Telomerase Activity in Somatic Tissues and Ovaries of Diploid and Triploid Rainbow Trout (Oncorhynchus mykiss) Females
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rainbow Trout Stocks, Origin and Maintenance
2.2. RNA Extraction and Analysis of Rainbow Trout Telomerase (Tert) Expression
2.3. Histological Preparation
2.4. Ploidy Confirmation: Preparation of the Metaphase Spreads and Microscopic Analysis
2.5. Statistical Analysis
3. Results
3.1. Confirmation of Ploidy—Cytogenetic Examination
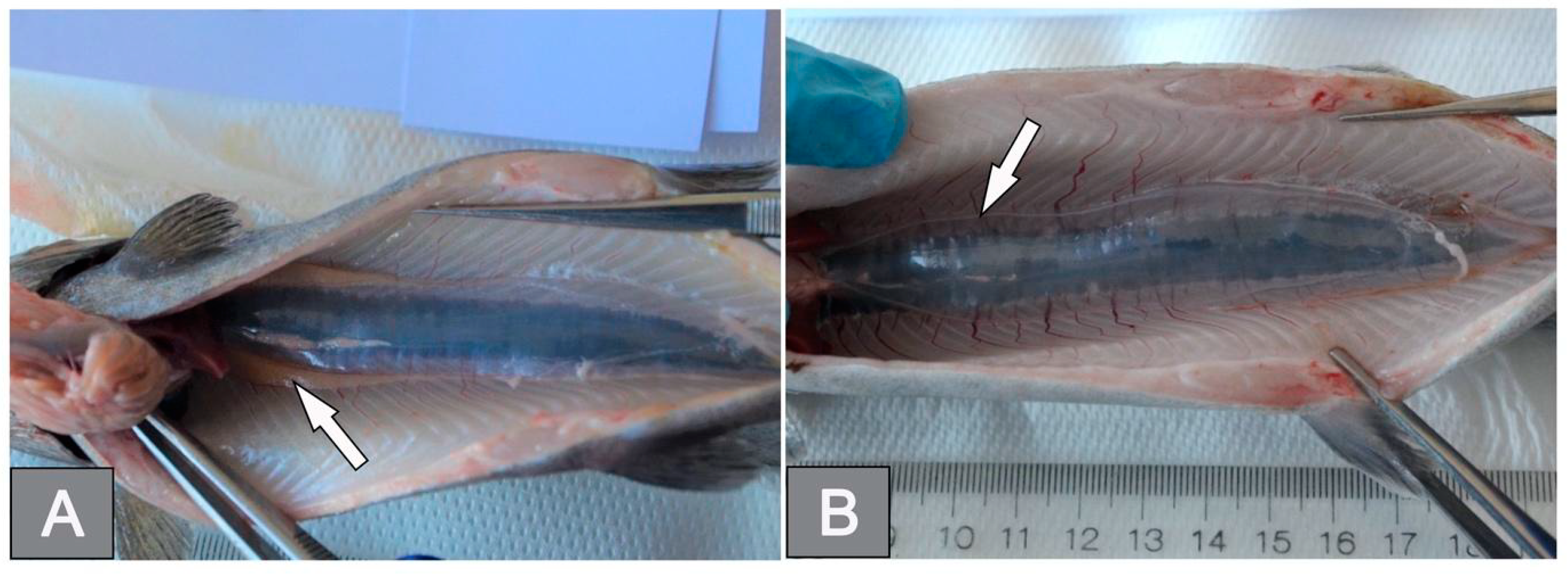
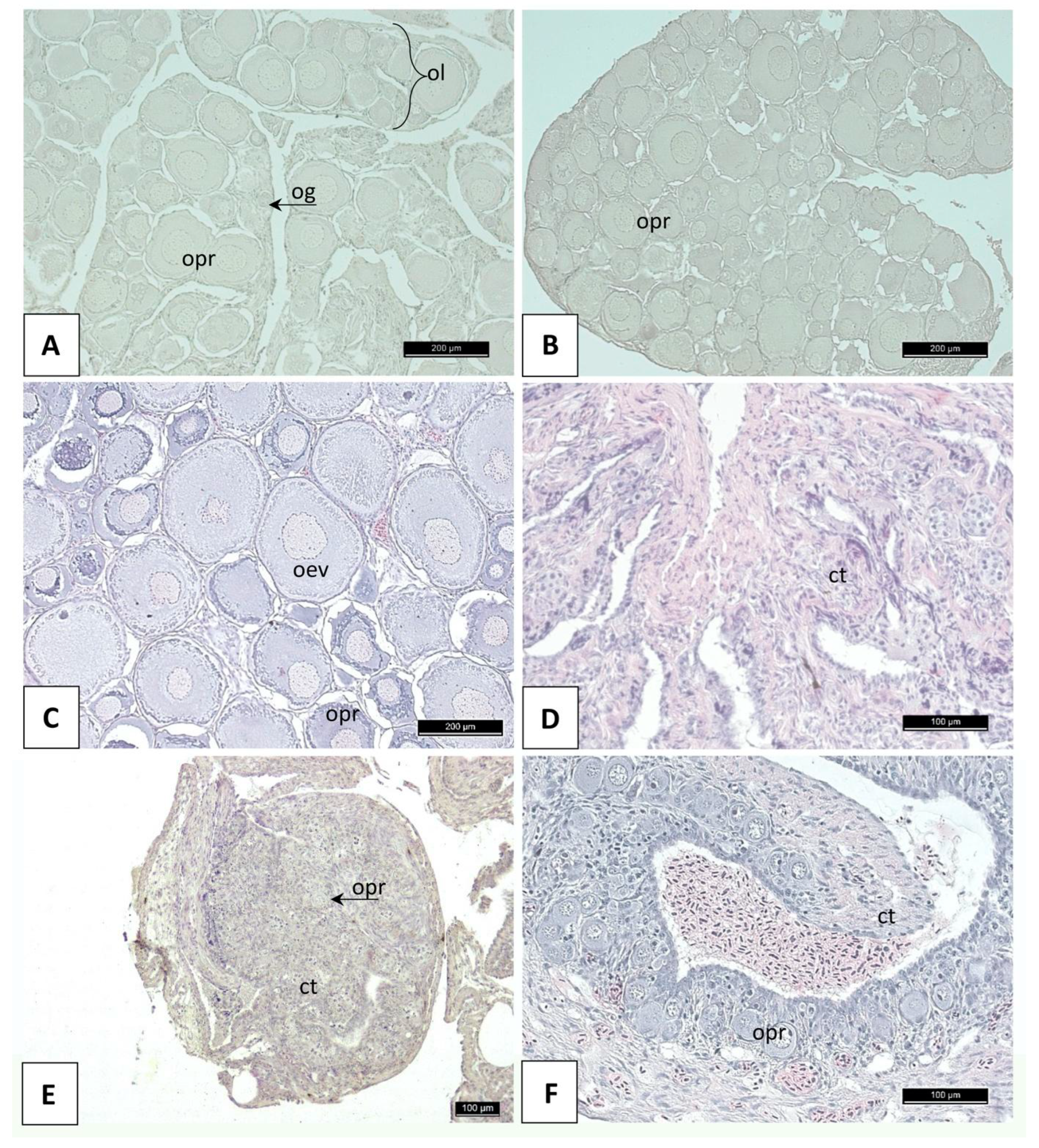
3.2. Histology of Gonads
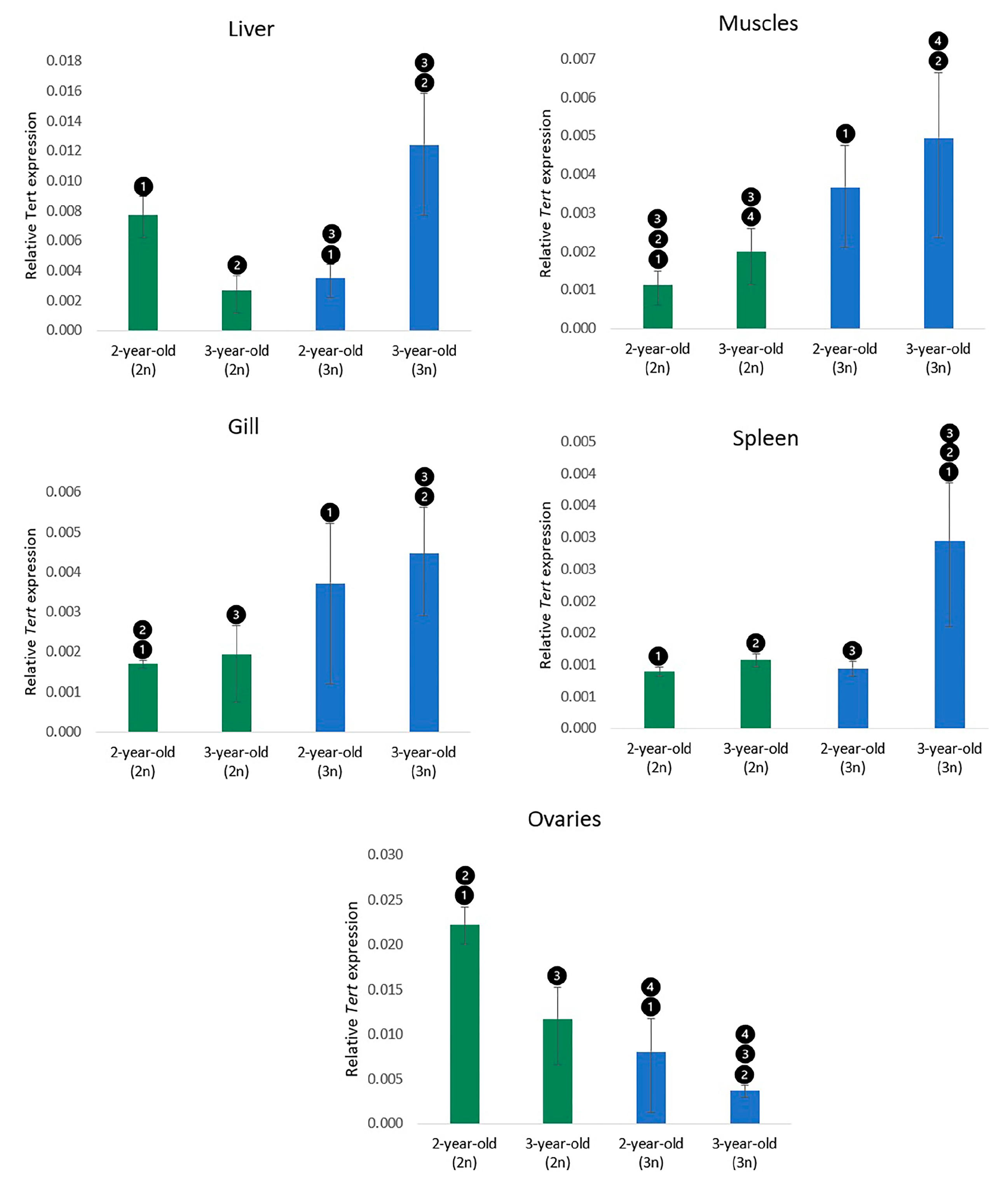
3.3. Expression of the Tert Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blackburn, E.H. Structure and Function of Telomeres. Nature 1991, 350, 569–573. [Google Scholar] [CrossRef]
- Shay, J.; Wright, W. Hallmarks of Telomeres in Ageing Research. J. Pathol. 2007, 211, 114–123. [Google Scholar] [CrossRef]
- Kolquist, K.A.; Ellisen, L.W.; Counter, C.M.; Meyerson, M.M.; Tan, L.K.; Weinberg, R.A.; Haber, D.A.; Gerald, W.L. Expression of TERT in Early Premalignant Lesions and a Subset of Cells in Normal Tissues. Nat. Genet. 1998, 19, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Hiyama, E.; Hiyama, K.; Yokoyama, T.; Shay, J.W. Immunohistochemical Detection of Telomerase (HTERT) Protein in Human Cancer Tissues and a Subset of Cells in Normal Tissues. Neoplasia 2001, 3, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Hornsby, P.J. Telomerase and the Aging Process. Exp. Gerontol. 2007, 42, 575–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anchelin, M.; Murcia, L.; Alcaraz-Pérez, F.; García-Navarro, E.M.; Cayuela, M.L. Behaviour of Telomere and Telomerase during Aging and Regeneration in Zebrafish. PLoS ONE 2011, 6, e16955. [Google Scholar] [CrossRef]
- Lund, T.C.; Glass, T.J.; Tolar, J.; Blazar, B.R. Expression of Telomerase and Telomere Length Are Unaffected by Either Age or Limb Regeneration in Danio Rerio. PLoS ONE 2009, 4, e7688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panasiak, L.; Dobosz, S.; Ocalewicz, K. Telomere Dynamics in the Diploid and Triploid Rainbow Trout (Oncorhynchus mykiss) Assessed by Q-FISH Analysis. Genes 2020, 11, 786. [Google Scholar] [CrossRef]
- Hatakeyama, H.; Yamazaki, H.; Nakamura, K.-I.; Izumiyama-Shimomura, N.; Aida, J.; Suzuki, H.; Tsuchida, S.; Matsuura, M.; Takubo, K.; Ishikawa, N. Telomere Attrition and Restoration in the Normal Teleost Oryzias Latipes Are Linked to Growth Rate and Telomerase Activity at Each Life Stage. Aging 2016, 8, 62–76. [Google Scholar] [CrossRef] [Green Version]
