Molecular Mechanisms of Colorectal Liver Metastases
Abstract
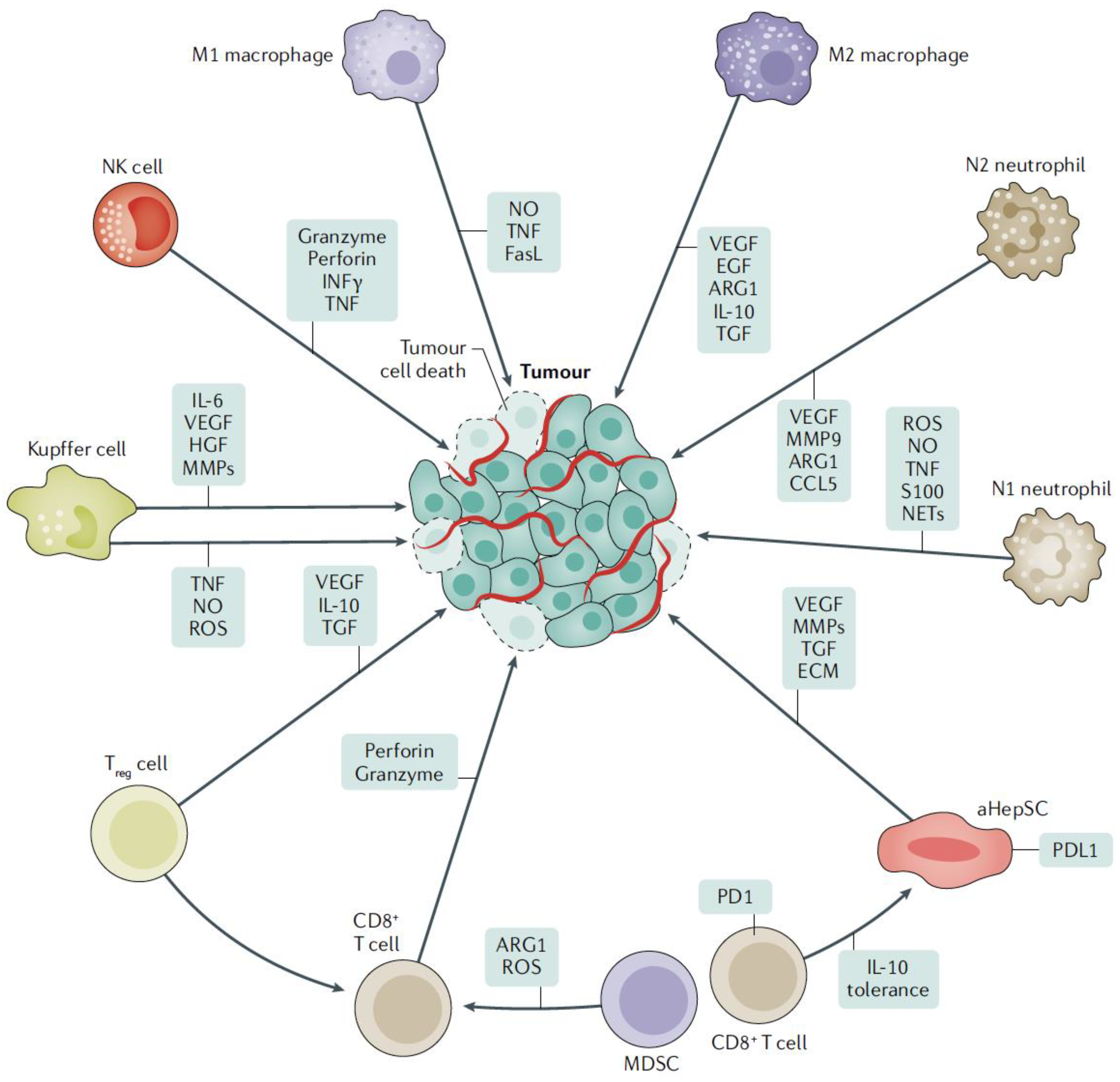
:1. Introduction
2. Molecular Mechanisms of Liver Metastases
3. Cellular Approach to Liver Metastasis
3.1. Hepatocytes
3.2. LSECs
3.3. Kupffer Cells
3.4. NK Cells
3.5. HepSCs
3.6. Dendritic Cells
3.7. Monocytes and Neutrophils
4. Formation of a Pre-Metastatic Niche in the Liver
5. Interactions of Cancer Cells with Liver Microenvironment
5.1. Tumor Cells First Encounter LSEC and Kupffer Cells
5.2. Tumor Cell Interaction with Neutrophils
5.3. Tumor Cell Interaction with Recruited Macrophages
5.4. MDSCs Have a Pro-Metastatic Potential
5.5. Tumor Cell Interaction with HepSCs
5.6. Tumor Cell Interaction with Hepatocytes
6. Role of Exosomes in Establishing Pre-Metastatic Niche in the Liver
7. Role of Angiogenesis in Developing of Liver Metastases
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.; de Gramont, A.; Figueras, J.; Kokudo, N.; Kunstlinger, F.; Loyer, E.; Poston, G.; Rougier, P.; Rubbia-Brandt, L.; Sobrero, A.; et al. Managing synchronous liver metastases from colorectal cancer: A multidisciplinary international consensus. Cancer Treat. Rev. 2015, 41, 729–741. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.M.; Pawlik, T.M.; Rodriguez-Bigas, M.A.; Monson, J.R.T.; Chang, G.J.; Larson, D.W. Timing of Surgical Resection for Curative Colorectal Cancer with Liver Metastasis. Ann. Surg. Oncol. 2018, 25, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Tsilimigras, D.I.; Brodt, P.; Clavien, P.A.; Muschel, R.J.; D’Angelica, M.I.; Endo, I.; Parks, R.W.; Doyle, M.; de Santibanes, E.; Pawlik, T.M. Liver metastases. Nat. Rev. Dis. Prim. 2021, 7, 27. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Hyer, J.M.; Bagante, F.; Guglielmi, A.; Ruzzenente, A.; Alexandrescu, S.; Poultsides, G.; Sasaki, K.; Aucejo, F.; Pawlik, T.M. Resection of Colorectal Liver Metastasis: Prognostic Impact of Tumor Burden vs KRAS Mutational Status. J. Am. Coll. Surg. 2021, 232, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Tsilimigras, D.I.; Ntanasis-Stathopoulos, I.; Bagante, F.; Moris, D.; Cloyd, J.; Spartalis, E.; Pawlik, T.M. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg. Oncol. 2018, 27, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Z.; Wang, Y.; Wen, X.; Amador, E.H.; Yuan, L.; Ran, X.; Xiong, L.; Ran, Y.; Chen, W.; et al. Colorectal liver metastasis: Molecular mechanism and interventional therapy. Signal Transduct. Target. Ther. 2022, 7, 70. [Google Scholar] [CrossRef]
- Chandra, R.; Karalis, J.D.; Liu, C.; Murimwa, G.Z.; Voth Park, J.; Heid, C.A.; Reznik, S.I.; Huang, E.; Minna, J.D.; Brekken, R.A. The Colorectal Cancer Tumor Microenvironment and Its Impact on Liver and Lung Metastasis. Cancers 2021, 13, 6206. [Google Scholar] [CrossRef]
- Brodt, P. Role of the Microenvironment in Liver Metastasis: From Pre- to Prometastatic Niches. Clin. Cancer Res. 2016, 22, 5971–5982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Eynden, G.G.; Majeed, A.W.; Illemann, M.; Vermeulen, P.B.; Bird, N.C.; Hoyer-Hansen, G.; Eefsen, R.L.; Reynolds, A.R.; Brodt, P. The multifaceted role of the microenvironment in liver metastasis: Biology and clinical implications. Cancer Res. 2013, 73, 2031–2043. [Google Scholar] [CrossRef] [Green Version]
