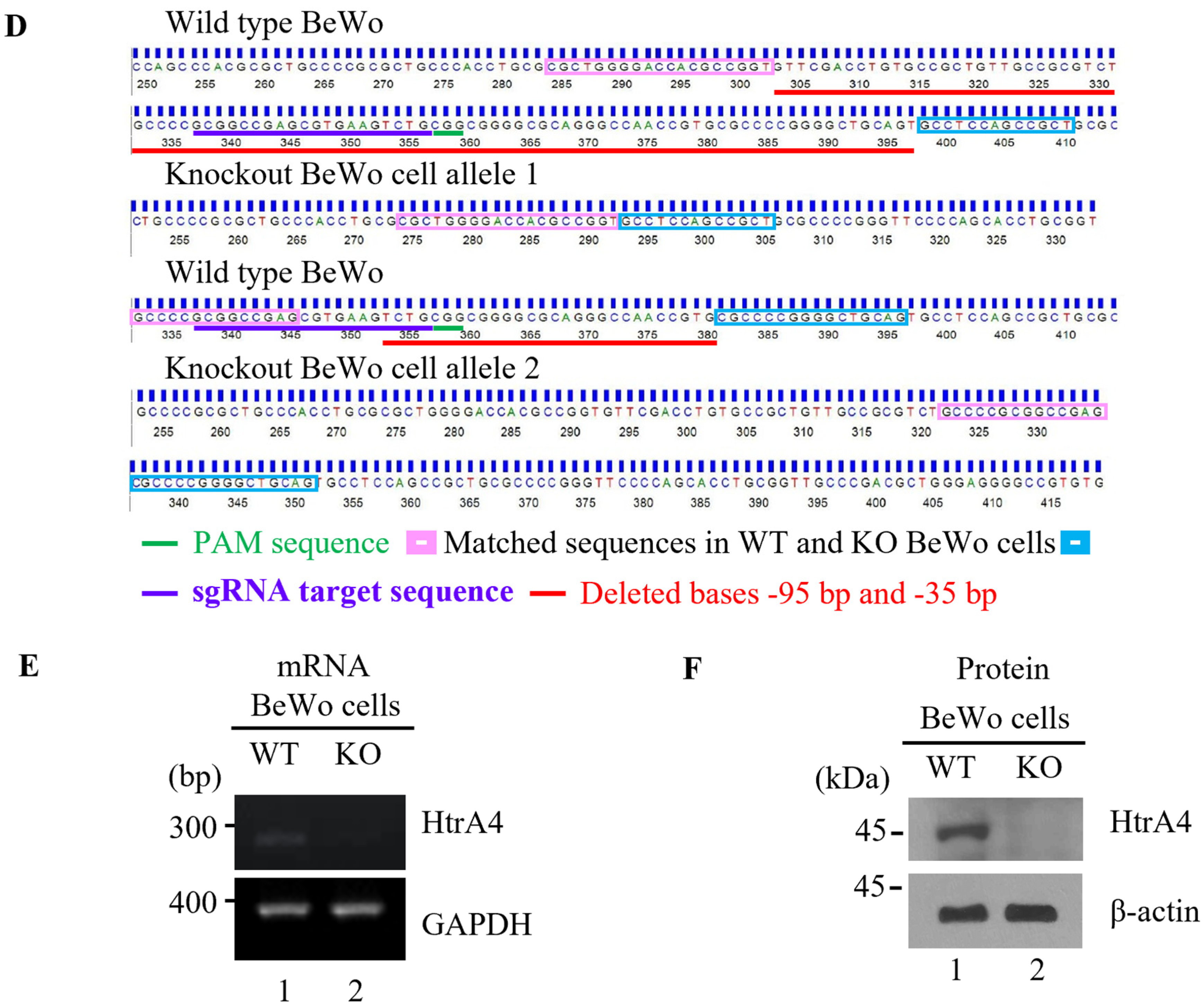
2.4. Generating HtrA4 Knockout in BeWo Cell Line by the CRISPR/Cas9 System
2.5. Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
2.6. Cell-Cell Fusion Analysis
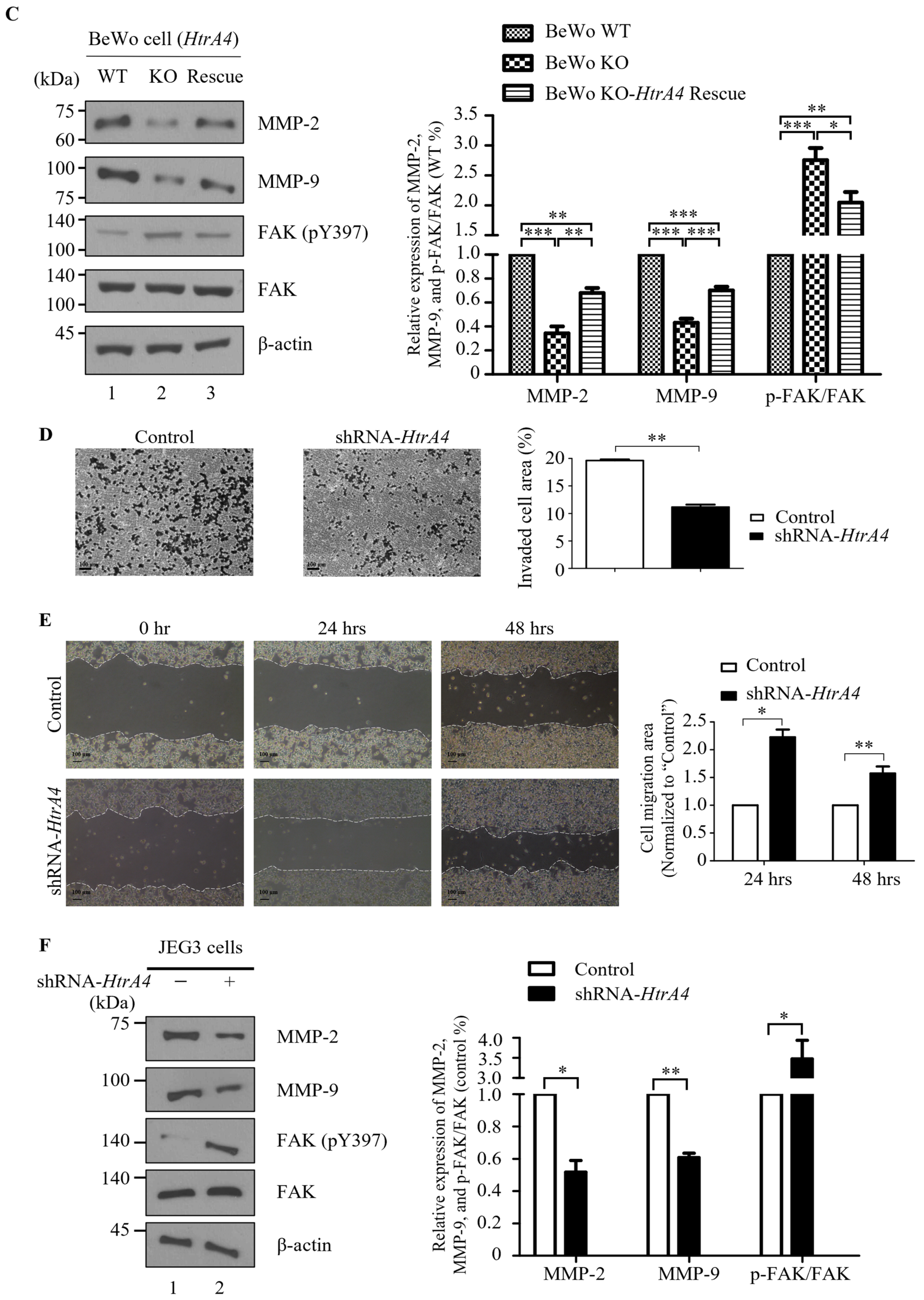
2.7. Invasion Assay In Vitro
2.8. Scratch Wound Assay In Vitro
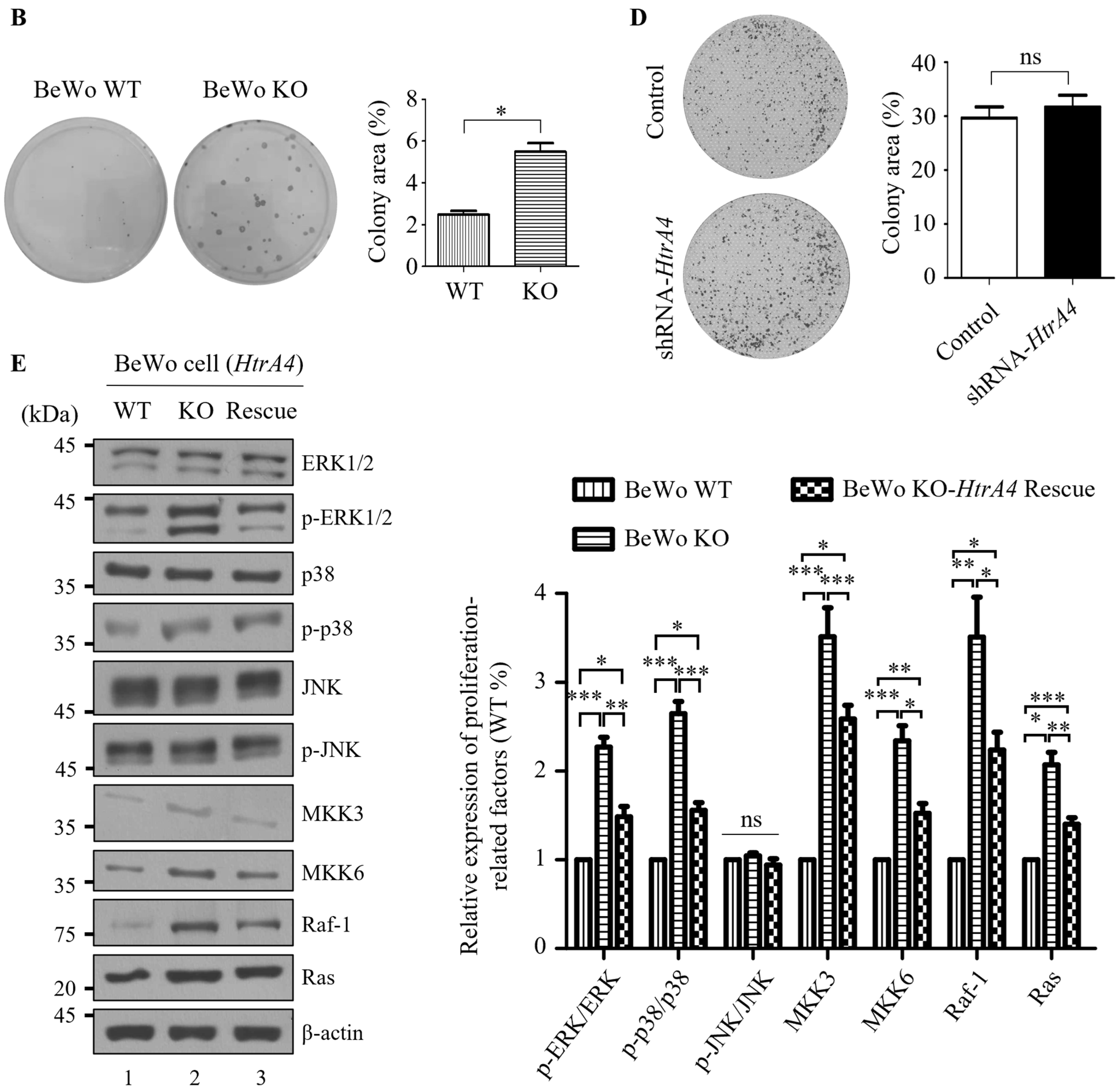
2.9. Cell Proliferation Assays
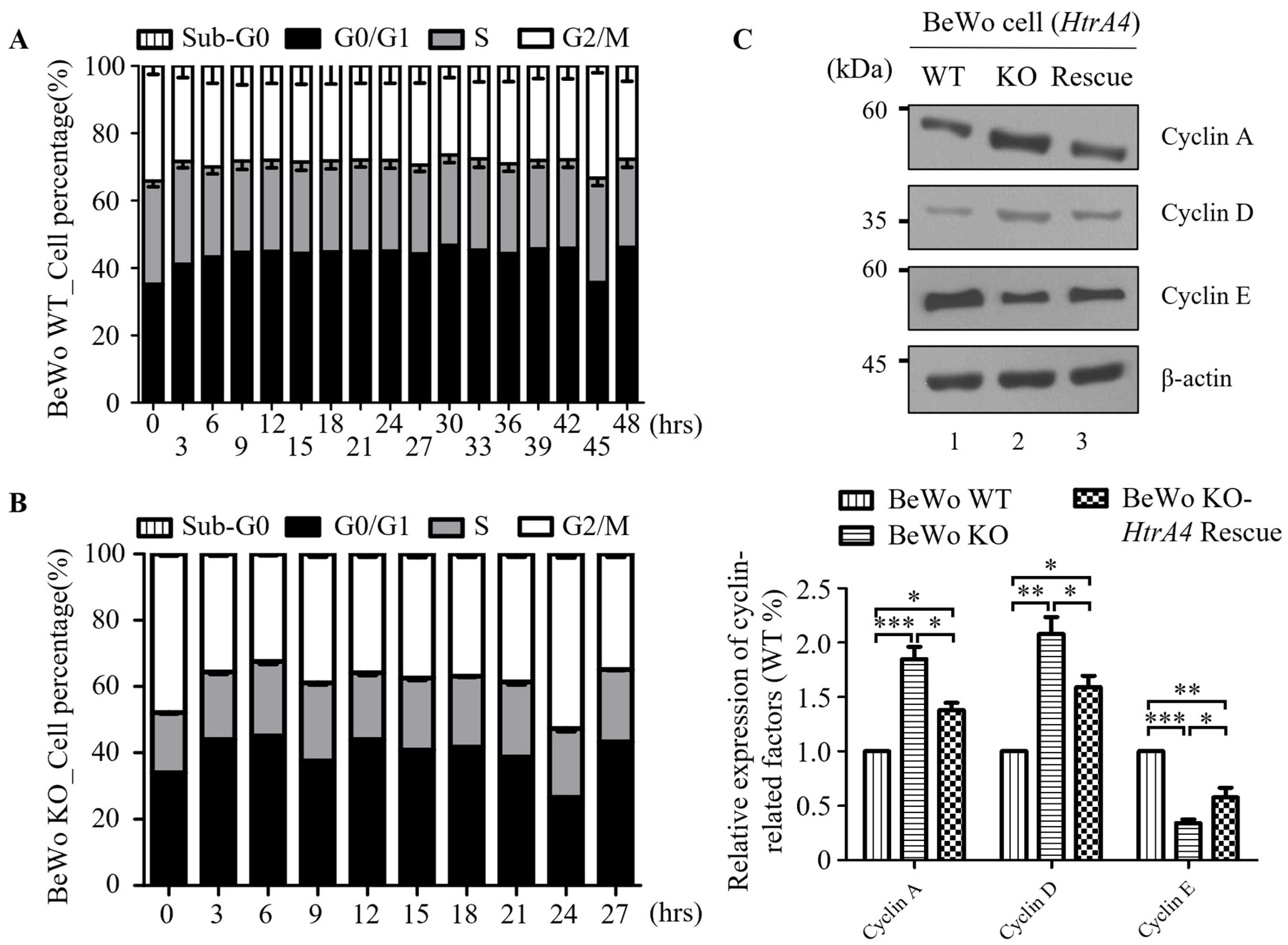
2.10. Cell Cycle Analysis and Flow Cytometry
2.11. Western Blot Analysis
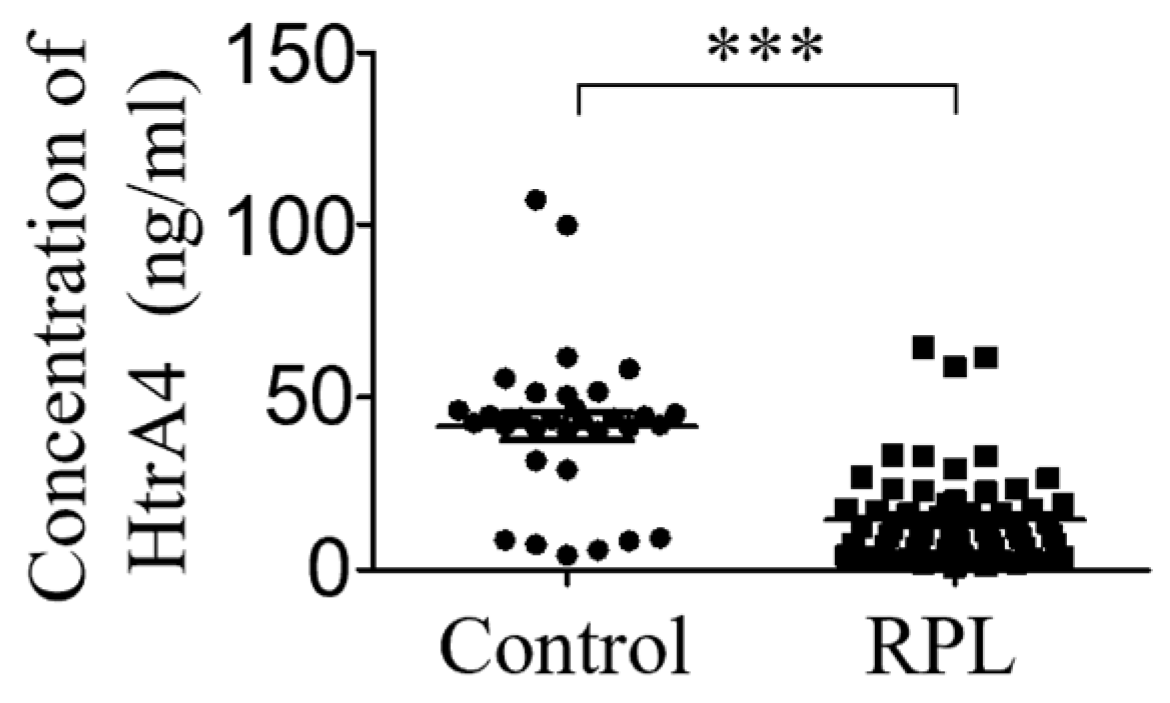
2.12. ELISA for HtrA4
2.13. Statistical Analysis
3. Results
3.1. Generation of HtrA4 Knockout in BeWo Cell Line Using CRISPR/Cas9 System
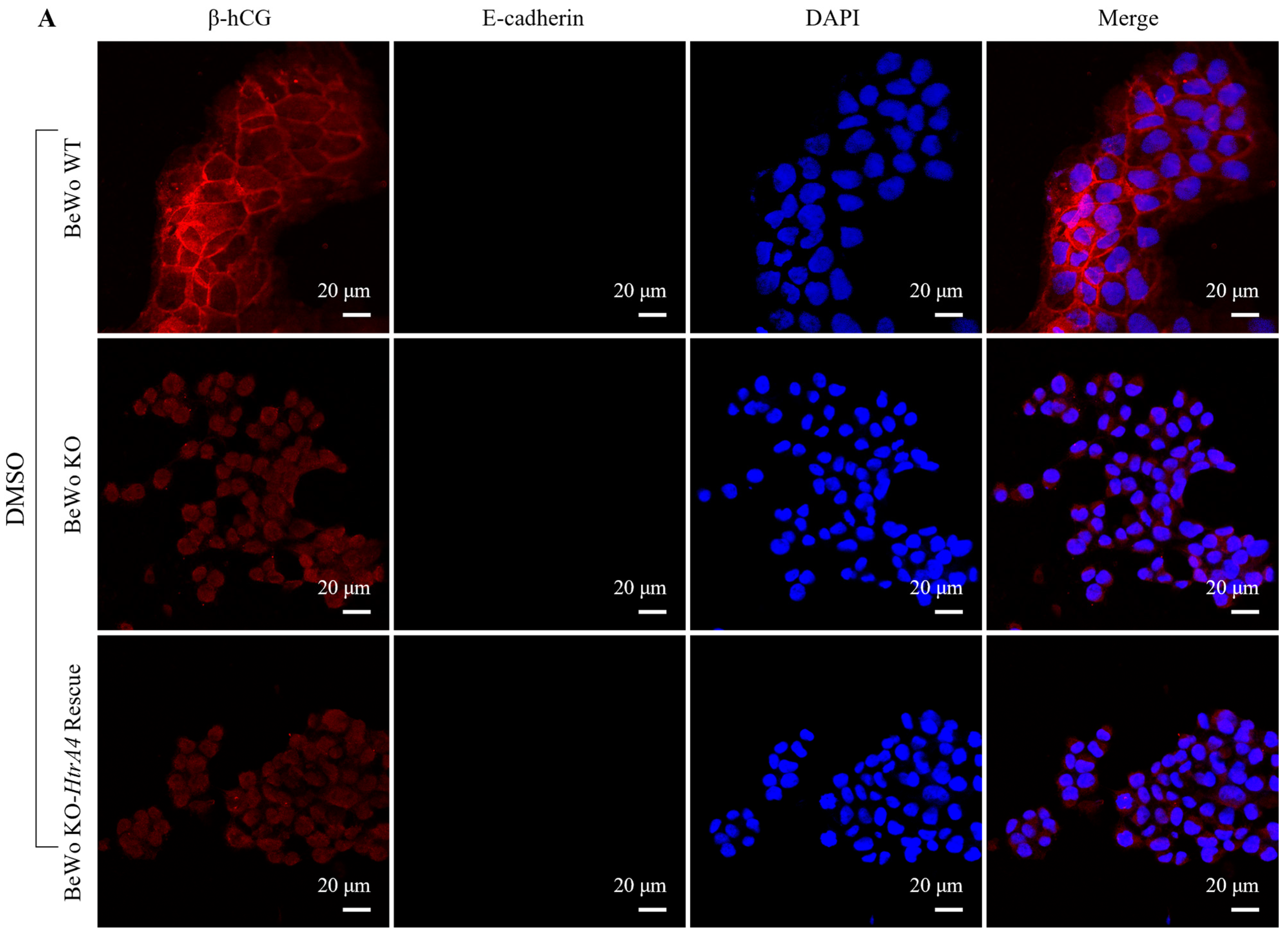
3.2. Effect of HtrA4 on Cell Fusion
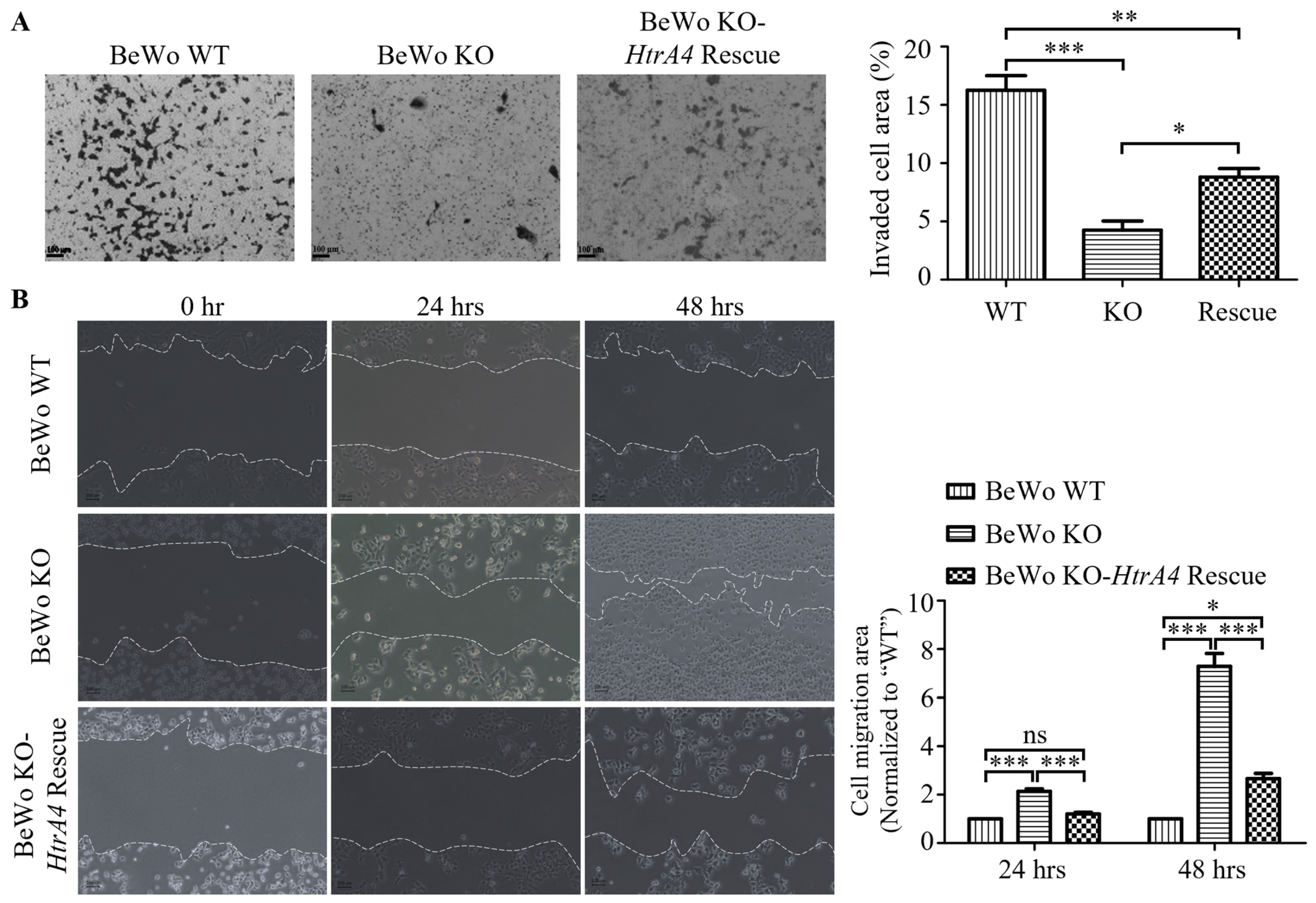
3.3. Effect of HtrA4 on Cell Invasion and Migration
3.4. Effect of HtrA4 on Cell Proliferation
3.5. Effect of HtrA4 on Cell Cycle
3.6. HtrA4 Expression Is Lower in Sera of RPL Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tossetta, G.; Fantone, S.; Licini, C.; Marzioni, D.; Mattioli-Belmonte, M. The multifaced role of HtrA1 in the development of joint and skeletal disorders. Bone 2022, 157, 116350. [Google Scholar] [CrossRef]
- Wenta, T.; Jarzab, M.; Rychlowski, M.; Borysiak, M.; Latala, A.; Zurawa-Janicka, D.; Filipek, A.; Lipinska, B. Cellular substrates and pro-apoptotic function of the human HtrA4 protease. J. Proteom. 2019, 209, 103505. [Google Scholar] [CrossRef] [PubMed]
- Kummari, R.; Dutta, S.; Chaganti, L.K.; Bose, K. Discerning the mechanism of action of HtrA4: A serine protease implicated in the cell death pathway. Biochem. J. 2019, 476, 1445–1463. [Google Scholar] [CrossRef] [PubMed]
- Wenta, T.; Rychlowski, M.; Jarzab, M.; Lipinska, B. HtrA4 Protease Promotes Chemotherapeutic-Dependent Cancer Cell Death. Cells 2019, 8, 1112. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; La, M.; Pham, T.; Lovrecz, G.O.; Nie, G. High levels of HtrA4 detected in preeclamptic circulation may disrupt endothelial cell function by cleaving the main VEGFA receptor KDR. FASEB J. 2019, 33, 5058–5066. [Google Scholar] [CrossRef] [PubMed]
