Collagen VI Is a Gi-Biased Ligand of the Adhesion GPCR GPR126/ADGRG6
Abstract
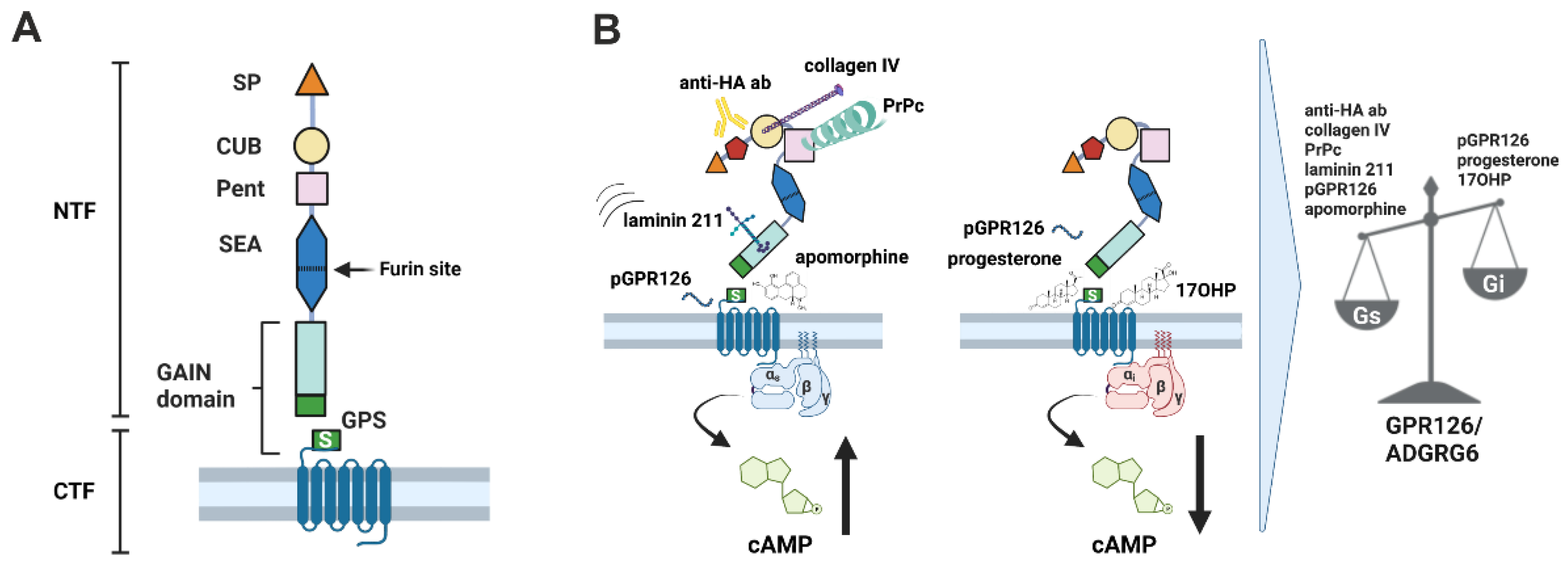
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. GPR126 Plasmid Construction and Fusion Protein Generation
- Forward Primer 31: 5′-CACGAATTCGGTTCCTCTCTCAGTGTGCGG-3′;
- Reverse Primer 807: 5′-GTTAGATCT GCA CAG ACA AAT GGT CTC ACC-3′
- Reverse Primer 438: 5′-GTTAGATCT GACTTTCATCCT GTCCTC TCC-3′
- Forward Primer 446F: 5′-CACGAATTCG GACAAAAGG TTG GTG CTC TGC-3′
- mFc 31-438 F (AscI): 5′-GCCAGGCGCGCCcctctctcagtgtgcg gatgtggcag-3′
- mFc 31-438 R (BamHI): 5′-CTCTGGATCCGAGACTTTCATCCTGTCCTCTTAC-3′
- mFc448-807 F (AscI): 5′-GCCAGGCGCGCCTTGGTGCTCTGGGCCCTTCTAGTC-3′
- mFc 448-807 R (KpnI): 5′-CTCTGGTACCTTGCACAGACAAATGGTCTCACC-3′
2.2.2. Generation of mFc and hFc Fusion Protein
2.2.3. In Vitro Biotinylation of mGPR126-mFc Fusion Proteins
2.2.4. Purification of GPR126 Immunocomplexes and Tandem MS and Sequencing
2.2.5. Co-Immunoprecipitation (co-IP) and Western Blot Analysis
2.2.6. In Vitro Functional Assays
2.2.7. Data Analysis
3. Results
3.1. Collagen VI Is a Novel Binding Partner of GPR126
3.2. Collagen VI Is a Gi-Biased Ligand of GPR126
3.3. Shaking Force Application Does Not Increase Gi-Signaling of GPR126
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Monk, K.R.; Oshima, K.; Jors, S.; Heller, S.; Talbot, W.S. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development 2011, 138, 2673–2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mogha, A.; Benesh, A.E.; Patra, C.; Engel, F.B.; Schöneberg, T.; Liebscher, I.; Monk, K.R. Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J. Neurosci. 2013, 33, 17976–17985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebscher, I.; Schön, J.; Petersen, S.C.; Fischer, L.; Auerbach, N.; Demberg, L.M.; Mogha, A.; Cöster, M.; Simon, K.-U.; Rothemund, S.; et al. A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 2014, 9, 2018–2026. [Google Scholar] [CrossRef] [Green Version]
- Liebscher, I.; Schöneberg, T.; Prömel, S. Progress in demystification of adhesion G protein-coupled receptors. J. Biol. Chem. 2013, 394, 937–950. [Google Scholar] [CrossRef]
- Leon, K.; Cunningham, R.L.; Riback, J.A.; Feldman, E.; Li, J.; Sosnick, T.R.; Zhao, M.; Monk, K.R.; Araç, D. Structural basis for adhesion G protein-coupled receptor Gpr126 function. Nat. Commun. 2020, 11, 194. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.-H.; Chang, G.-W.; Davies, J.Q.; Stacey, M.; Harris, J.; Gordon, S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004, 279, 31823–31832. [Google Scholar] [CrossRef] [Green Version]
