Novel Isolation Method Reveals Sex-Specific Composition and Neurotoxicity of Small Extracellular Vesicles in a Mouse Model of Alzheimer’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
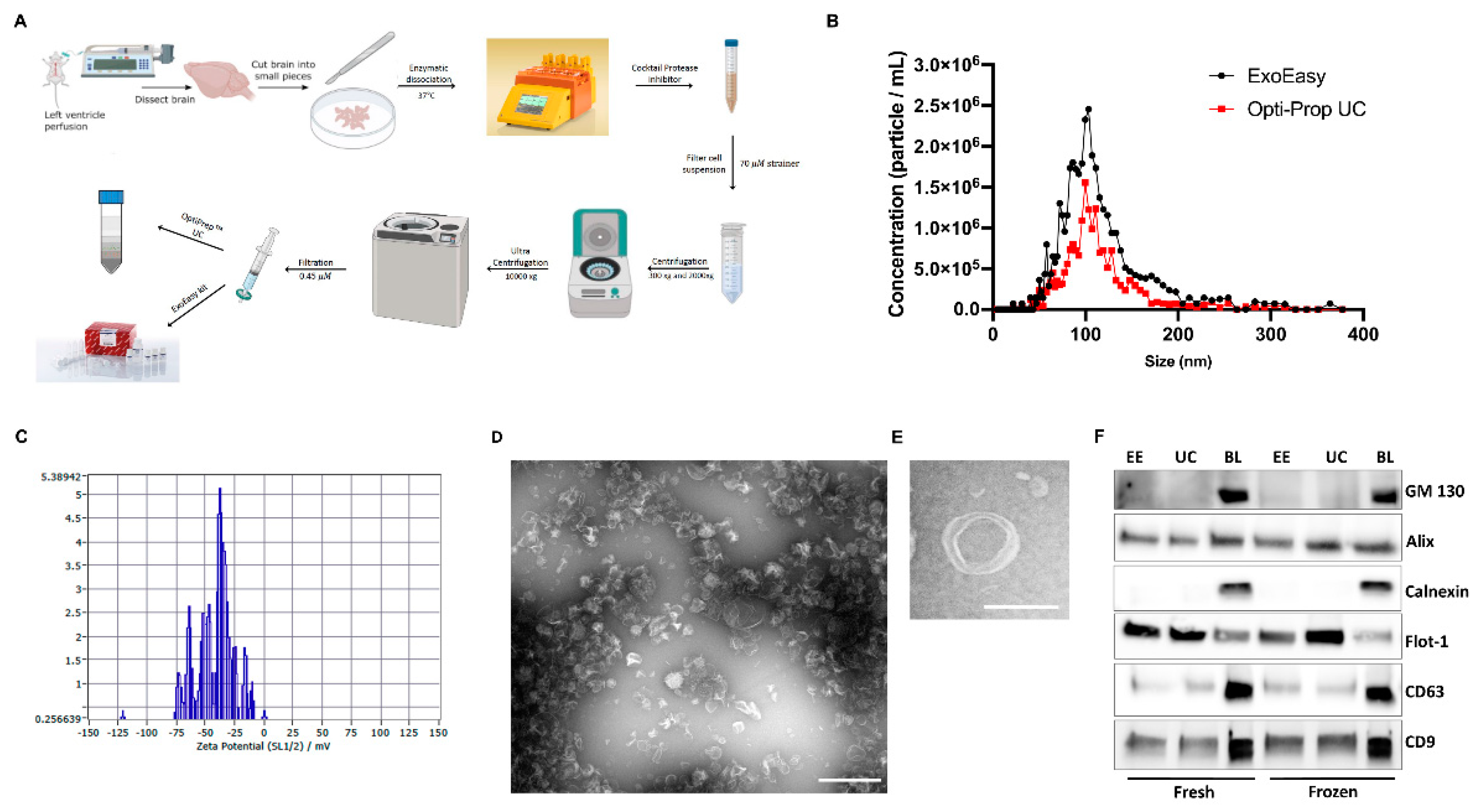
2.2. Brain EVs Isolation
2.3. Nanoparticle Tracking Analysis
2.4. Immunoblot Analysis
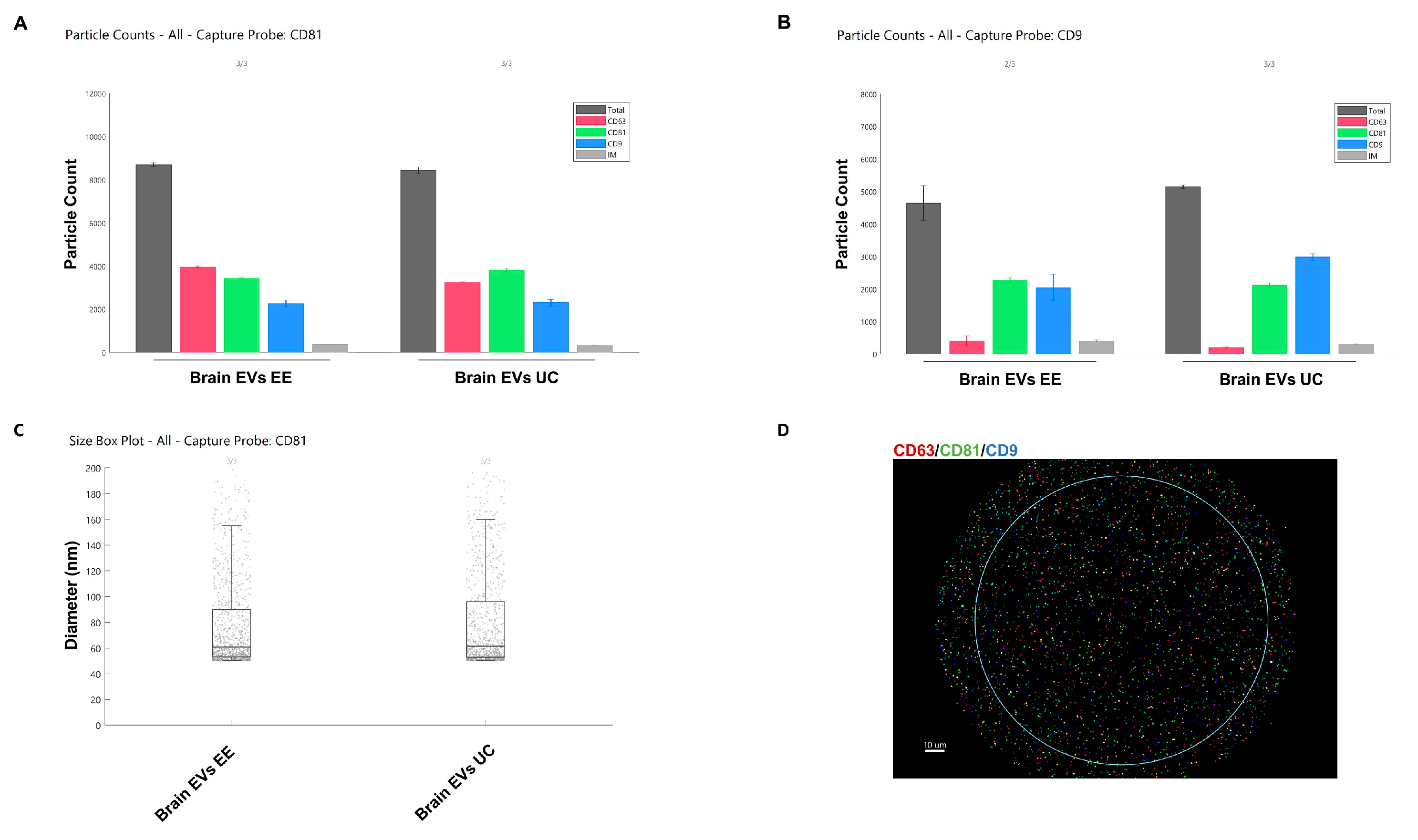
2.5. ExoView
2.6. ASM Activity Assay
2.7. Cytotoxicity Assay
2.8. Preparation and Treatment of Primary Astrocytes Cultures
2.9. Seahorse Assay
2.10. Transmission Electron Microscopy (TEM)
2.11. Proximity Ligation Assay
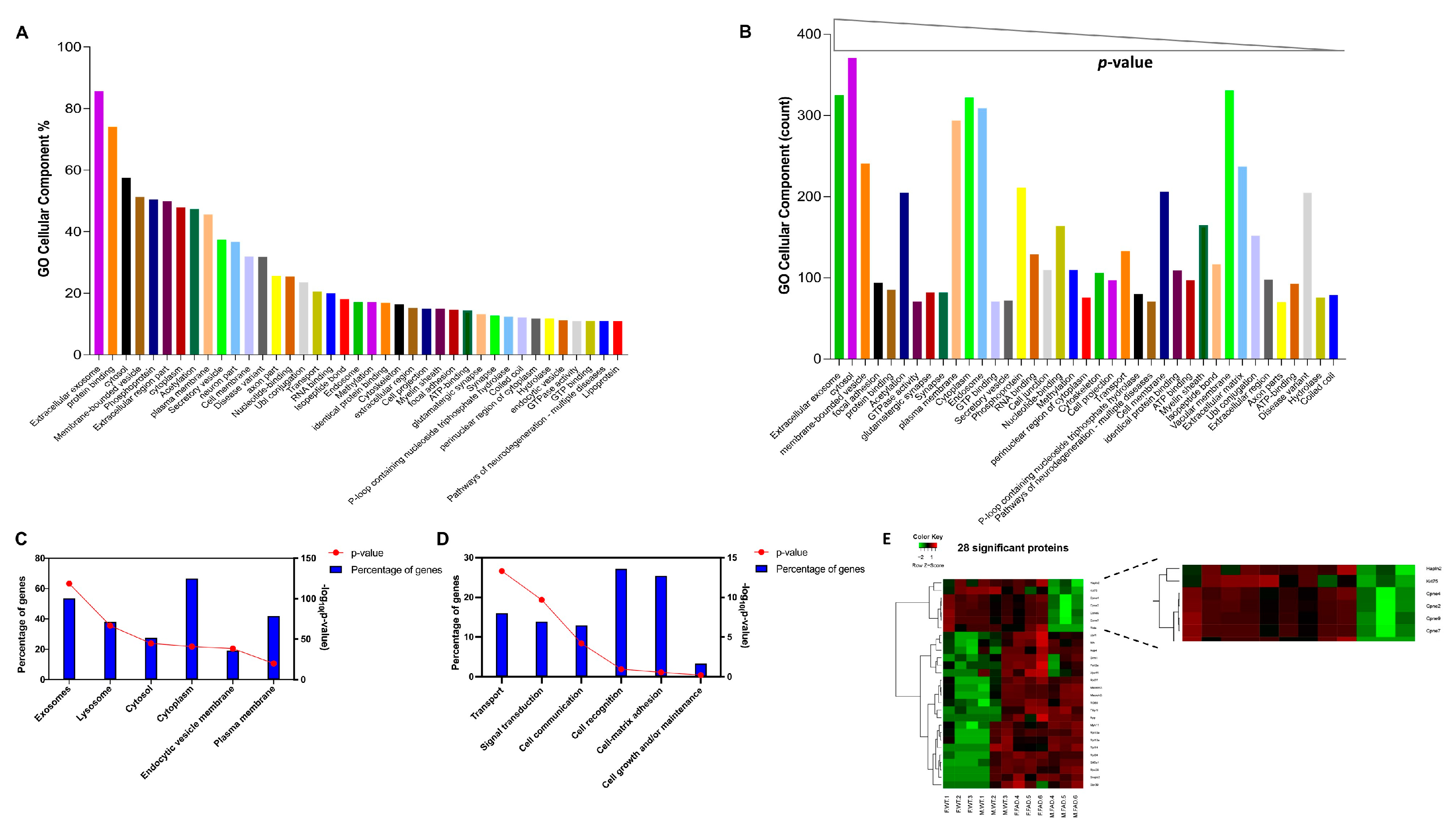
2.12. Mass Spectrometric (Proteomic) Analysis of EV Proteins
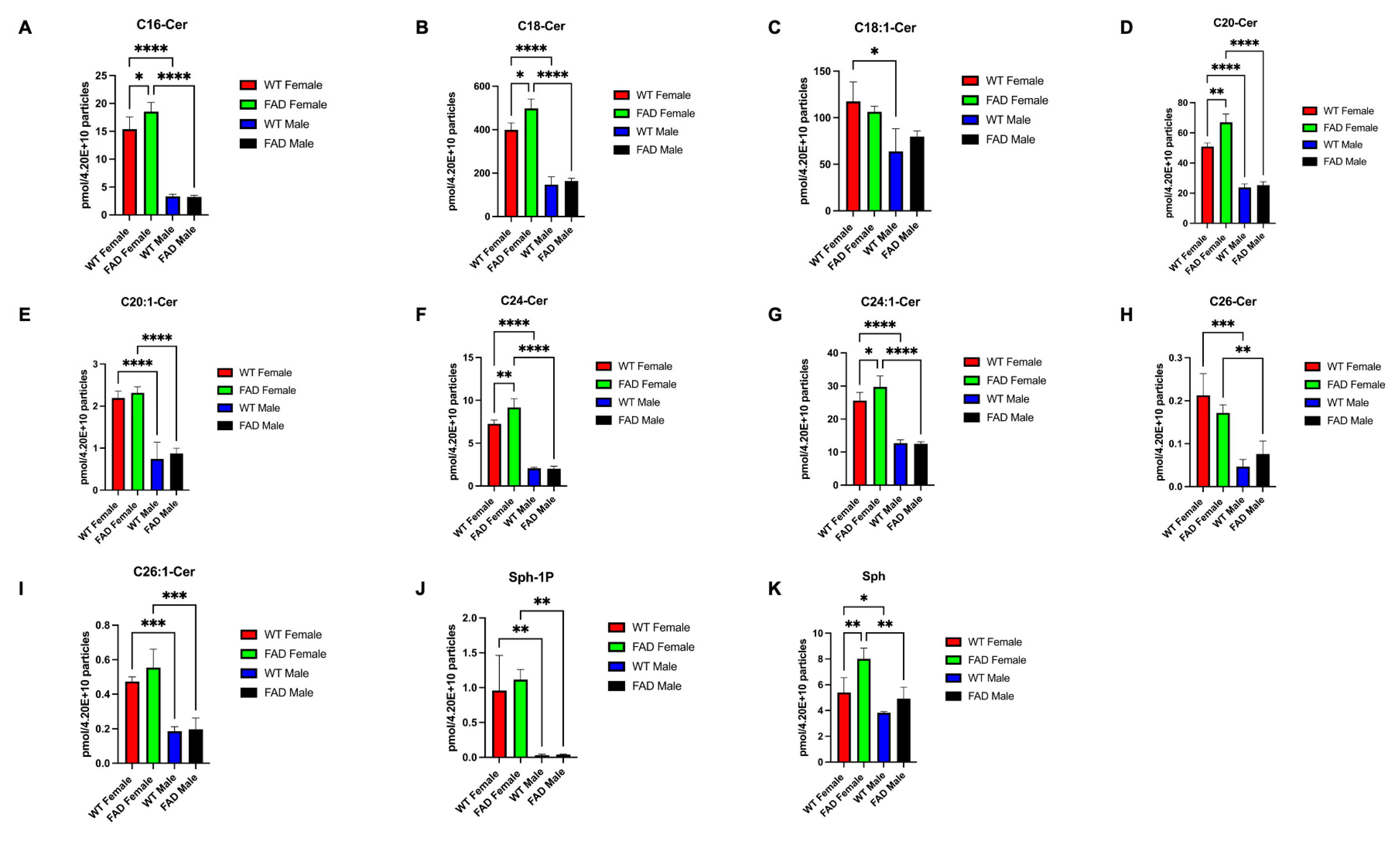
2.13. Sphingolipid Analysis
2.14. Statistical Analysis
3. Results
3.1. Characterization of Brain-Derived EVs
3.2. Proteomic Analysis Elucidates EVs Sexual Dimorphism
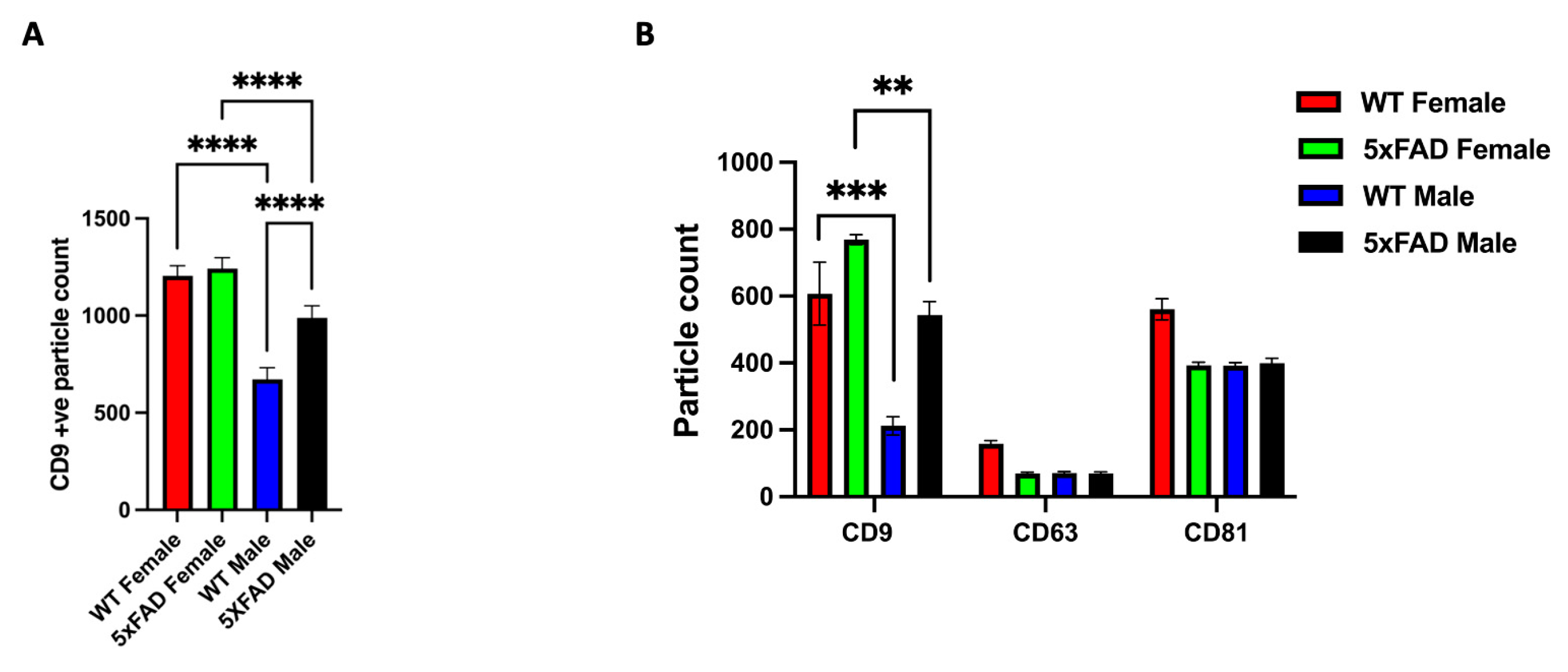
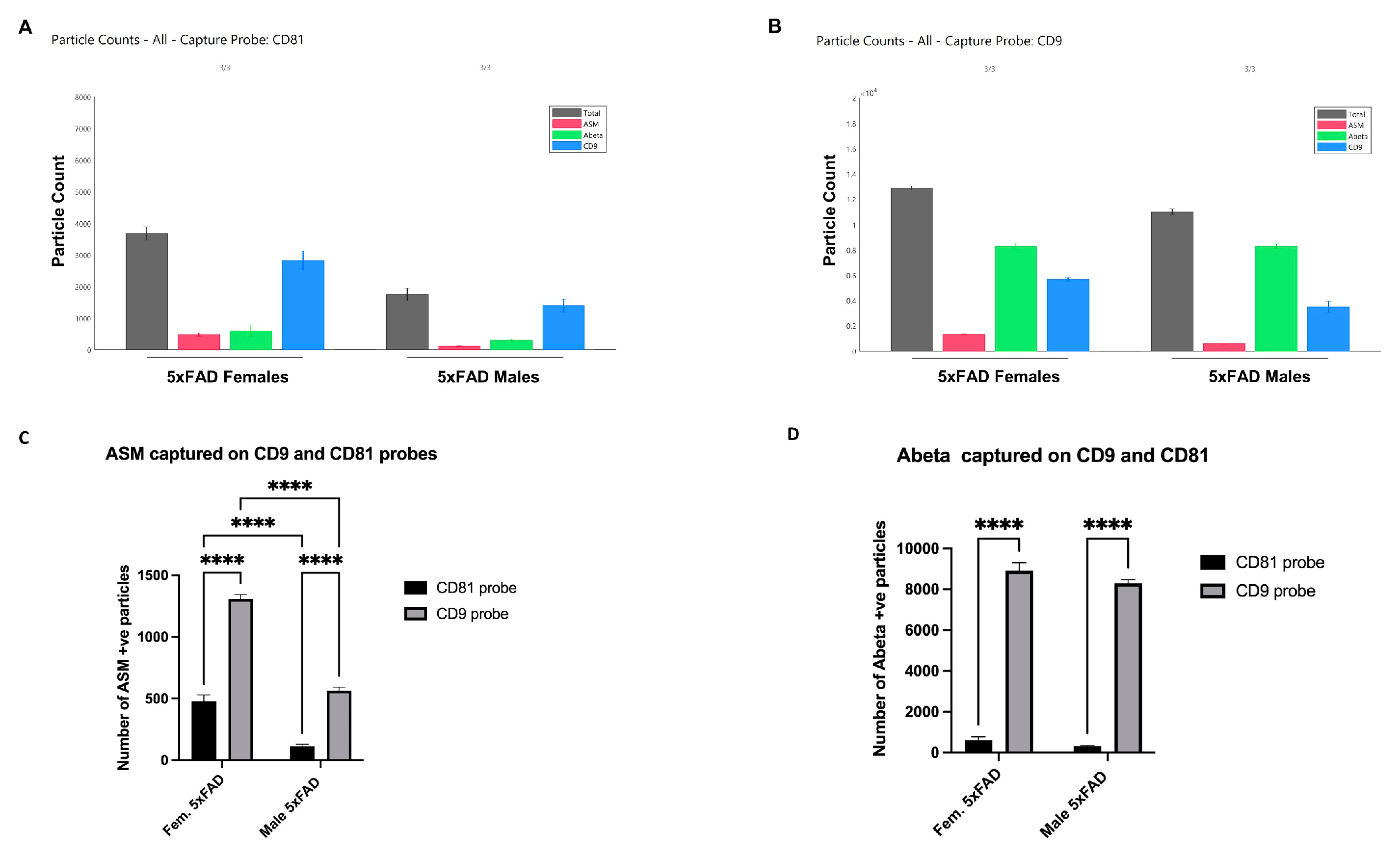
3.3. CD9 Is Differentially Expressed in Male and Female sEVs
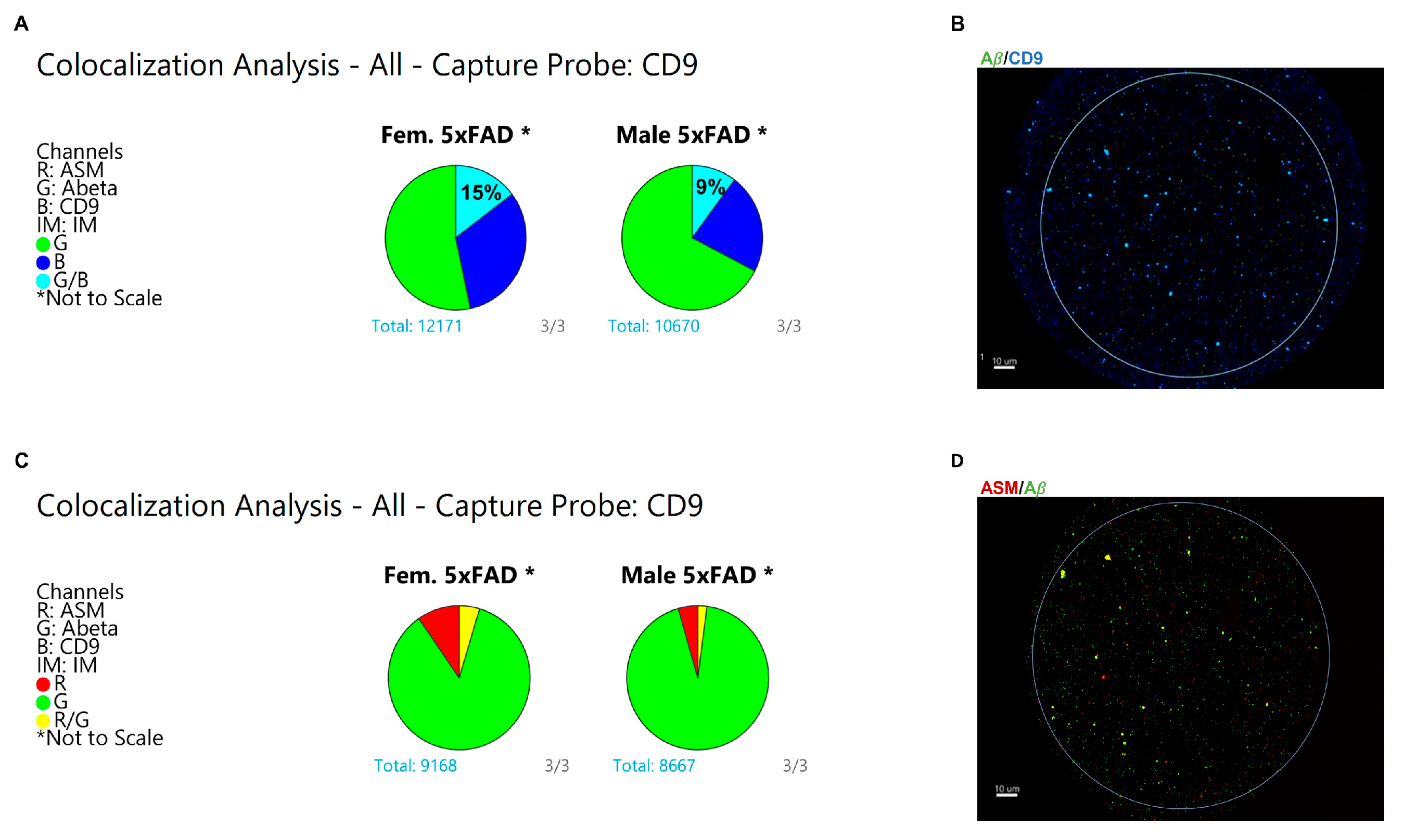
3.4. Specific Enrichment of ASM on Female CD9(+) EVs and Colocalization with Aβ
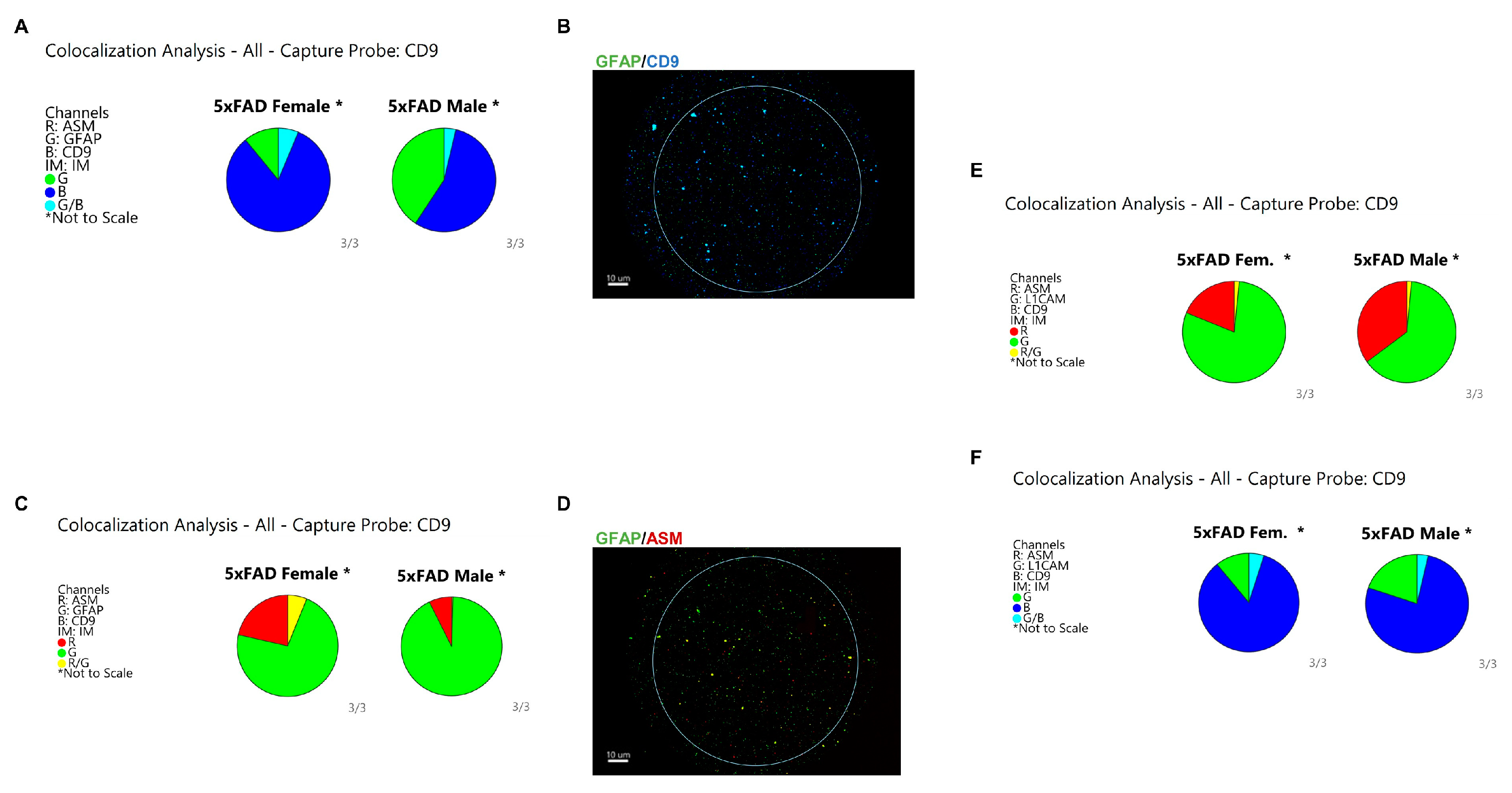
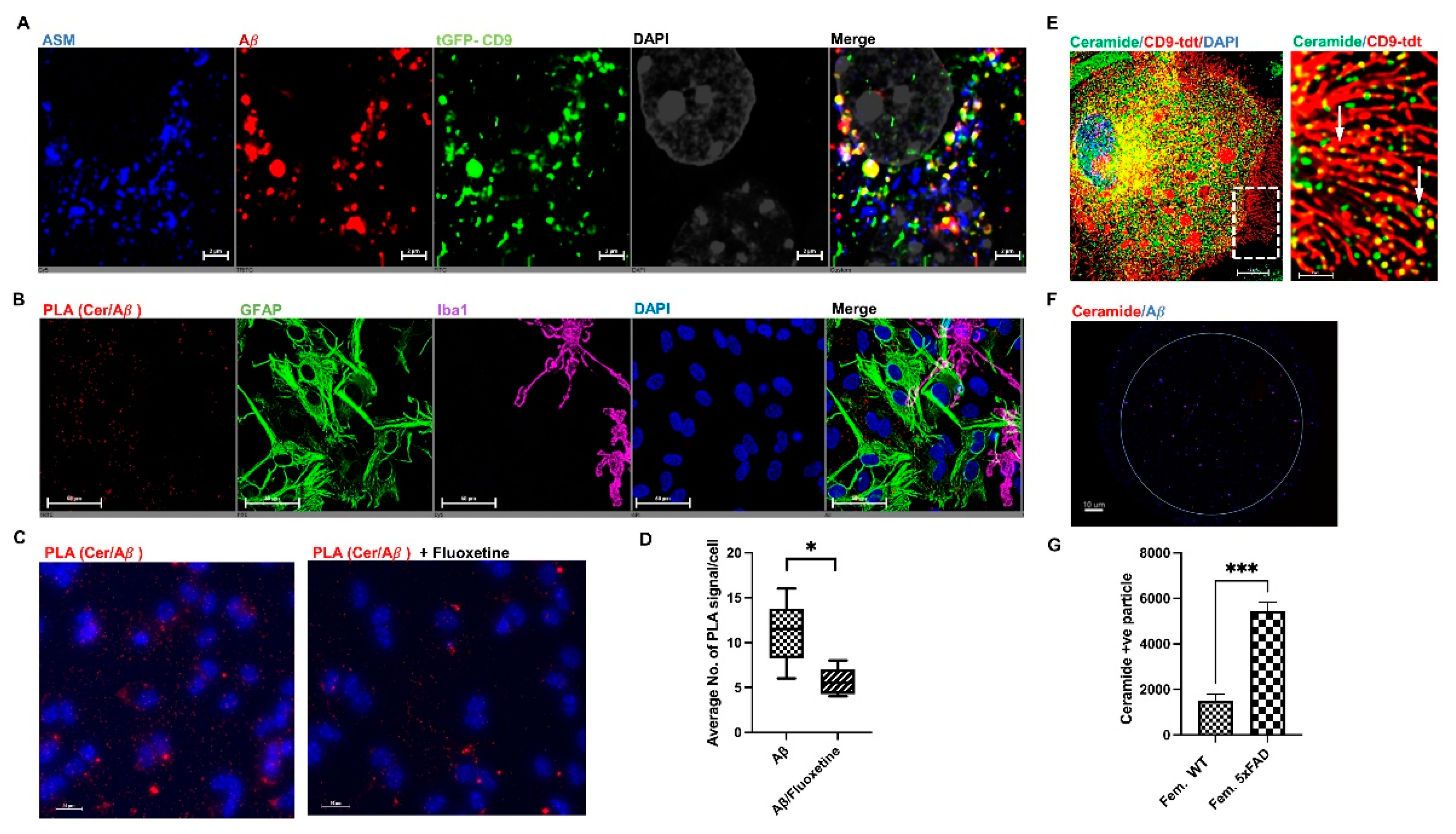
3.5. Astrocytes Are the Major Origin of CD9/ASM Vesicles in Female 5xFAD Brains
3.6. ASM Inhibitors Prevent Complex Formation between Ceramide and Aβ in Astrocytes
3.7. Female sEVs Exert Increased Mitochondrial Bioenergetic Disturbance and Elevated Neurotoxicity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 1–19. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Song, Z.; Xu, Y.; Deng, W.; Zhang, L.; Zhu, H.; Yu, P.; Qu, Y.; Zhao, W.; Han, Y.; Qin, C. Brain Derived Exosomes Are a Double-Edged Sword in Alzheimer’s Disease. Front. Mol. Neurosci. 2020, 13, 79. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Huo, L.; Du, X.; Li, X.; Liu, S.; Xu, Y. The Emerging Role of Neural Cell-Derived Exosomes in Intercellular Communication in Health andNeuro-degenerative Diseases. Front. Neurosci. 2021, 15, 738442. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Hareendran, S.; Loh, Y.P. Function of exosomes in neurological disorders and brain tumors. Extracell. Vesicles Circ. Nucleic Acids 2021, 2, 55–79. [Google Scholar] [CrossRef]