- Tseng, E.; Teoh, S.S.Y.; Wang, Y.; Nie, G. Elevated protease HtrA4 in the maternal circulation of preeclampsia may contribute to endothelial barrier disruption by cleaving key junctional protein VE-cadherin. Placenta 2019, 76, 51–53. [Google Scholar] [CrossRef]
- Wang, Y.; Nie, G. High levels of HtrA4 observed in preeclamptic circulation drastically alter endothelial gene expression and induce inflammation in human umbilical vein endothelial cells. Placenta 2016, 47, 46–55. [Google Scholar] [CrossRef]
- Wang, Y.; Nie, G. Overview of Human HtrA Family Proteases and Their Distinctive Physiological Roles and Unique Involvement in Diseases, Especially Cancer and Pregnancy Complications. Int. J. Mol. Sci. 2021, 22, 10756. [Google Scholar] [CrossRef]
- Wang, L.-J.; Cheong, M.-L.; Lee, Y.-S.; Lee, M.-T.; Chen, H. High-Temperature Requirement Protein A4 (HtrA4) Suppresses the Fusogenic Activity of Syncytin-1 and Promotes Trophoblast Invasion. Mol. Cell. Biol. 2012, 32, 3707–3717. [Google Scholar] [CrossRef]
- Wang, C.; Lo, H.; Lin, S.; Chen, H. RACK1 (receptor for activated C-kinase 1) interacts with FBW2 (F-box and WD-repeat domain-containing 2) to up-regulate GCM1 (glial cell missing 1) stability and placental cell migration and invasion. Biochem. J. 2013, 453, 201–208. [Google Scholar] [CrossRef]
- Chiu, Y.H.; Chen, H. GATA3 inhibits GCM1 activity and trophoblast cell invasion. Sci. Rep. 2016, 6, 21630. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Zhao, M.; Chen, Q.; Wang, Y.; Li, Y.; Kaitu’U-Lino, T.J.; Tong, S.; Nie, G. Human HtrA4 Expression Is Restricted to the Placenta, Is Significantly Up-Regulated in Early-Onset Preeclampsia, and High Levels of HtrA4 Cause Endothelial Dysfunction. J. Clin. Endocrinol. Metab. 