- Vidal-Vanaclocha, F. Architectural and Functional Aspects of the Liver with Implications for Cancer Metastasis. In Liver Metastasis: Biology and Clinical Management; Brodt, P., Ed.; Springer Science+Business Media B.V.: Dordrecht, The Netherlands, 2011; p. 1. [Google Scholar]
- Wisse, E.; De Zanger, R.B.; Charels, K.; Van Der Smissen, P.; McCuskey, R.S. The liver sieve: Considerations concerning the structure and function of endothelial fenestrae, the sinusoidal wall and the space of Disse. Hepatology 1985, 5, 683–692. [Google Scholar] [CrossRef]
- Wisse, E.; Knook, D.L. Kupffer Cells and Other Liver Sinusoidal Cells. In Proceedings of the International Kupffer Cell Symposium Held in Noordwijkerhout, Noordwijkerhout, The Netherlands, 4–7 September 1977; Elsevier: Amsterdam, The Netherlands; North-Holland Biomedical Press: New York, NY, USA, 1977. [Google Scholar]
- Li, X.; Ramadori, P.; Pfister, D.; Seehawer, M.; Zender, L.; Heikenwalder, M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer 2021, 21, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Stanger, B.Z. Cellular homeostasis and repair in the mammalian liver. Annu. Rev Physiol 2015, 77, 179–200. [Google Scholar] [CrossRef] [Green Version]
- Malato, Y.; Naqvi, S.; Schurmann, N.; Ng, R.; Wang, B.; Zape, J.; Kay, M.A.; Grimm, D.; Willenbring, H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Investig. 2011, 121, 4850–4860. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Pan, C.; Hu, H.; Zheng, S.; Ding, L. Osteopontin-enhanced hepatic metastasis of colorectal cancer cells. PLoS ONE 2012, 7, e47901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crispe, I.N. Liver antigen-presenting cells. J. Hepatol. 2011, 54, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Kolios, G.; Valatas, V.; Kouroumalis, E. Role of Kupffer cells in the pathogenesis of liver disease. World J. Gastroenterol. 2006, 12, 7413–7420. [Google Scholar] [CrossRef]
- Kubes, P.; Jenne, C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018, 36, 247–277. [Google Scholar] [CrossRef]
- Crispe, I.N. The liver as a lymphoid organ. Annu. Rev. Immunol. 2009, 27, 147–163. [Google Scholar] [CrossRef]
- Doherty, D.G.; O’Farrelly, C. Innate and adaptive lymphoid cells in the human liver. Immunol. Rev. 2000, 174, 5–20. [Google Scholar] [CrossRef]
- Benlagha, K.; Kyin, T.; Beavis, A.; Teyton, L.; Bendelac, A. A thymic precursor to the NK T cell lineage. Science 2002, 296, 553–555. [Google Scholar] [CrossRef]
- Dashtsoodol, N.; Shigeura, T.; Aihara, M.; Ozawa, R.; Kojo, S.; Harada, M.; Endo, T.A.; Watanabe, T.; Ohara, O.; Taniguchi, M. Alternative pathway for the development of Valpha14+ NKT cells directly from CD4−CD8− thymocytes that bypasses the CD4+CD8+ stage. Nat. Immunol. 2017, 18, 274–282. [Google Scholar] [CrossRef]
- Friedman, S.L. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008, 88, 125–172. [Google Scholar] [CrossRef]
- Gressner, A.M.; Bachem, M.G. Molecular mechanisms of liver fibrogenesis—A homage to the role of activated fat-storing cells. Digestion 1995, 56, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Muhanna, N.; Doron, S.; Wald, O.; Horani, A.; Eid, A.; Pappo, O.; Friedman, S.L.; Safadi, R. Activation of hepatic stellate cells after phagocytosis of lymphocytes: A novel pathway of fibrogenesis. Hepatology 2008, 48, 963–977. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Zhang, L.; Xu, Y.; Zhang, Z.; Ren, G.; Tang, K.; Kuang, P.; Zhao, B.; Yin, Z.; Wang, X. Hepatic stellate cells promote tumor progression by enhancement of immunosuppressive cells in an orthotopic liver tumor mouse model. Lab. Investig. A J. Tech. Methods Pathol. 2014, 94, 182–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doherty, D.G. Immunity, tolerance and autoimmunity in the liver: A comprehensive review. J. Autoimmun. 2016, 66, 60–75. [Google Scholar] [CrossRef]