- Moriguchi, T.; Haraguchi, K.; Ueda, N.; Okada, M.; Furuya, T.; Akiyama, T. DREG, a developmentally regulated G protein-coupled receptor containing two conserved proteolytic cleavage sites. Genes Cells 2004, 9, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Arac, D.; Boucard, A.A.; Bolliger, M.F.; Nguyen, J.; Soltis, S.M.; Südhof, T.C.; Brunger, A.T. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012, 31, 1364–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monk, K.R.; Naylor, S.G.; Glenn, T.D.; Mercurio, S.; Perlin, J.R.; Dominguez, C.; Moens, C.B.; Talbot, W.S. A G Protein-Coupled Receptor Is Essential for Schwann Cells to Initiate Myelination. Science 2009, 325, 1402–1405. [Google Scholar] [CrossRef] [Green Version]
- Ravenscroft, G.; Nolent, F.; Rajagopalan, S.; Meireles, A.M.; Paavola, K.J.; Gaillard, D.; Alanio, E.; Buckland, M.; Arbuckle, S.; Krivanek, M.; et al. Mutations of GPR126 are responsible for severe arthrogryposis multiplex congenita. Am. J. Hum. Genet. 2015, 96, 955–961. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; He, L.; Jia, K.; Yue, Z.; Li, S.; Jin, Y.; Li, Z.; Siwko, S.; Xue, F.; Su, J.; et al. Regulation of body length and bone mass by Gpr126/Adgrg6. Sci. Adv. 2020, 6, eaaz0368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suchý, T.; Zieschang, C.; Popkova, Y.; Kaczmarek, I.; Weiner, J.; Liebing, A.-D.; Çakir, M.V.; Landgraf, K.; Gericke, M.; Pospisilik, J.A.; et al. The repertoire of Adhesion G protein-coupled receptors in adipocytes and their functional relevance. Int. J. Obes. 2020, 44, 2124–2136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, W.; Lin, H.; Ma, L.; Zhang, C.; Zheng, Y.; Cheng, Q.; Ma, C.; Wu, X.; Zhang, Z.; Zhong, Y.; et al. Progesterone activates GPR126 to promote breast cancer development via the Gi pathway. Proc. Natl. Acad. Sci. USA 2022, 119, e2117004119. [Google Scholar] [CrossRef] [PubMed]
- Paavola, K.J.; Sidik, H.; Zuchero, J.B.; Eckart, M.; Talbot, W.S. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci. Signal 2014, 7, ra76. [Google Scholar] [CrossRef] [Green Version]
- Küffer, A.; Lakkaraju, A.K.K.; Mogha, A.; Petersen, S.C.; Airich, K.; Doucerain, C.; Marpakwar, R.; Bakirci, P.; Senatore, A.; Monnard, A.; et al. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 2016, 536, 464–468. [Google Scholar] [CrossRef] [Green Version]
- Petersen, S.C.; Luo, R.; Liebscher, I.; Giera, S.; Jeong, S.-J.; Mogha, A.; Ghidinelli, M.; Feltri, M.L.; Schöneberg, T.; Piao, X.; et al. The Adhesion GPCR GPR126 Has Distinct, Domain-Dependent Functions in Schwann Cell Development Mediated by Interaction with Laminin-211. Neuron 2015, 85, 755–769. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Jin, Z.; Koirala, S.; Bu, L.; Xu, L.; Hynes, R.O.; Walsh, C.A.; Corfas, G.; Piao, X. GPR56 regulates pial basement membrane integrity and cortical lamination. J. Neurosci. 2008, 28, 5817–5826. [Google Scholar] [CrossRef] [Green Version]