- Danzer, K.M.; Kranich, L.R.; Ruf, W.P.; Cagsal-Getkin, O.; Winslow, A.R.; Zhu, L.; Vanderburg, C.R.; McLean, P.J. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol. Neurodegener. 2012, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Taha, H.B.; Kearney, B.; Bitan, G. A minute fraction of α-synuclein in extracellular vesicles may be a major contributor to α-synuclein spreading following autophagy inhibition. Front. Mol. Neurosci. 2022, 15, 1001382. [Google Scholar] [CrossRef]
- Wang, Y.; Balaji, V.; Kaniyappan, S.; Krüger, L.; Irsen, S.; Tepper, K.; Mandelkow, E.M. The release and trans-synaptic transmission of Tau via exosomes. Mol. Neurodegener. 2017, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, E.; Bilousova, T.; Melnik, M.; Fakhrutdinov, D.; Poon, W.W.; Vinters, H.V.; Miller, C.A.; Corrada, M.; Kawas, C.; Bohannan, R.; et al. Exosomal tau with seeding activity is released from Alzheimer’s disease synapses, and seeding potential is associated with amyloid beta. Lab. Investig. 2021, 101, 1605–1617. [Google Scholar] [CrossRef]
- Jackson, N.A.; Guerrero-Muñoz, M.J.; Castillo-Carranza, D.L. The prion-like transmission of tau oligomers via exosomes. Front. Aging Neurosci. 2022, 14, 974414. [Google Scholar] [CrossRef] [PubMed]
- Elsherbini, A.; Kirov, A.S.; Dinkins, M.B.; Wang, G.; Qin, H.; Zhu, Z.; Tripathi, P.; Crivelli, S.M.; Bieberich, E. Association of Aβ with ceramide-enriched astrosomes mediates Aβ neurotoxicity. Acta Neuropathol. Commun. 2020, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Dinkins, M.B.; Dasgupta, S.; Wang, G.; Zhu, G.; Bieberich, E. Exosome reduction in vivo is associated with lower amyloid plaque load in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 2014, 35, 1792–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Huang, Y.; Li, B.; Gong, C.