- Tiegs, G.; Lohse, A.W. Immune tolerance: What is unique about the liver. J. Autoimmun. 2010, 34, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bae, J.S. Tumor-Associated Macrophages and Neutrophils in Tumor Microenvironment. Mediat. Inflamm 2016, 2016, 6058147. [Google Scholar] [CrossRef] [Green Version]
- Shaul, M.E.; Fridlender, Z.G. Cancer-related circulating and tumor-associated neutrophils—Subtypes, sources and function. FEBS J. 2018, 285, 4316–4342. [Google Scholar] [CrossRef] [PubMed]
- Grossman, J.G.; Nywening, T.M.; Belt, B.A.; Panni, R.Z.; Krasnick, B.A.; DeNardo, D.G.; Hawkins, W.G.; Goedegebuure, S.P.; Linehan, D.C.; Fields, R.C. Recruitment of CCR2+ tumor associated macrophage to sites of liver metastasis confers a poor prognosis in human colorectal cancer. Oncoimmunology 2018, 7, e1470729. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yao, W.; Yuan, Y.; Chen, P.; Li, B.; Li, J.; Chu, R.; Song, H.; Xie, D.; Jiang, X.; et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut 2017, 66, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lim, S.Y.; Gordon-Weeks, A.N.; Tapmeier, T.T.; Im, J.H.; Cao, Y.; Beech, J.; Allen, D.; Smart, S.; Muschel, R.J. Recruitment of a myeloid cell subset (CD11b/Gr1 mid) via CCL2/CCR2 promotes the development of colorectal cancer liver metastasis. Hepatology 2013, 57, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Aalto, K.; Autio, A.; Kiss, E.A.; Elima, K.; Nymalm, Y.; Veres, T.Z.; Marttila-Ichihara, F.; Elovaara, H.; Saanijoki, T.; Crocker, P.R.; et al. Siglec-9 is a novel leukocyte ligand for vascular adhesion protein-1 and can be used in PET imaging of inflammation and cancer. Blood 2011, 118, 3725–3733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kivi, E.; Elima, K.; Aalto, K.; Nymalm, Y.; Auvinen, K.; Koivunen, E.; Otto, D.M.; Crocker, P.R.; Salminen, T.A.; Salmi, M.; et al. Human Siglec-10 can bind to vascular adhesion protein-1 and serves as its substrate. Blood 2009, 114, 5385–5392. [Google Scholar] [CrossRef] [Green Version]
- Heymann, F.; Tacke, F. Immunology in the liver—From homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 88–110. [Google Scholar] [CrossRef]
- Bilen, M.A.; Shabto, J.M.; Martini, D.J.; Liu, Y.; Lewis, C.; Collins, H.; Akce, M.; Kissick, H.; Carthon, B.C.; Shaib, W.L.; et al. Sites of metastasis and association with clinical outcome in advanced stage cancer patients treated with immunotherapy. BMC Cancer 2019, 19, 857. [Google Scholar] [CrossRef] [Green Version]
- Shiroyama, T.; Suzuki, H.; Tamiya, M.; Tamiya, A.; Tanaka, A.; Okamoto, N.; Nakahama, K.; Taniguchi, Y.; Isa, S.I.; Inoue, T.; et al. Clinical Characteristics of Liver Metastasis in Nivolumab-treated Patients with Non-small Cell Lung Cancer. Anticancer Res. 2018, 38, 4723–4729. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Hellmann, M.D.; Hamid, O.; Tsai, K.K.; Loo, K.L.; Gubens, M.A.; Rosenblum, M.; Harview, C.L.; Taube, J.M.; Handley, N.; et al. Liver Metastasis and Treatment Outcome with Anti-PD-1 Monoclonal Antibody in Patients with Melanoma and NSCLC. Cancer Immunol. Res. 2017, 5, 417–424. [Google Scholar] [CrossRef] [Green Version]