- Klapper, W.; Heidorn, K.; Kühne, K.; Parwaresch, R.; Krupp, G. Telomerase Activity in “Immortal” Fish 1. FEBS Lett. 1998, 434, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Elmore, L.W.; Norris, M.W.; Sircar, S.; Bright, A.T.; McChesney, P.A.; Winn, R.N.; Holt, S.E. Upregulation of Telomerase Function during Tissue Regeneration. Exp. Biol. Med. 2008, 233, 958–967. [Google Scholar] [CrossRef]
- Olsson, M.; Wapstra, E.; Friesen, C. Ectothermic Telomeres: It’s Time They Came in from the Cold. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160449. [Google Scholar] [CrossRef] [Green Version]
- Pfennig, F.; Kind, B.; Zieschang, F.; Busch, M.; Gutzeit, H.O. Tert Expression and Telomerase Activity in Gonads and Somatic Cells of the Japanese Medaka (Oryzias latipes). Dev. Growth Differ. 2008, 50, 131–141. [Google Scholar] [CrossRef]
- Peterson, D.R.; Mok, H.O.L.; Au, D.W.T. Modulation of Telomerase Activity in Fish Muscle by Biological and Environmental Factors. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2015, 178, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Lau, B.W.-M.; Wong, A.O.-L.; Tsao, G.S.-W.; So, K.-F.; Yip, H.K.-F. Molecular Cloning and Characterization of the Zebrafish (Danio rerio) Telomerase Catalytic Subunit (Telomerase Reverse Transcriptase, TERT). J. Mol. Neurosci. 2007, 34, 63–75. [Google Scholar] [CrossRef]
- McChesney, P.A.; Elmore, L.W.; Holt, S.E. Vertebrate Marine Species as Model Systems for Studying Telomeres and Telomerase. Zebrafish 2005, 1, 349–355. [Google Scholar] [CrossRef]
- Kishi, S.; Uchiyama, J.; Baughman, A.M.; Goto, T.; Lin, M.C.; Tsai, S.B. The Zebrafish as a Vertebrate Model of Functional Aging and Very Gradual Senescence. Exp. Gerontol. 2003, 38, 777–786. [Google Scholar] [CrossRef]
- Harel, I.; Benayoun, B.A.; Machado, B.; Singh, P.P.; Hu, C.-K.; Pech, M.F.; Valenzano, D.R.; Zhang, E.; Sharp, S.C.; Artandi, S.E.; et al. A Platform for Rapid Exploration of Aging and Diseases in a Naturally Short-Lived Vertebrate. Cell 2015, 160, 1013–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The State of World Fisheries 2022. FAO|Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/publications/sofia/2022/en/ (accessed on 5 May 2023).
- Thorgaard, G.H.; Bailey, G.S.; Williams, D.; Buhler, D.R.; Kaattari, S.L.; Ristow, S.S.; Hansen, J.D.; Winton, J.R.; Bartholomew, J.L.; Nagler, J.J.; et al. Status and Opportunities for Genomics Research with Rainbow Trout. Comp. Biochem. Physiol. B Biochem. Mol. Bio. 2002, 133, 609–646. [Google Scholar] [CrossRef]
- Pérez-Cerezales, S.; Gutiérrez-Adán, A.; Martínez-Páramo, S.; Beirão, J.; Herráez, M.P. Altered Gene Transcription and Telomere Length in Trout Embryo and Larvae Obtained with DNA Cryodamaged Sperm. Theriogenology 2011, 76, 1234–1245. [Google Scholar] [CrossRef]