-X.; Schuchman, E. Deregulation of sphingolipid metabolism in Alzheimer’s disease. Neurobiol. Aging 2010, 31, 398–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crivelli, S.M.; Giovagnoni, C.; Visseren, L.; Scheithauer, A.L.; de Wit, N.; den Hoedt, S.; Losen, M.; Mulder, M.T.; Walter, J.; de Vries, H.E.; et al. Sphingolipids in Alzheimer’s disease, how can we target them? Adv. Drug. Deliv. Rev. 2020, 159, 214–231. [Google Scholar] [CrossRef]
- Crivelli, S.M.; Giovagnoni, C.; Zhu, Z.; Tripathi, P.; Elsherbini, A.; Quadri, Z.; Pu, J.; Zhang, L.; Ferko, B.; Berkes, D.; et al. Function of ceramide transfer protein for biogenesis and sphingolipid composition of extracellular vesicles. J. Extracell. Vesicles 2022, 11, 12233. [Google Scholar] [CrossRef]
- Krishnamurthy, K.; Dasgupta, S.; Bieberich, E. Development and characterization of a novel anti-ceramide antibody. J. Lipid Res. 2007, 48, 968–975. [Google Scholar] [CrossRef] [Green Version]
- Bielawski, J.; Pierce, J.S.; Snider, J.; Rembiesa, B.; Szulc, Z.M.; Bielawska, A. Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatog-raphy-tandem mass spectrometry. Methods Mol. Biol. 2009, 579, 443–467. [Google Scholar]
- Clos-Sansalvador, M.; Monguió-Tortajada, M.; Roura, S.; Franquesa, M.; Borràs, F.E. Commonly used methods for extracellular vesicles’ enrichment: Implications in downstream analyses and use. Eur. J. Cell Biol. 2022, 101, 151227. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [Green Version]
- Xavier, L.D.L.; Hanekamp, S.; Simonyan, K. Sexual Dimorphism Within Brain Regions Controlling Speech Production. Front. Neurosci. 2019, 13, 795. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Luo, Q.; Huang, C.-C.; Lo, C.-Y.Z.; Langley, C.; Desrivières, S.; Quinlan, E.B.; Banaschewski, T.; Millenet, S.; Bokde, A.L.W.; et al. The Human Brain Is Best Described as Being on a Female/Male Continuum: Evidence from a Neuroimaging Connectivity Study. Cereb. Cortex 2021, 31, 3021–3033. [Google Scholar] [CrossRef]