- Tsilimigras, D.I.; Ntanasis-Stathopoulos, I.; Moris, D.; Pawlik, T.M. Liver Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1296, 227–241. [Google Scholar] [CrossRef]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; MacDonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawada, K.; Hasegawa, S.; Murakami, T.; Itatani, Y.; Hosogi, H.; Sonoshita, M.; Kitamura, T.; Fujishita, T.; Iwamoto, M.; Matsumoto, T.; et al. Molecular mechanisms of liver metastasis. Int. J. Clin. Oncol. 2011, 16, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, M. Role of platelets and platelet receptors in cancer metastasis. J. Hematol. Oncol. 2018, 11, 125. [Google Scholar] [CrossRef] [Green Version]
- Foss, A.; Munoz-Sagredo, L.; Sleeman, J.; Thiele, W. The contribution of platelets to intravascular arrest, extravasation, and outgrowth of disseminated tumor cells. Clin. Exp. Metastasis 2020, 37, 47–67. [Google Scholar] [CrossRef]
- Valcarcel, M.; Carrascal, T.; Crende, O.; Vidal-Vanaclocha, F. IL-18 regulates melanoma VLA-4 integrin activation through a Hierarchized sequence of inflammatory factors. J. Investig. Dermatol. 2014, 134, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucotti, S.; Cerutti, C.; Soyer, M.; Gil-Bernabe, A.M.; Gomes, A.L.; Allen, P.D.; Smart, S.; Markelc, B.; Watson, K.; Armstrong, P.C.; et al. Aspirin blocks formation of metastatic intravascular niches by inhibiting platelet-derived COX-1/thromboxane A2. J. Clin. Investig. 2019, 129, 1845–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, A.R.; Wang, S.E. Cancer Tills the Premetastatic Field: Mechanistic Basis and Clinical Implications. Clin. Cancer Res. 2016, 22, 3725–3733. [Google Scholar] [CrossRef] [Green Version]
- Kruger, A. Premetastatic niche formation in the liver: Emerging mechanisms and mouse models. J. Mol. Med. 2015, 93, 1193–1201. [Google Scholar] [CrossRef]
- Paiva, A.E.; Lousado, L.; Guerra, D.A.P.; Azevedo, P.O.; Sena, I.F.G.; Andreotti, J.P.; Santos, G.S.P.; Goncalves, R.; Mintz, A.; Birbrair, A. Pericytes in the Premetastatic Niche. Cancer Res. 2018, 78, 2779–2786. [Google Scholar] [CrossRef] [Green Version]
- Matsumura, H.; Kondo, T.; Ogawa, K.; Tamura, T.; Fukunaga, K.; Murata, S.; Ohkohchi, N. Kupffer cells decrease metastasis of colon cancer cells to the liver in the early stage. Int. J. Oncol. 2014, 45, 2303–2310. [Google Scholar] [CrossRef] [Green Version]
- Vidal-Vanaclocha, F. The Prometastatic Microenvironment of the Liver. Cancer Microenviron. 2008, 1, 113–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramadori, G.; Moriconi, F.; Malik, I.; Dudas, J. Physiology and pathophysiology of liver inflammation, damage and repair. J. Physiol. Pharmacol. 2008, 59 (Suppl. 1), 107–117. [Google Scholar] [PubMed]
- Keirsse, J.; Van Damme, H.; Geeraerts, X.; Beschin, A.; Raes, G.; Van Ginderachter, J.A. The role of hepatic macrophages in liver metastasis. Cell. Immunol. 2018, 330, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Peng, Y.; Deng, J.; Miao, H.; Zhou, J.; Zha, L.; Zhou, R.; Yu, L.; Shi, H.; Liang, H. Endothelial cell-derived fibronectin extra domain A promotes colorectal cancer metastasis via inducing epithelial-mesenchymal transition. Carcinogenesis 2014, 35, 1661–1670. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.T.; Guo, L.L.; Feng, N.; Zhang, L.; Zhou, N.; Ma, L.L.; Shen, L.; Tong, G.H.; Yan, Q.W.; Zhu, S.J.; et al. MIF, secreted by human hepatic sinusoidal endothelial cells, promotes chemotaxis and outgrowth of colorectal cancer in liver prometastasis. Oncotarget 2015, 6, 22410–22423. [Google Scholar] [CrossRef] [Green Version]