- Luo, R.; Jeong, S.-J.; Jin, Z.; Strokes, N.; Li, S.; Piao, X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc. Natl. Acad. Sci. USA 2011, 108, 12925–12930. [Google Scholar] [CrossRef] [Green Version]
- Mogha, A.; Harty, B.L.; Carlin, D.; Joseph, J.; Sanchez, N.E.; Suter, U.; Piao, X.; Cavalli, V.; Monk, K.R. Gpr126/Adgrg6 Has Schwann Cell Autonomous and Nonautonomous Functions in Peripheral Nerve Injury and Repair. J. Neurosci. 2016, 36, 12351–12367. [Google Scholar] [CrossRef] [Green Version]
- Monk, K.R.; Feltri, M.L.; Taveggia, C. New insights on Schwann cell development. Glia 2015, 63, 1376–1393. [Google Scholar] [CrossRef] [Green Version]
- Braghetta, P.; Fabbro, C.; Piccolo, S.; Marvulli, D.; Bonaldo, P.; Volpin, D.; Bressan, G.M. Distinct regions control transcriptional activation of the alpha1(VI) collagen promoter in different tissues of transgenic mice. J. Cell Biol. 1996, 135, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Vitale, P.; Braghetta, P.; Volpin, D.; Bonaldo, P.; Bressan, G.M. Mechanisms of transcriptional activation of the col6a1 gene during Schwann cell differentiation. Mech. Dev. 2001, 102, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Cescon, M.; Megighian, A.; Bonaldo, P. Collagen VI regulates peripheral nerve myelination and function. FASEB J. 2014, 28, 1145–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conklin, B.R.; Farfel, Z.; Lustig, K.D.; Julius, D.; Bourne, H.R. Substitution of three amino acids switches receptor specificity of Gqα to that of Giα. Nature 1993, 363, 274–276. [Google Scholar] [CrossRef]
- Wishart, A.L.; Conner, S.J.; Guarin, J.R.; Fatherree, J.P.; Peng, Y.; McGinn, R.A.; Crews, R.; Naber, S.P.; Hunter, M.; Greenberg, A.S.; et al. Decellularized extracellular matrix scaffolds identify full-length collagen VI as a driver of breast cancer cell invasion in obesity and metastasis. Sci. Adv. 2020, 6, eabc3175. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilde, C.; Chaudhry, P.M.; Luo, R.; Simon, K.-U.; Piao, X.; Liebscher, I. Collagen VI Is a Gi-Biased Ligand of the Adhesion GPCR GPR126/ADGRG6. Cells 2023, 12, 1551. https://doi.org/10.3390/cells12111551
Wilde C, Chaudhry PM, Luo R, Simon K-U, Piao X, Liebscher I. Collagen VI Is a Gi-Biased Ligand of the Adhesion GPCR GPR126/ADGRG6. Cells. 2023; 12(11):1551. https://doi.org/10.3390/cells12111551
Chicago/Turabian StyleWilde, Caroline, Paulomi Mehta Chaudhry, Rong Luo, Kay-Uwe Simon, Xianhua Piao, and Ines Liebscher. 2023. "Collagen VI Is a Gi-Biased Ligand of the Adhesion GPCR GPR126/ADGRG6" Cells 12, no. 11: 1551. https://doi.org/10.3390/cells12111551
APA StyleWilde, C., Chaudhry, P. M., Luo, R., Simon, K.-U., Piao, X., & Liebscher, I. (2023). Collagen VI Is a Gi-Biased Ligand of the Adhesion GPCR GPR126/ADGRG6. Cells, 12(11), 1551. https://doi.org/10.3390/cells12111551