- Wilfredo López-Ojeda, M.S.; Robin, A.; Hurley, M.D. Sexual Dimorphism in Brain Development: Influence on Affective Dis-orders. J. Neuropsychiatry Clin. Neurosci. 2021, 33, 85–89. [Google Scholar]
- Sandu, A.-L.; Waiter, G.D.; Staff, R.T.; Nazlee, N.; Habota, T.; McNeil, C.J.; Chapko, D.; Williams, J.H.; Fall, C.H.D.; Chandak, G.R.; et al. Sexual dimorphism in the relationship between brain complexity, volume and general intelligence (g): A cross-cohort study. Sci. Rep. 2022, 12, 11025. [Google Scholar] [CrossRef]
- Fisher, D.W.; Bennett, D.A.; Dong, H. Sexual dimorphism in predisposition to Alzheimer’s disease. Neurobiol. Aging 2018, 70, 308–324. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Jackson, H.; Hristov, H.; Isaacson, R.S.; Saif, N.; Shetty, T.; Etingin, O.; Henchcliffe, C.; Brinton, R.D.; Mosconi, L. Sex and Gender Driven Modifiers of Alzheimer’s: The Role for Estrogenic Control Across Age, Race, Medical, and Lifestyle Risks. Front. Aging Neurosci. 2019, 11, 315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghav, A.; Singh, M.; Jeong, G.-B.; Giri, R.; Agarwal, S.; Kala, S.; Gautam, K.A. Extracellular vesicles in neurodegenerative diseases: A systematic review. Front. Mol. Neurosci. 2022, 15. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.-S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Blue, E.; Thornton, T.; Kooperberg, C.; Liu, S.; Wactawski-Wende, J.; Manson, J.; Kuller, L.; Hayden, K.; Reiner, A. Non-coding variants in MYH11, FZD3, and SORCS3 are associated with dementia in women. Alzheimers Dement. 2021, 17, 215–225. [Google Scholar] [CrossRef]
- Mathieu, M.; Névo, N.; Jouve, M.; Valenzuela, J.I.; Maurin, M.; Verweij, F.; Palmulli, R.; Lankar, D.; Dingli, F.; Loew, D.; et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 2021, 12, 4389. [Google Scholar] [CrossRef]