- Wen, S.W.; Ager, E.I.; Christophi, C. Bimodal role of Kupffer cells during colorectal cancer liver metastasis. Cancer Biol. Ther. 2013, 14, 606–613. [Google Scholar] [CrossRef] [Green Version]
- Ciner, A.T.; Jones, K.; Muschel, R.J.; Brodt, P. The unique immune microenvironment of liver metastases: Challenges and opportunities. Semin. Cancer Biol. 2020, 71, 143–156. [Google Scholar] [CrossRef]
- Yu, X.; Chen, L.; Liu, J.; Dai, B.; Xu, G.; Shen, G.; Luo, Q.; Zhang, Z. Immune modulation of liver sinusoidal endothelial cells by melittin nanoparticles suppresses liver metastasis. Nat. Commun. 2019, 10, 574. [Google Scholar] [CrossRef]
- Mizuno, R.; Kawada, K.; Itatani, Y.; Ogawa, R.; Kiyasu, Y.; Sakai, Y. The Role of Tumor-Associated Neutrophils in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 529. [Google Scholar] [CrossRef] [Green Version]
- Spicer, J.; Brodt, P.; Ferri, L.E. Role of Inflammation in the Early Stages of Liver Metastasis. In Liver Metastasis: Biology and Clinical Management; Brodt, P., Ed.; Springer: New York, NY, USA, 2011; pp. 155–185. [Google Scholar]
- Cools-Lartigue, J.; Spicer, J.; McDonald, B.; Gowing, S.; Chow, S.; Giannias, B.; Bourdeau, F.; Kubes, P.; Ferri, L. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Investig. 2013, 123, 3446–3458. [Google Scholar] [CrossRef]
- Giese, M.A.; Hind, L.E.; Huttenlocher, A. Neutrophil plasticity in the tumor microenvironment. Blood 2019, 133, 2159–2167. [Google Scholar] [CrossRef] [PubMed]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Rayes, R.F.; Milette, S.; Fernandez, M.C.; Ham, B.; Wang, N.; Bourdeau, F.; Perrino, S.; Yakar, S.; Brodt, P. Loss of neutrophil polarization in colon carcinoma liver metastases of mice with an inducible, liver-specific IGF-I deficiency. Oncotarget 2018, 9, 15691–15704. [Google Scholar] [CrossRef] [Green Version]
- Gordon-Weeks, A.N.; Lim, S.Y.; Yuzhalin, A.E.; Jones, K.; Markelc, B.; Kim, K.J.; Buzzelli, J.N.; Fokas, E.; Cao, Y.; Smart, S.; et al. Neutrophils promote hepatic metastasis growth through fibroblast growth factor 2-dependent angiogenesis in mice. Hepatology 2017, 65, 1920–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leach, J.; Morton, J.P.; Sansom, O.J. Neutrophils: Homing in on the myeloid mechanisms of metastasis. Mol. Immunol. 2019, 110, 69–76. [Google Scholar] [CrossRef]
- Coffelt, S.B.; Wellenstein, M.D.; de Visser, K.E. Neutrophils in cancer: Neutral no more. Nat. Rev. Cancer 2016, 16, 431–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, T.; Fujishita, T.; Loetscher, P.; Revesz, L.; Hashida, H.; Kizaka-Kondoh, S.; Aoki, M.; Taketo, M.M. Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses colon cancer liver metastasis by blocking accumulation of immature myeloid cells in a mouse model. Proc. Natl. Acad. Sci. USA 2010, 107, 13063–13068. [Google Scholar] [CrossRef] [Green Version]
- Mills, C.D. Anatomy of a discovery: m1 and m2 macrophages. Front. Immunol. 2015, 6, 212. [Google Scholar] [CrossRef]
- Ehling, J.; Bartneck, M.; Wei, X.; Gremse, F.; Fech, V.; Mockel, D.; Baeck, C.; Hittatiya, K.; Eulberg, D.; Luedde, T.; et al. CCL2-dependent infiltrating macrophages promote angiogenesis in progressive liver fibrosis. Gut 2014, 63, 1960–1971. [Google Scholar] [CrossRef] [Green Version]