2015, 100, E936–E945. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xing, F.; He, Y.; Zong, S.; Luo, C.; Li, C.; Duan, T.; Wang, K.; Zhou, Q. Elevated HTRA1 and HTRA4 in severe preeclampsia and their roles in trophoblast functions. Mol. Med. Rep. 2018, 18, 2937–2944. [Google Scholar] [CrossRef]
- Richardson, A.L.; Wang, Z.C.; De Nicolo, A.; Lu, X.; Brown, M.; Miron, A.; Liao, X.; Iglehart, J.D.; Livingston, D.M.; Ganesan, S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006, 9, 121–132. [Google Scholar] [CrossRef]
- Sun, L.; Hui, A.-M.; Su, Q.; Vortmeyer, A.; Kotliarov, Y.; Pastorino, S.; Passaniti, A.; Menon, J.; Walling, J.; Bailey, R.; et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006, 9, 287–300. [Google Scholar] [CrossRef]
- Maser, R.S.; Choudhury, B.; Campbell, P.J.; Feng, B.; Wong, K.-K.; Protopopov, A.; O’neil, J.; Gutierrez, A.; Ivanova, E.; Perna, I.; et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 2007, 447, 966–971. [Google Scholar] [CrossRef]
- Renke, J.Z.; Kędzierska-Mieszkowska, S.; Lange, M.; Nedoszytko, B.; Liberek, A.; Plata-Nazar, K.; Renke, M.; Wenta, T.; Zurawa-Janicka, D.; Skorko-Glonek, J.; et al. Immune response against HtrA proteases in children with cutaneous mastocytosis. Acta Biochim. Pol. 2018, 65, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Yu, J.; Laxman, B.; Rhodes, D.R.; Mehra, R.; Tomlins, S.A.; Shah, R.B.; Chandran, U.; Monzon, F.A.; Becich, M.J.; et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005, 8, 393–406. [Google Scholar] [CrossRef]
- Baek, K.-H.; Choi, B.C.; Lee, J.-H.; Choi, H.-K.; Lee, S.-H.; Kim, J.-W.; Hill, J.A.; Chung, H.-M.; Ko, J.J.; Cha, K.Y. Comparison of gene expression at the feto–maternal interface between normal and recurrent pregnancy loss patients. Reprod. Fertil. Dev. 2002, 14, 235–240. [Google Scholar] [CrossRef]