- Santos, M.F.; Rappa, G.; Fontana, S.; Karbanová, J.; Aalam, F.; Tai, D.; Li, Z.; Pucci, M.; Alessandro, R.; Morimoto, C.; et al. Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells. Cells 2022, 11, 2474. [Google Scholar] [CrossRef]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E.; et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Goetzl, E.; Mustapic, M.; Kapogiannis, D.; Eitan, E.; Lobach, I.; Goetzl, L.; Schwartz, J.; Miller, B. Cargo proteins of plasma astrocyte-derived exosomes in Alzheimer’s disease. FASEB J. 2016, 30, 3853–3859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Zhang, H.; Liu, S.; Hou, Y.; Chi, G. The Dual Role of Astrocyte-Derived Exosomes and Their Contents in the Process of Alzheimer’s Disease. J. Alzheimer’s Dis. 2023, 91, 33–42. [Google Scholar] [CrossRef]
- Eitan, E.; Hutchison, E.R.; Marosi, K.; Comotto, J.; Mustapic, M.; Nigam, S.M.; Suire, C.; Maharana, C.; A Jicha, G.; Liu, D.; et al. Extracellular vesicle-associated Aβ mediates trans-neuronal bioenergetic and Ca2+-handling deficits in Alzheimer’s disease models. npj Aging Mech. Dis. 2016, 2, 16019. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Shu, J.; Yang, X.; Wei, W.; Yan, A. Exosomes Derived from M2 Microglia Cells Attenuates Neuronal Impairment and Mitochondrial Dysfunction in Alzheimer’s Disease through the PINK1/Parkin Pathway. Front. Cell. Neurosci. 2022, 16, 874102. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the In-ternational Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [Green Version]
- Dekker, M.; Waissi, F.; van Bennekom, J.; Silvis, M.; Timmerman, N.; Bank, I.; Walter, J.; Mueller, C.; Schoneveld, A.H.; Schiffelers, R.; et al. Plasma extracellular vesicle proteins are associated with stress-induced myocardial ischemia in women pre-senting with chest pain. Sci. Rep. 2020, 10, 12257. [Google Scholar] [CrossRef]
- Kolhe, R.; Hunter, M.; Liu, S.; Jadeja, R.N.; Pundkar, C.; Mondal, A.K.; Mendhe, B.; Drewry, M.; Rojiani, M.V.; Liu, Y.; et al. Gender-specific differential expression of exosomal miRNA in synovial fluid of patients with osteoarthritis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kolhe, R.; Owens, V.; Sharma, A.; Lee, T.J.; Zhi, W.; Ghilzai, U.; Mondal, A.K.; Liu, Y.; Isales, C.M.; Hamrick, M.W.; et al. Sex-Specific Differences in Extracellular Vesicle Protein Cargo in Synovial Fluid of Patients with Osteoarthritis. Life 2020, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Navas, C.; Morselli, E.; Clegg, D.J. Sexually dimorphic brain fatty acid composition in low and high fat diet-fed mice. Mol. Metab. 