- Karlmark, K.R.; Weiskirchen, R.; Zimmermann, H.W.; Gassler, N.; Ginhoux, F.; Weber, C.; Merad, M.; Luedde, T.; Trautwein, C.; Tacke, F. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 2009, 50, 261–274. [Google Scholar] [CrossRef]
- Milette, S.; Sicklick, J.K.; Lowy, A.M.; Brodt, P. Molecular Pathways: Targeting the Microenvironment of Liver Metastases. Clin. Cancer Res. 2017, 23, 6390–6399. [Google Scholar] [CrossRef] [Green Version]
- Condamine, T.; Gabrilovich, D.I. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011, 32, 19–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, H.; Wang, D.; Daikoku, T.; Sun, H.; Dey, S.K.; Dubois, R.N. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 2013, 24, 631–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabrilovich, D.I. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keskinov, A.A.; Shurin, M.R. Myeloid regulatory cells in tumor spreading and metastasis. Immunobiology 2014, 220, 236–242. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef] [Green Version]
- Kusmartsev, S.; Nefedova, Y.; Yoder, D.; Gabrilovich, D.I. Antigen-specific inhibition of CD8+ T cell response by immature myeloid cells in cancer is mediated by reactive oxygen species. J. Immunol. 2004, 172, 989–999. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Pan, P.Y.; Li, Q.; Sato, A.I.; Levy, D.E.; Bromberg, J.; Divino, C.M.; Chen, S.H. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006, 66, 1123–1131. [Google Scholar] [CrossRef] [Green Version]
- Sade-Feldman, M.; Kanterman, J.; Ish-Shalom, E.; Elnekave, M.; Horwitz, E.; Baniyash, M. Tumor necrosis factor-alpha blocks differentiation and enhances suppressive activity of immature myeloid cells during chronic inflammation. Immunity 2013, 38, 541–554. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Ren, L.; Jian, M.; Xu, P.; Li, J.; Zheng, P.; Feng, Q.; Yang, L.; Ji, M.; Wei, Y.; et al. The mechanism of the premetastatic niche facilitating colorectal cancer liver metastasis generated from myeloid-derived suppressor cells induced by the S1PR1-STAT3 signaling pathway. Cell Death Dis. 2019, 10, 693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milette, S.; Hashimoto, M.; Perrino, S.; Qi, S.; Chen, M.; Ham, B.; Wang, N.; Istomine, R.; Lowy, A.M.; Piccirillo, C.A.; et al. Sexual dimorphism and the role of estrogen in the immune microenvironment of liver metastases. Nat. Commun. 2019, 10, 5745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal-Vanaclocha, F. The Tumor Microenvironment at Different Stages of Hepatic Metastasis. In Liver Metastasis: Biology and Clinical Management; Brodt, P., Ed.; Springer Science+Business Media B.V.: Dordrecht, The Netherlands, 2011; p. 1. [Google Scholar]
- Olaso, E.; Santisteban, A.; Bidaurrazaga, J.; Gressner, A.M.; Rosenbaum, J.; Vidal-Vanaclocha, F. Tumor-dependent activation of rodent hepatic stellate cells during experimental melanoma metastasis. Hepatology 1997, 26, 634–642. [Google Scholar] [CrossRef]
- Nielsen, S.R.; Quaranta, V.; Linford, A.; Emeagi, P.; Rainer, C.; Santos, A.; Ireland, L.; Sakai, T.; Sakai, K.; Kim, Y.S.; et al. Macrophage-secreted granulin supports pancreatic cancer metastasis by inducing liver fibrosis. Nat. Cell Biol. 2016, 18, 549–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology 2008, 134, 1655–1669. [Google Scholar] [CrossRef] [Green Version]