- Luo, N.; Cheng, W.; Zhou, Y.; Gu, B.; Zhao, Z.; Zhao, Y. Screening Candidate Genes Regulating Placental Development from Trophoblast Transcriptome at Early Pregnancy in Dazu Black Goats (Capra hircus). Animals 2021, 11, 2132. [Google Scholar] [CrossRef]
- Adu-Gyamfi, E.A.; Czika, A.; Gorleku, P.N.; Ullah, A.; Panhwar, Z.; Ruan, L.-L.; Ding, Y.-B.; Wang, Y.-X. The Involvement of Cell Adhesion Molecules, Tight Junctions, and Gap Junctions in Human Placentation. Reprod. Sci. 2020, 28, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, Y.; Sun, X.; Kang, M.; Zhao, M.; Wan, J.; Chen, Q. Exporting Proteins Associated with Senescence Repair via Extracellular Vesicles May Be Associated with Early Pregnancy Loss. Cells 2022, 11, 2772. [Google Scholar] [CrossRef] [PubMed]
- Manna, C.; Lacconi, V.; Rizzo, G.; De Lorenzo, A.; Massimiani, M. Placental Dysfunction in Assisted Reproductive Pregnancies: Perinatal, Neonatal and Adult Life Outcomes. Int. J. Mol. Sci. 2022, 23, 659. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Kim, S.-Y.; Cho, H.-J.; Lee, S.-Y.; Baek, K.-H. YOD1 Deubiquitinates NEDD4 Involved in the Hippo Signaling Pathway. Cell. Physiol. Biochem. 2020, 54, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, M.; Wang, Y.; Lim, R.; Palmer, K.; Nie, G. HtrA4 is up-regulated during trophoblast syncytialization and BeWo cells fail to syncytialize without HtrA4. Sci. Rep. 2021, 11, 14363. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.; Turco, M.Y.; Burton, G.J.; Moffett, A. Investigation of human trophoblast invasion in vitro. Hum. Reprod. Updat. 2020, 26, 501–513. [Google Scholar] [CrossRef]
- Chen, X.; Qi, L.; Zhao, C.; Xue, J.; Chen, M.; Diao, L.; He, W.; Lv, B.; Zeng, Y.; Xue, Z. Decreased expression of SEMA4D in recurrent implantation failure induces reduction of trophoblast invasion and migration via the Met/PI3K/Akt pathway. J. Reprod. Immunol. 2022, 153, 103657. [Google Scholar] [CrossRef]