2016, 5, 680–689. [Google Scholar] [CrossRef]
- Ibáñez, F.; Ureña-Peralta, J.; Costa-Alba, P.; Torres, J.-L.; Laso, F.-J.; Marcos, M.; Guerri, C.; Pascual, M. Circulating MicroRNAs in Extracellular Vesicles as Potential Biomarkers of Alcohol-Induced Neuroinflammation in Adolescence: Gender Differences. Int. J. Mol. Sci. 2020, 21, 6730. [Google Scholar] [CrossRef] [PubMed]
- Tomsig, J.L.; Creutz, C.E. Copines: A ubiquitous family of Ca2+ -dependent phospholipid-binding proteins. Cell. Mol. Life Sci. 2002, 59, 1467–1477. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.-M.; Park, S.-J.; Lee, H.-K.; Park, J.-C. Copine-7 binds to the cell surface receptor, nucleolin, and regulates ciliogenesis and Dspp expression during odontoblast differentiation. Sci. Rep. 2017, 7, 11283. [Google Scholar] [CrossRef] [Green Version]
- Coppotelli, G.; Ross, J.; Ling, A.; Kim, K.; Sinclair, D. CPNE2: A New Regulator of Lysosomal-Mitochondrial Function Involved in Aging and Disease. Innov. Aging 2018, 2, 387–388. [Google Scholar] [CrossRef]
- Creutz, C.E.; Edwardson, J.M. Organization and synergistic binding of copine I and annexin A1 on supported lipid bilayers observed by atomic force microscopy. Biochim. Et Biophys. Acta (BBA) Biomembr. 2009, 1788, 1950–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigri, J.; Leca, J.; Tubiana, S.-S.; Finetti, P.; Guillaumond, F.; Martinez, S.; Lac, S.; Iovanna, J.L.; Audebert, S.; Camoin, L.; et al. CD9 mediates the uptake of extracellular vesicles from cancer-associated fibroblasts that promote pancreatic cancer cell aggressiveness. Sci. Signal. 2022, 15, eabg8191. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.; Akbar, A.; Thakor, A.S. An emerging role of CD9 in stemness and chemoresistance. Oncotarget 2019, 10, 4000–4001. [Google Scholar] [CrossRef]