- Cuellar, O.; Uyeno, T. Triploidy in Rainbow Trout. Cytogenet. Genome Res. 1972, 11, 508–515. [Google Scholar] [CrossRef]
- Thorgaard, G.H.; Gall, G.A.E. Adult Triploids in a Rainbow Trout Family. Genetics 1979, 93, 961–973. [Google Scholar] [CrossRef]
- Thorgaard, G.H.; Rabinovitch, P.S.; Shen, M.W.; Gall, G.A.E.; Propp, J.; Utter, F.M. Triploid Rainbow Trout Identified by Flow Cytometry. Aquaculture 1982, 29, 305–309. [Google Scholar] [CrossRef]
- Aegerter, S.; Jalabert, B. Effects of Post-Ovulatory Oocyte Ageing and Temperature on Egg Quality and on the Occurrence of Triploid Fry in Rainbow Trout. Oncorhynchus Mykiss. Aquac. 2004, 231, 59–71. [Google Scholar] [CrossRef]
- Ocalewicz, K.; Dobosz, S. Karyotype Variation in the Albino Rainbow Trout (Oncorhynchus mykiss (Walbaum)). Genome 2009, 52, 347–352. [Google Scholar] [CrossRef]
- Ueda, T.; Kobayashi, M.; Sato, R. Triploid Rainbow Trouts Induced by Polyethylene Glycol. Proc. Jpn. Acad. B 1986, 62, 161–164. [Google Scholar] [CrossRef] [Green Version]
- Chourrout, D.; Chevassus, B.; Krieg, F.; Happe, A.; Burger, G.; Renard, P. Production of Second Generation Triploid and Tetraploid Rainbow Trout by Mating Tetraploid Males and Diploid Females ? Potential of Tetraploid Fish. Theor. Appl. Genet. 1986, 72, 193–206. [Google Scholar] [CrossRef]
- Blanc, J.-M.; Chourrout, D.; Krieg, F. Evaluation of Juvenile Rainbow Trout Survival and Growth in Half-Sib Families from Diploid and Tetraploid Sires. Aquaculture 1987, 65, 215–220. [Google Scholar] [CrossRef]
- Myers, J.M.; Hershberger, W.K. Early Growth and Survival of Heat-Shocked and Tetraploid-Derived Triploid Rainbow Trout (Oncorhynchus mykiss). Aquaculture 1991, 96, 97–107. [Google Scholar] [CrossRef]
- Thorgaard, G.H. Ploidy Manipulation and Performance. Aquaculture 1986, 57, 57–64. [Google Scholar] [CrossRef]
- Pandian, T.J.; Koteeswaran, R. Ploidy Induction and Sex Control in Fish. Hydrobiologia 1998, 384, 167–243. [Google Scholar] [CrossRef]
- Arai, K.; Fujimoto, T. Genomic Constitution and Atypical Reproduction in Polyploid and Unisexual Lineages of the Misgurnus Loach, a Teleost Fish. Cytogenet. Genome Res. 2013, 140, 226–240. [Google Scholar] [CrossRef] [Green Version]
- Benfey, T.J. The Physiology and Behavior of Triploid Fishes. Rev. Fish. Sci. 1999, 7, 39–67. [Google Scholar] [CrossRef]
- Piferrer, F.; Beaumont, A.; Falguière, J.-C.; Flajšhans, M.; Haffray, P.; Colombo, L. Polyploid Fish and Shellfish: Production, Biology and Applications to Aquaculture for Performance Improvement and Genetic Containment. Aquaculture 2009, 293, 125–156. [Google Scholar] [CrossRef] [Green Version]