- Copple, B.L.; Bai, S.; Burgoon, L.D.; Moon, J.-O.K. Hypoxia-inducible Factor-1α Regulates Expression of Genes in Hypoxic Hepatic Stellate Cells Important for Collagen Deposition and Angiogenesis. Liver Int. 2011, 31, 230–244. [Google Scholar] [CrossRef] [Green Version]
- Taura, K.; De Minicis, S.; Seki, E.; Hatano, E.; Iwaisako, K.; Osterreicher, C.H.; Kodama, Y.; Miura, K.; Ikai, I.; Uemoto, S.; et al. Hepatic Stellate Cells Secrete Angiopoietin 1 That Induces Angiogenesis in Liver Fibrosis. Gastroenterology 2008, 135, 1729–1738. [Google Scholar] [CrossRef]
- Smedsrod, B.; Le Couteur, D.; Ikejima, K.; Jaeschke, H.; Kawada, N.; Naito, M.; Knolle, P.; Nagy, L.; Senoo, H.; Vidal-Vanaclocha, F.; et al. Hepatic sinusoidal cells in health and disease: Update from the 14th International Symposium. Liver Int. 2009, 29, 490–501. [Google Scholar] [CrossRef]
- Kang, N.; Shah, V.H.; Urrutia, R. Membrane-to-Nucleus Signals and Epigenetic Mechanisms for Myofibroblastic Activation and Desmoplastic Stroma: Potential Therapeutic Targets for Liver Metastasis? Mol. Cancer Res. 2015, 13, 604–612. [Google Scholar] [CrossRef] [Green Version]
- Olaso, E.; Salado, C.; Egilegor, E.; Gutierrez, V.; Santisteban, A.; Sancho-Bru, P.; Friedman, S.L.; Vidal-Vanaclocha, F. Proangiogenic role of tumor-activated hepatic stellate cells in experimental melanoma metastasis. Hepatology 2003, 37, 674–685. [Google Scholar] [CrossRef]
- Charles, R.; Chou, H.S.; Wang, L.; Fung, J.J.; Lu, L.; Qian, S. Human hepatic stellate cells inhibit T-cell response through B7-H1 pathway. Transplantation 2013, 96, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.; Yang, H.R.; Wang, L.; Wildey, G.M.; Fung, J.; Qian, S.; Lu, L. Hepatic stellate cells preferentially expand allogeneic CD4+ CD25+ FoxP3+ regulatory T cells in an IL-2-dependent manner. Transplantation 2008, 86, 1492–1502. [Google Scholar] [CrossRef] [Green Version]
- Hochst, B.; Schildberg, F.A.; Sauerborn, P.; Gabel, Y.A.; Gevensleben, H.; Goltz, D.; Heukamp, L.C.; Turler, A.; Ballmaier, M.; Gieseke, F.; et al. Activated human hepatic stellate cells induce myeloid derived suppressor cells from peripheral blood monocytes in a CD44-dependent fashion. J. Hepatol. 2013, 59, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Yamada, N.; Sawada, T.; Ikeda, K.; Nakatani, K.; Seki, S.; Kaneda, K.; Hirakawa, K. Ultrastructure of early phase hepatic metastasis of human colon carcinoma cells with special reference to desmosomal junctions with hepatocytes. Pathol. Int. 2000, 50, 953–959. [Google Scholar] [CrossRef]
- Mook, O.R.F.; van Marie, J.; Jonges, R.; Vreeling-Sindelarova, H.; Frederiks, W.M.; Van Noorden, C.J.F. Interactions between colon cancer cells and hepatocytes in rats in relation to metastasis. J. Cell. Mol. Med. 2008, 12, 2052–2061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabaries, S.; Dupuy, F.; Dong, Z.; Monast, A.; Annis, M.G.; Spicer, J.; Ferri, L.E.; Omeroglu, A.; Basik, M.; Amir, E.; et al. Claudin-2 promotes breast cancer liver metastasis by facilitating tumor cell interactions with hepatocytes. Mol. Cell Biol. 2012, 32, 2979–2991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georges, R.; Bergmann, F.; Hamdi, H.; Zepp, M.; Eyol, E.; Hielscher, T.; Berger, M.R.; Adwan, H. Sequential biphasic changes in claudin1 and claudin4 expression are correlated to colorectal cancer progression and liver metastasis. J. Cell. Mol. Med. 2012, 16, 260–272. [Google Scholar] [CrossRef]