- Poveda, E.; Tabernilla, A.; Fitzgerald, W.; Salgado-Barreira, A.; Grandal, M.; Pérez, A.; Mariño, A.; Álvarez, H.; Valcarce, N.; González-García, J.; et al. Massive Release of CD9+ Microvesicles in Human Immunodeficiency Virus Infection, Regardless of Virologic Control. J. Infect. Dis. 2020, 225, 1040–1049. [Google Scholar] [CrossRef]
- Böker, K.O.; Lemus-Diaz, N.; Ferreira, R.R.; Schiller, L.; Schneider, S.; Gruber, J. The Impact of the CD9 Tetraspanin on Lentivirus Infectivity and Exosome Secretion. Mol. Ther. 2018, 26, 634–647. [Google Scholar] [CrossRef] [Green Version]
- Suárez, H.; Andreu, Z.; Mazzeo, C.; Toribio, V.; Pérez-Rivera, A.E.; López-Martín, S.; García-Silva, S.; Hurtado, B.; Morato, E.; Peláez, L.; et al. CD9 inhibition reveals a functional connection of extracellular vesicle secretion with mitophagy in melanoma cells. J. Extracell. Vesicles 2021, 10, e12082. [Google Scholar] [CrossRef]
- Lucchetti, D.; Tenore, C.R.; Colella, F.; Sgambato, A. Extracellular Vesicles and Cancer: A Focus on Metabolism, Cytokines, and Immunity. Cancers 2020, 12, 171. [Google Scholar] [CrossRef] [Green Version]
- Heinrichs, A. Ceramide buds in. Nat. Rev. Mol. Cell Biol. 2008, 9, 265. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Umeda, R.; Satouh, Y.; Takemoto, M.; Nakada-Nakura, Y.; Liu, K.; Yokoyama, T.; Shirouzu, M.; Iwata, S.; Nomura, N.; Sato, K.; et al. Structural insights into tetraspanin CD9 function. Nat. Commun. 2020, 11, 1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dharan, R.; Goren, S.; Cheppali, S.K.; Shendrik, P.; Brand, G.; Vaknin, A.; Yu, L.; Kozlov, M.M.; Sorkin, R. Transmembrane proteins tetraspanin 4 and CD9 sense membrane curvature. Proc. Natl. Acad. Sci. USA 2022, 119. [Google Scholar] [CrossRef] [PubMed]
- Breiden, B.; Sandhoff, K. Acid Sphingomyelinase, a Lysosomal and Secretory Phospholipase C, Is Key for Cellular Phospholipid Catabolism. Int. J. Mol. Sci. 2021, 22, 9001. [Google Scholar] [CrossRef]