- Hulata, G. Genetic Manipulations in Aquaculture: A Review of Stock Improvement by Classical and Modern Technologies. Genetica 2001, 111, 155–173. [Google Scholar] [CrossRef]
- Rothbard, S. A Review of Ploidy Manipulations in Aquaculture: The Israeli Experience. Isr. J. Aquac. Bamidgeh. 2006, 58, 266–279. [Google Scholar] [CrossRef]
- Koenig, M.K.; Kozfkay, J.R.; Meyer, K.A.; Schill, D.J. Performance of Diploid and Triploid Rainbow Trout Stocked in Idaho Alpine Lakes. N. Am. J. Fish. Manag. 2011, 31, 124–133. [Google Scholar] [CrossRef]
- Razak, S.A.; Hwang, G.-L.; Rahman, M.A.; Maclean, N. Growth Performance and Gonadal Development of Growth Enhanced Transgenic Tilapia Oreochromis niloticus (L.) Following Heat-Shock-Induced Triploidy. Mar. Biotechnol. 1999, 1, 533–544. [Google Scholar] [CrossRef]
- Nam, Y.K.; Park, I.-S.; Kim, D.S. Triploid Hybridization of Fast-Growing Transgenic Mud Loach Misgurnus Mizolepis Male to Cyprinid Loach Misgurnus anguillicaudatus Female: The First Performance Study on Growth and Reproduction of Transgenic Polyploid Hybrid Fish. Aquaculture 2004, 231, 559–572. [Google Scholar] [CrossRef]
- Zajicek, P.; Goodwin, A.E.; Weier, T. Triploid Grass Carp: Triploid Induction, Sterility, Reversion, and Certification. N. Am. J. Fish. Manag. 2011, 31, 614–618. [Google Scholar] [CrossRef]
- Hliwa, P.; Panasiak, L.; Ziomek, E.; Rożyński, R.; Leonowicz, Ł.; Grudniewska, J.; Dobosz, S.; Ocalewicz, K. Application of High Hydrostatic Pressure (HHP) Shock to Induce Triploid Development in the European Grayling (Thymallus thymallus L.). Anim. Reprod. Sci. 2022, 237, 106929. [Google Scholar] [CrossRef]
- Opstad, I.; Fjelldal, P.G.; Karlsen, Ø.; Thorsen, A.; Hansen, T.J.; Taranger, G.L. The Effect of Triploidization of Atlantic Cod (Gadus morhua L.) on Survival, Growth and Deformities during Early Life Stages. Aquaculture 2013, 388–391, 54–59. [Google Scholar] [CrossRef]
- Hattori, R.S.; Yoshinaga, T.T.; Katayama, N.; Hattori-Ihara, S.; Tsukamoto, R.Y.; Takahashi, N.S.; Tabata, Y.A. Surrogate Production of Salmo Salar Oocytes and Sperm in Triploid Oncorhynchus Mykiss by Germ Cell Transplantation Technology. Aquaculture 2019, 506, 238–245. [Google Scholar] [CrossRef]
- Nynca, J.; Słowińska, M.; Wiśniewska, J.; Jastrzębski, J.; Dobosz, S.; Ciereszko, A. Ovarian Transcriptome Analysis of Diploid and Triploid Rainbow Trout Revealed New Pathways Related to Gonadal Development and Fertility. Animal 2022, 16, 100594. [Google Scholar] [CrossRef]
- Benfey, T.J.; Devlin, R.H. Ploidy Has Minimal Effect on Hypoxia Tolerance at High Temperature in Rainbow Trout (Oncorhynchus mykiss). Physiol. Biochem. Zool. 2018, 91, 1091–1101. [Google Scholar] [CrossRef]