- Alfaqih, A.M.; Khader, Y.S.; Al-Dwairi, A.N.; Alzoubi, A.; Othman, A.-S.; Hatim, A. Lower Levels of Serum Adiponectin and the T Allele of rs1501299 of the ADIPOQ Gene Are Protective against Polycystic Ovarian Syndrome in Jordan. Korean J. Fam. Med. 2018, 39, 108–113. [Google Scholar] [CrossRef]
- Du, Y.; Cai, Z.; Zhang, H.; Liang, W.; Wang, H.; Man, Q.; Wang, W. Nitric oxide mediates disruption of human placental trophoblast invasion induced by perfluorobutane sulfonate. Environ. Pollut. 2021, 283, 117137. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Liu, C.; Chen, S.; Liu, X.; Fan, S. TAGLN2-Regulated Trophoblast Migration, Invasion and Fusion are Impaired in Preeclampsia. Front. Cell Dev. Biol. 2022, 10, 810633. [Google Scholar] [CrossRef]
- Adler, R.R.; Ng, A.-K.; Rote, N.S. Monoclonal Antiphosphatidylserine Antibody Inhibits Intercellular Fusion of the Choriocarcinoma Line, JAR1. Biol. Reprod. 1995, 53, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Wice, B.; Menton, D.; Geuze, H.; Schwartz, A.L. Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp. Cell Res. 1990, 186, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Duff, D.; Long, A. Roles for RACK1 in cancer cell migration and invasion. Cell. Signal. 2017, 35, 250–255. [Google Scholar] [CrossRef]
- Li, G.; Ji, X.-D.; Gao, H.; Zhao, J.-S.; Xu, J.-F.; Sun, Z.-J.; Deng, Y.-Z.; Shi, S.; Feng, Y.-X.; Zhu, Y.-Q.; et al. EphB3 suppresses non-small-cell lung cancer metastasis via a PP2A/RACK1/Akt signalling complex. Nat. Commun. 2012, 3, 667. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Chuang, P.-Y.; Chen, C.-P.; Chiu, Y.-H.; Lo, H.-F.; Cheong, M.-L.; Huang, J.-Y.; Kuo, P.-L.; Chen, H. Functional Antagonism between High Temperature Requirement Protein A (HtrA) Family Members Regulates Trophoblast Invasion. J. Biol. Chem. 2014, 289, 22958–22968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Luo, Q.; Chen, Z.; Sun, J.; Xu, B.; Ju, Y.; Song, G. Cyclic mechanical stretching promotes migration but inhibits invasion of rat bone marrow stromal cells. Stem Cell Res. 2015, 14, 155–164. [Google Scholar] [CrossRef]