- Pieragostino, D.; Cicalini, I.; Lanuti, P.; Ercolino, E.; Di Ioia, M.; Zucchelli, M.; Zappacosta, R.; Miscia, S.; Marchisio, M.; Sacchetta, P.; et al. Enhanced release of acid sphingomyelinase-enriched exosomes generates a lipidomics signature in CSF of Multiple Sclerosis patients. Sci. Rep. 2018, 8, 3071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornhuber, J.; Tripal, P.; Reichel, M.; Terfloth, L.; Bleich, S.; Wiltfang, J.; Gulbins, E. Identification of New Functional Inhibitors of Acid Sphingomyelinase Using a Structure−Property−Activity Relation Model. J. Med. Chem. 2007, 51, 219–237. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elsherbini, A.; Zhu, Z.; Quadri, Z.; Crivelli, S.M.; Ren, X.; Vekaria, H.J.; Tripathi, P.; Zhang, L.; Zhi, W.; Bieberich, E. Novel Isolation Method Reveals Sex-Specific Composition and Neurotoxicity of Small Extracellular Vesicles in a Mouse Model of Alzheimer’s Disease. Cells 2023, 12, 1623. https://doi.org/10.3390/cells12121623
Elsherbini A, Zhu Z, Quadri Z, Crivelli SM, Ren X, Vekaria HJ, Tripathi P, Zhang L, Zhi W, Bieberich E. Novel Isolation Method Reveals Sex-Specific Composition and Neurotoxicity of Small Extracellular Vesicles in a Mouse Model of Alzheimer’s Disease. Cells. 2023; 12(12):1623. https://doi.org/10.3390/cells12121623
Chicago/Turabian StyleElsherbini, Ahmed, Zhihui Zhu, Zainuddin Quadri, Simone M. Crivelli, Xiaojia Ren, Hemendra J. Vekaria, Priyanka Tripathi, Liping Zhang, Wenbo Zhi, and Erhard Bieberich. 2023. "Novel Isolation Method Reveals Sex-Specific Composition and Neurotoxicity of Small Extracellular Vesicles in a Mouse Model of Alzheimer’s Disease" Cells 12, no. 12: 1623. https://doi.org/10.3390/cells12121623
APA StyleElsherbini, A., Zhu, Z., Quadri, Z., Crivelli, S. M., Ren, X., Vekaria, H. J., Tripathi, P., Zhang, L., Zhi, W., & Bieberich, E. (2023). Novel Isolation Method Reveals Sex-Specific Composition and Neurotoxicity of Small Extracellular Vesicles in a Mouse Model of Alzheimer’s Disease. Cells, 12(12), 1623. https://doi.org/10.3390/cells12121623