- Han, B.; Meng, Y.; Tian, H.; Li, C.; Li, Y.; Gongbao, C.; Fan, W.; Ma, R. Effects of Acute Hypoxic Stress on Physiological and Hepatic Metabolic Responses of Triploid Rainbow Trout (Oncorhynchus mykiss). Front. Physiol. 2022, 13, 921709. [Google Scholar] [CrossRef]
- Cleveland, B.M.; Weber, G.M. Ploidy Effects on Genes Regulating Growth Mechanisms during Fasting and Refeeding in Juvenile Rainbow Trout (Oncorhynchus mykiss). Mol. Cell. Endocrinol. 2014, 382, 139–149. [Google Scholar] [CrossRef]
- Carrizo, V.; Valenzuela, C.A.; Zuloaga, R.; Aros, C.; Altamirano, C.; Valdés, J.A.; Molina, A. Effect of Cortisol on the Immune-like Response of Rainbow Trout (Oncorhynchus mykiss) Myotubes Challenged with Piscirickettsia Salmonis. Vet. Immunol. Immunopathol. 2021, 237, 110240. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zawistowski, S. Histological Technique, Histology and the Principles of Histopathology; PZWL: Warsaw, Poland, 1983. [Google Scholar]
- Brown-Peterson, N.J.; Wyanski, D.M.; Saborido-Rey, F.; Macewicz, B.J.; Lowerre-Barbieri, S.K. A Standardized Terminology for Describing Reproductive Development in Fishes. Mar. Coast. Fish. 2011, 3, 52–70. [Google Scholar] [CrossRef]
- Ocalewicz, K. Cytogenetic Markers for X Chromosome in a Karyotype of Rainbow Trout from Rutki Strain (Poland). Folia Biol. 2002, 50, 13–16. [Google Scholar]
- Phillips, R.; Ráb, P. Chromosome Evolution in the Salmonidae (Pisces): An Update. Biol. Rev. 2001, 76, 1–25. [Google Scholar] [CrossRef]
- Maxime, V. The Physiology of Triploid Fish: Current Knowledge and Comparisons with Diploid Fish. Fish Fish. 2008, 9, 67–78. [Google Scholar] [CrossRef]
- Szarski, H. Cell Size and Nuclear DNA Content in Vertebrates. Int. Rev. Cytol. 1976, 44, 93–111. [Google Scholar] [CrossRef]
- Fraser, T.W.K.; Hansen, T.; Skjæraasen, J.E.; Mayer, I.; Sambraus, F.; Fjelldal, P.G. The Effect of Triploidy on the Culture Performance, Deformity Prevalence, and Heart Morphology in Atlantic Salmon. Aquaculture 2013, 416–417, 255–264. [Google Scholar] [CrossRef]
- Vera, L.M.; Metochis, C.; Taylor, J.F.; Clarkson, M.; Skjærven, K.H.; Migaud, H.; Tocher, D.R. Early Nutritional Programming Affects Liver Transcriptome in Diploid and Triploid Atlantic Salmon, Salmo Salar. BMC Genom. 2017, 18, 886. [Google Scholar] [CrossRef] [Green Version]
- Kozlowski, J.; Konarzewski, M.; Gawelczyk, A.T. Cell Size as a Link between Noncoding DNA and Metabolic Rate Scaling. Proc. Natl. Acad. Sci. USA 2003, 100, 14080–14085. [Google Scholar] [CrossRef]
- Christensen, K.A.; Sakhrani, D.; Rondeau, E.B.; Richards, J.; Koop, B.F.; Devlin, R.H. Effect of Triploidy on Liver Gene Expression in Coho Salmon (Oncorhynchus kisutch) under Different Metabolic States. BMC Genom. 2019, 20, 336. [Google Scholar] [CrossRef] [Green Version]