- Newby, A.C. Matrix metalloproteinases regulate migration, proliferation, and death of vascular smooth muscle cells by degrading matrix and non-matrix substrates. Cardiovasc. Res. 2006, 69, 614–624. [Google Scholar] [CrossRef]
- Qi, D.; Lu, J.; Fu, Z.; Lv, S.; Hou, L. Psoralen Promotes Proliferation, Migration, and Invasion of Human Extravillous Trophoblast Derived HTR-8/Svneo Cells in vitro by NF-κB Pathway. Front. Pharmacol. 2022, 13, 804400. [Google Scholar] [CrossRef]
- Natarajan, M.; Hecker, T.P.; Gladson, C.L. FAK Signaling in Anaplastic Astro cytoma and Glioblastoma Tumors. Cancer J. 2003, 9, 126–133. [Google Scholar] [CrossRef]
- Howe, A. Anchorage-dependent ERK signaling–mechanisms and consequences. Curr. Opin. Genet. Dev. 2002, 12, 30–35. [Google Scholar] [CrossRef]
- Li, Z.; Li, C.; Du, L.; Zhou, Y.; Wu, W. Human Chorionic Gonadotropin β Induces Migration and Invasion via Activating ERK1/2 and MMP-2 in Human Prostate Cancer DU145 Cells. PLoS ONE 2013, 8, e54592. [Google Scholar] [CrossRef] [PubMed]
- Hsia, D.A.; Mitra, S.K.; Hauck, C.R.; Streblow, D.N.; Nelson, J.A.; Ilic, D.; Huang, S.; Li, E.; Nemerow, G.R.; Leng, J.; et al. Differential regulation of cell motility and invasion by FAK. J. Cell Biol. 2003, 160, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Stricker, J.; Falzone, T.; Gardel, M.L. Mechanics of the F-actin cytoskeleton. J. Biomech. 2010, 43, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Xu, E.; Luo, Q.; Song, G. Rac1: A Regulator of Cell Migration and a Potential Target for Cancer Therapy. Molecules 2023, 28, 2976. [Google Scholar] [CrossRef]
- Zhong, J.; Paul, A.; Kellie, S.J.; O’Neill, G.M. Mesenchymal Migration as a Therapeutic Target in Glioblastoma. J. Oncol. 2010, 2010, 430142. [Google Scholar] [CrossRef]