- Zvibel, I.; Wagner, A.; Pasmanik-Chor, M.; Varol, C.; Oron-Karni, V.; Santo, E.M.; Halpern, Z.; Kariv, R. Transcriptional profiling identifies genes induced by hepatocyte-derived extracellular matrix in metastatic human colorectal cancer cell lines. Clin. Exp. Metastasis 2013, 30, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Fan, X.; Stoicov, C.; Liu, J.H.; Zubair, S.; Tsai, E.; Ste Marie, R.; Wang, T.C.; Lyle, S.; Kurt-Jones, E.; et al. Human and mouse colon cancer utilizes CD95 signaling for local growth and metastatic spread to liver. Gastroenterology 2009, 137, 934–944.e934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagh, P.; Peace, B.E.; Waltz, S.E. The Met-Related Receptor Tyrosine Kinase Ron in Tumor Growth and Metastasis. Adv. Cancer Res. 2008, 100, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, T.; Nishikawa, Y.; Ito, R.; Kawamata, M.; Doi, Y.; Yamamoto, Y.; Yoshida, M.; Omori, Y.; Kotanagi, H.; Masuko, T.; et al. Significance of integrin alphavbeta5 and erbB3 in enhanced cell migration and liver metastasis of colon carcinomas stimulated by hepatocyte-derived heregulin. Cancer Sci. 2010, 101, 2011–2018. [Google Scholar] [CrossRef] [PubMed]
- Dome, B.; Hendrix, M.J.; Paku, S.; Tovari, J.; Timar, J. Alternative vascularization mechanisms in cancer: Pathology and therapeutic implications. Am. J. Pathol. 2007, 170, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Chen, T.; Zheng, X.; Yang, S.; Xu, K.; Chen, X.; Xu, F.; Wang, L.; Shen, Y.; Wang, T.; et al. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis 2018, 39, 1368–1379. [Google Scholar] [CrossRef]
- Rahbari, N.N.; Kedrin, D.; Incio, J.; Liu, H.; Ho, W.W.; Nia, H.T.; Edrich, C.M.; Jung, K.; Daubriac, J.; Chen, I.; et al. Anti-VEGF therapy induces ECM remodeling and mechanical barriers to therapy in colorectal cancer liver metastases. Sci. Transl. Med. 2016, 8, 360ra135. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Zhang, Y.; Iwamoto, H.; Hosaka, K.; Seki, T.; Andersson, P.; Lim, S.; Fischer, C.; Nakamura, M.; Abe, M.; et al. Discontinuation of anti-VEGF cancer therapy promotes metastasis through a liver revascularization mechanism. Nat. Commun. 2016, 7, 12680. [Google Scholar] [CrossRef] [Green Version]
- Frentzas, S.; Simoneau, E.; Bridgeman, V.L.; Vermeulen, P.B.; Foo, S.; Kostaras, E.; Nathan, M.; Wotherspoon, A.; Gao, Z.H.; Shi, Y.; et al. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat. Med. 2016, 22, 1294–1302. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsilimigras, D.I.; Ntanasis-Stathopoulos, I.; Pawlik, T.M. Molecular Mechanisms of Colorectal Liver Metastases. Cells 2023, 12, 1657. https://doi.org/10.3390/cells12121657
Tsilimigras DI, Ntanasis-Stathopoulos I, Pawlik TM. Molecular Mechanisms of Colorectal Liver Metastases. Cells. 2023; 12(12):1657. https://doi.org/10.3390/cells12121657
Chicago/Turabian StyleTsilimigras, Diamantis I., Ioannis Ntanasis-Stathopoulos, and Timothy M. Pawlik. 2023. "Molecular Mechanisms of Colorectal Liver Metastases" Cells 12, no. 12: 1657. https://doi.org/10.3390/cells12121657
APA StyleTsilimigras, D. I., Ntanasis-Stathopoulos, I., & Pawlik, T. M. (2023). Molecular Mechanisms of Colorectal Liver Metastases. Cells, 12(12), 1657. https://doi.org/10.3390/cells12121657