- Zhong, H.; Zhou, Y.; Liu, S.; Tao, M.; Long, Y.; Liu, Z.; Zhang, C.; Duan, W.; Hu, J.; Song, C.; et al. Elevated Expressions of GH/IGF Axis Genes in Triploid Crucian Carp. Gen. Comp. Endocrinol. 2012, 178, 291–300. [Google Scholar] [CrossRef]
- Harvey, A.; Fjelldal, P.G.; Solberg, M.F.; Hansen, T.W.; Glover, K.A. Ploidy Elicits a Whole-Genome Dosage Effect: Growth of Triploid Atlantic Salmon Is Linked to the Genetic Origin of the Second Maternal Chromosome Set. BMC Genet. 2017, 18, 34. [Google Scholar] [CrossRef] [Green Version]
- Chatchaiphan, S.; Srisapoome, P.; Kim, J.-H.; Devlin, R.H.; Na-Nakorn, U. De Novo Transcriptome Characterization and Growth-Related Gene Expression Profiling of Diploid and Triploid Bighead Catfish (Clarias macrocephalus Günther, 1864). Mar. Biotechnol. 2017, 19, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Shrimpton, J.M.; Heath, J.W.; Devlin, R.H.; Heath, D.D. Effect of Triploidy on Growth and Ionoregulatory Performance in Ocean-Type Chinook Salmon: A Quantitative Genetics Approach. Aquaculture 2012, 362–363, 248–254. [Google Scholar] [CrossRef]
- Meiler, K.A.; Cleveland, B.; Radler, L.; Kumar, V. Oxidative Stress-Related Gene Expression in Diploid and Triploid Rainbow Trout (Oncorhynchus mykiss) Fed Diets with Organic and Inorganic Zinc. Aquaculture 2021, 533, 736149. [Google Scholar] [CrossRef]
- Ching, B.; Jamieson, S.; Heath, J.W.; Heath, D.D.; Hubberstey, A. Transcriptional Differences between Triploid and Diploid Chinook Salmon (Oncorhynchus tshawytscha) during Live Vibrio Anguillarum Challenge. Heredity 2009, 104, 224–234. [Google Scholar] [CrossRef] [Green Version]
- Henriques, C.M.; Carneiro, M.C.; Tenente, I.M.; Jacinto, A.; Ferreira, M.G. Telomerase Is Required for Zebrafish Lifespan. PLoS Genet. 2013, 9, e1003214. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.M.; Chen, E.X.; Kong, R.Y.; Ng, P.K.; Mok, H.O.; Au, D.W. Hypoxia Induces Telomerase Reverse Transcriptase (TERT) Gene Expression in Non-Tumor Fish Tissues in Vivo: The Marine Medaka (Oryzias melastigma) Model. BMC Mol. Biol. 2006, 7, 27. [Google Scholar] [CrossRef] [Green Version]
- Au, D.W.T.; Mok, H.O.L.; Elmore, L.W.; Holt, S.E. Japanese Medaka: A New Vertebrate Model for Studying Telomere and Telomerase Biology. Comp. Biochem. Physiol. Part—C Toxicol. Pharmacol. 2009, 149, 161–167. [Google Scholar] [CrossRef]
- López de Abechuco, E.; Bilbao, E.; Soto, M.; Díez, G. Molecular Cloning and Measurement of Telomerase Reverse Transcriptase (TERT) Transcription Patterns in Tissues of European Hake (Merluccius merluccius) and Atlantic Cod (Gadus morhua) during Aging. Gene 2014, 541, 8–18. [Google Scholar] [CrossRef]