- Legrand, M.; Mousson, A.; Carl, P.; Rossé, L.; Justiniano, H.; Gies, J.-P.; Bouvard, D.; Sick, E.; Dujardin, D.; Rondé, P. Protein dynamics at invadopodia control invasion–migration transitions in melanoma cells. Cell Death Dis. 2023, 14, 190. [Google Scholar] [CrossRef]
- Graham, K.; Chandrasekaran, A.; Wang, L.; Ladak, A.; Lafer, E.M.; Rangamani, P.; Stachowiak, J.C. Liquid-like VASP condensates drive actin polymerization and dynamic bundling. Nat. Phys. 2023, 19, 574–585. [Google Scholar] [CrossRef]
- Xie, Y.; Yue, L.; Shi, Y.; Su, X.; Gan, C.; Liu, H.; Xue, T.; Ye, T. Application and Study of ROCK Inhibitors in Pulmonary Fibrosis: Recent Developments and Future Perspectives. J. Med. Chem. 2023, 66, 4342–4360. [Google Scholar] [CrossRef]
- Deng, N.; Qiao, M.; Li, Y.; Liang, F.; Li, J.; Liu, Y. Anticancer effects of licochalcones: A review of the mechanisms. Front. Pharmacol. 2023, 14, 1074506. [Google Scholar] [CrossRef]
- Schmidt, N.; Irle, I.; Ripkens, K.; Lux, V.; Nelles, J.; Johannes, C.; Parry, L.; Greenow, K.; Amir, S.; Campioni, M.; et al. Epigenetic silencing of serine protease HTRA1 drives polyploidy. BMC Cancer 2016, 16, 399. [Google Scholar] [CrossRef]
- Haneke, K.; Schott, J.; Lindner, D.; Hollensen, A.K.; Damgaard, C.K.; Mongis, C.; Knop, M.; Palm, W.; Ruggieri, A.; Stoecklin, G. CDK1 couples proliferation with protein synthesis. J. Cell Biol. 2020, 219, e201906147. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lim, R.; Nie, G. HtrA4 may play a major role in inhibiting endothelial repair in pregnancy complication preeclampsia. Sci. Rep. 2019, 9, 2728. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Zhang, C.; Parker, B.-T.; You, L.; Mathey-Prevot, B. Cyclin D/CDK4/6 activity controls G1 length in mammalian cells. PLoS ONE 2018, 13, e0185637. [Google Scholar] [CrossRef]
- Honda, R.; Lowe, E.D.; Dubinina, E.; Skamnaki, V.T.; Cook, A.; Brown, N.R.; Johnson, L.N. The structure of cyclin E1/CDK2: Implications for CDK2 activation and CDK2-independent roles. EMBO J. 2005, 24, 452–463. [Google Scholar] [CrossRef]