- Panasiak, L.; Kuciński, M.; Błaszczyk, A.; Ocalewicz, K. Telomerase Activity in Androgenetic Rainbow Trout with Growth Deficiency and in Normally Developed Individuals. Zebrafish 2022, 19, 131–136. [Google Scholar] [CrossRef]
- Schoenfeld, B.J. The Mechanisms of Muscle Hypertrophy and Their Application to Resistance Training. J. Strength. Cond. Res. 2010, 24, 2857–2872. [Google Scholar] [CrossRef] [Green Version]
- Alonso, M.; Tabata, Y.A.; Rigolino, M.G.; Tsukamoto, R.Y. Effect of Induced Triploidy on Fin Regeneration of Juvenile Rainbow Trout, Oncorhynchus mykiss. J. Exp. Zoo. 2000, 287, 493–502. [Google Scholar] [CrossRef]
- Yegorov, Y.E. Healthy Aging: Antioxidants, Uncouplers And/or Telomerase? Mol. Biol. 2020, 54, 311–316. [Google Scholar] [CrossRef]
- Anatskaya, O.V.; Vinogradov, A.E. Genome Multiplication as Adaptation to Tissue Survival: Evidence from Gene Expression in Mammalian Heart and Liver. Genomics 2007, 89, 70–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaeschke, H. Reactive Oxygen and Mechanisms of Inflammatory Liver Injury. J. Gastroenterol. Hepatol. 2000, 15, 718–724. [Google Scholar] [CrossRef]
- Bernier, N.J.; Brauner, C.J.; Heath, J.W.; Randall, D.J. Oxygen and Carbon Dioxide Transport during Sustained Exercise in Diploid and Triploid Chinook Salmon (Oncorhynchus tshawytscha). Can. J. Fish. Aquat. Sci. 2004, 61, 1797–1805. [Google Scholar] [CrossRef]
- Kobayashi, T.; Fushiki, S.; Ueno, K. Physiological Response of Triploid Rainbow Trout to Exhaustive Exercise. Aquacult. Sci. 2009, 57, 361–370. [Google Scholar] [CrossRef]
- Betts, D.H.; King, W.A. Telomerase Activity and Telomere Detection during Early Bovine Development. Dev. Genet. 1999, 25, 397–403. [Google Scholar] [CrossRef]
- Liu, J.-P.; Li, H. Telomerase in the Ovary. Reproduction 2010, 140, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Kinugawa, C.; Murakami, T.; Okamura, K.; Yajima, A. Telomerase Activity in Normal Ovaries and Premature Ovarian Failure. Tohoku J. Exp. Med. 2000, 190, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Rodgers, R.J.; Irving-Rodgers, H.F.; van Wezel, I.L.; Krupa, M.; Lavranos, T.C. Dynamics of the Membrana Granulosa during Expansion of the Ovarian Follicular Antrum. Mol. Cell. Endocrinol. 2001, 171, 41–48. [Google Scholar] [CrossRef]
- Tománek, M.; Chronowska, E.; Kott, T.; Czerneková, V. Telomerase Activity in Pig Granulosa Cells Proliferating and Differentiating in Vitro. Anim. Reprod. Sci. 2008, 104, 284–298. [Google Scholar] [CrossRef]
- Hartmann, N.; Reichwald, K.; Lechel, A.; Graf, M.; Kirschner, J.; Dorn, A.; Terzibasi, E.; Wellner, J.; Platzer, M.; Rudolph, K.L.; et al. Telomeres Shorten While Tert Expression Increases during Ageing of the Short-Lived Fish Nothobranchius Furzeri. Mech. Ageing Dev. 2009, 130, 290–296. [Google Scholar] [CrossRef]