- Yam, C.H.; Fung, T.K.; Poon, R. Cyclin A in cell cycle control and cancer. Cell. Mol. Life Sci. 2002, 59, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- De Boer, L.; Oakes, V.; Beamish, H.; Giles, N.; Stevens, F.; Somodevilla-Torres, M.; DeSouza, C.; Gabrielli, B. Cyclin A/cdk2 coordinates centrosomal and nuclear mitotic events. Oncogene 2008, 27, 4261–4268. [Google Scholar] [CrossRef]


| Characteristics | Control (n = 32) | RPL (n = 60) | p-Value | |
|---|---|---|---|---|
| Age (y) | 31.8 ± 4.4 | 32.6 ± 5.2 | ns | |
| BMI | 24.9 ± 2.4 | 26.3 ± 1.7 | ns | |
| Gravidity (n) | 2.5 ± 1.6 | 5.3 ± 2.6 | <0.01 ** | |
| Parity (n) | 1.8 ± 0.5 | 1.4 ± 0.2 | <0.05 * | |
| Previous pregnancy losses (wk) | - | 7.8 ± 1.5 | - | |
| Spontaneous abortion (no.) | 0 ± 0 | 3.6 ± 0.8 | - | |
| Artificial abortion (no.) | ||||
| times | 0 | 20 (62.5%) | 0 | |
| 1~2 | 12 (37.5%) | 0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, C.-Z.; Choi, B.-C.; Park, J.-H.; Park, H.Y.; Paek, J.; Lee, K.-J.; Yun, B.-S.; Kim, Y.J.; Baek, K.-H. Cellular Functions of High-Temperature Requirement Factor A4 in Placenta. Cells 2023, 12, 1459. https://doi.org/10.3390/cells12111459
Pei C-Z, Choi B-C, Park J-H, Park HY, Paek J, Lee K-J, Yun B-S, Kim YJ, Baek K-H. Cellular Functions of High-Temperature Requirement Factor A4 in Placenta. Cells. 2023; 12(11):1459. https://doi.org/10.3390/cells12111459
Chicago/Turabian StylePei, Chang-Zhu, Bum-Chae Choi, Jun-Hyeok Park, Hyo Young Park, Jinyoung Paek, Kyung-Ju Lee, Bo-Seong Yun, Young Ju Kim, and Kwang-Hyun Baek. 2023. "Cellular Functions of High-Temperature Requirement Factor A4 in Placenta" Cells 12, no. 11: 1459. https://doi.org/10.3390/cells12111459
APA StylePei, C.-Z., Choi, B.-C., Park, J.-H., Park, H. Y., Paek, J., Lee, K.-J., Yun, B.-S., Kim, Y. J., & Baek, K.-H. (2023). Cellular Functions of High-Temperature Requirement Factor A4 in Placenta. Cells, 12(11), 1459. https://doi.org/10.3390/cells12111459