- Downs, K.P.; Shen, Y.; Pasquali, A.; Beldorth, I.; Savage, M.; Gallier, K.; Garcia, T.; Booth, R.E.; Walter, R.B. Characterization of Telomeres and Telomerase Expression in Xiphophorus. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2012, 155, 89–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrasco, L.A.P.; Doroshov, S.; Penman, D.J.; Bromage, N. Long-Term, Quantitative Analysis of Gametogenesis in Autotriploid Rainbow Trout, Oncorhynchus Mykiss. Reproduction 1998, 113, 197–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lincoln, R.F.; Scott, A.P. Sexual Maturation in Triploid Rainbow Trout, Salmo Gairdneri Richardson. J. Fish Biol. 1984, 25, 385–392. [Google Scholar] [CrossRef]
- Krisfalusi, M.; Cloud, J.G. Gonadal Sex Reversal in Triploid Rainbow Trout (Oncorhynchus mykiss). J. Exp. Zool. 1999, 284, 466–472. [Google Scholar] [CrossRef]
- Han, Y.; Liu, M.; Zhang, L.L.; Simpson, B.; Zhang, G.X. Comparison of Reproductive Development in Triploid and Diploid Female Rainbow Trout, Oncorhynchus Mykiss. J. Fish Biol. 2010, 76, 1742–1750. [Google Scholar] [CrossRef]
- Thomé, R.G.; Domingos, F.F.T.; Santos, H.B.; Martinelli, P.M.; Sato, Y.; Rizzo, E.; Bazzoli, N. Apoptosis, Cell Proliferation and Vitellogenesis during the Folliculogenesis and Follicular Growth in Teleost Fish. Tissue Cell 2012, 44, 54–62. [Google Scholar] [CrossRef]
- Kyo, S.; Takakura, M.; Kanaya, T.; Zhuo, W.; Fujimoto, K.; Nishio, Y.; Orimo, A.; Inoue, M. Estrogen Activates Telomerase. Cancer Res. 1999, 59, 5917–5921. [Google Scholar]
- Monaghan, P.; Ozanne, S.E. Somatic Growth and Telomere Dynamics in Vertebrates: Relationships, Mechanisms and Consequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160446. [Google Scholar] [CrossRef] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panasiak, L.; Kuciński, M.; Hliwa, P.; Pomianowski, K.; Ocalewicz, K. Telomerase Activity in Somatic Tissues and Ovaries of Diploid and Triploid Rainbow Trout (Oncorhynchus mykiss) Females. Cells 2023, 12, 1772. https://doi.org/10.3390/cells12131772
Panasiak L, Kuciński M, Hliwa P, Pomianowski K, Ocalewicz K. Telomerase Activity in Somatic Tissues and Ovaries of Diploid and Triploid Rainbow Trout (Oncorhynchus mykiss) Females. Cells. 2023; 12(13):1772. https://doi.org/10.3390/cells12131772
Chicago/Turabian StylePanasiak, Ligia, Marcin Kuciński, Piotr Hliwa, Konrad Pomianowski, and Konrad Ocalewicz. 2023. "Telomerase Activity in Somatic Tissues and Ovaries of Diploid and Triploid Rainbow Trout (Oncorhynchus mykiss) Females" Cells 12, no. 13: 1772. https://doi.org/10.3390/cells12131772





