Development of Woolly Hair and Hairlessness in a CRISPR−Engineered Mutant Mouse Model with KRT71 Mutations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Ethics Statement
2.2. CRISPR/Cas9 sgRNA Preparation, Embryo Microinjection and Embryo Transfer
2.3. Mutation Detection in Mice by PCR and Sequencing
2.4. RNA Extraction and Quantitative RT–PCR
2.5. Body Weight and Survival Curve
2.6. Flow Cytometry
2.7. Western Blotting
2.8. Histology Analysis
2.9. Scanning Electron Microscopy
2.10. Statistical Analysis
3. Results
3.1. Generation of Krt71 Mice the CRISPR/Cas9 System
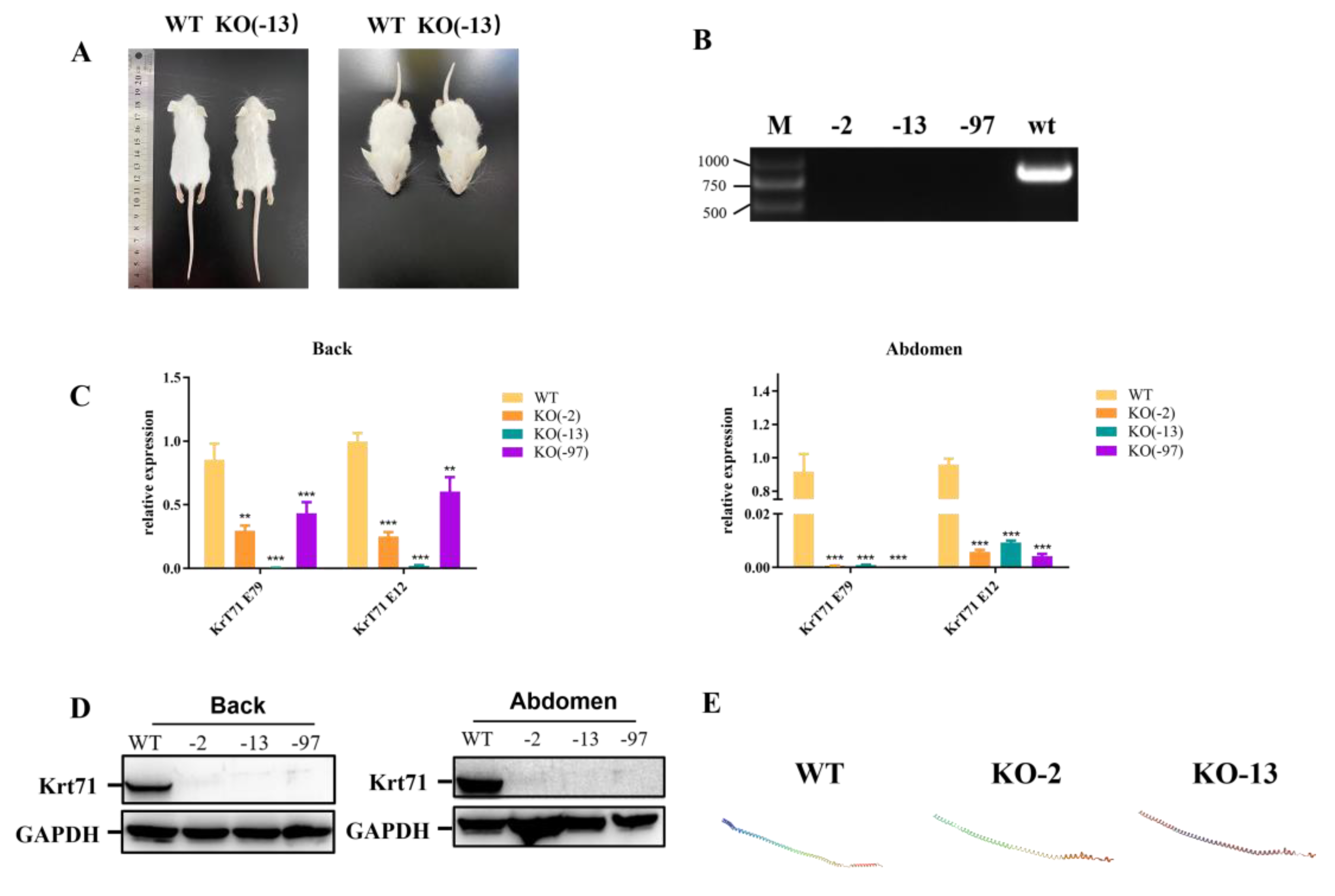
- To disrupt the function of Krt71, we designed a sgRNA targeting exon 6 for the mouse Krt71 gene (Figure 1A). The target site is shown in Figure 1A. Then, 126 injected zygotes were transferred into the oviducts of two surrogate mice. All surrogates were pregnant to term and gave birth to six live pups. The genomic DNA from each pup was extracted and tested for Krt71 mutations via a PCR, which was then analyzed by Sanger sequencing (Figure 1B). In total, five of the six (83.3%) newborn pups carried a Krt71 mutation (Figure 1C).
- To examine off–target effects in Krt71–KO mice, we predicted sequences similar to sgRNAs through an online website and selected the five most likely potential off–target sites (POTs) before these sites were detected by a PCR and Sanger sequencing. There were no off–target mutations at potential off–target sites (POTs) in Krt71 mutant mice (Supplementary Figure S1A,B). These results demonstrated that mutations in Krt71 could be achieved via the CRISPR/Cas9 system with high efficiency in mice.
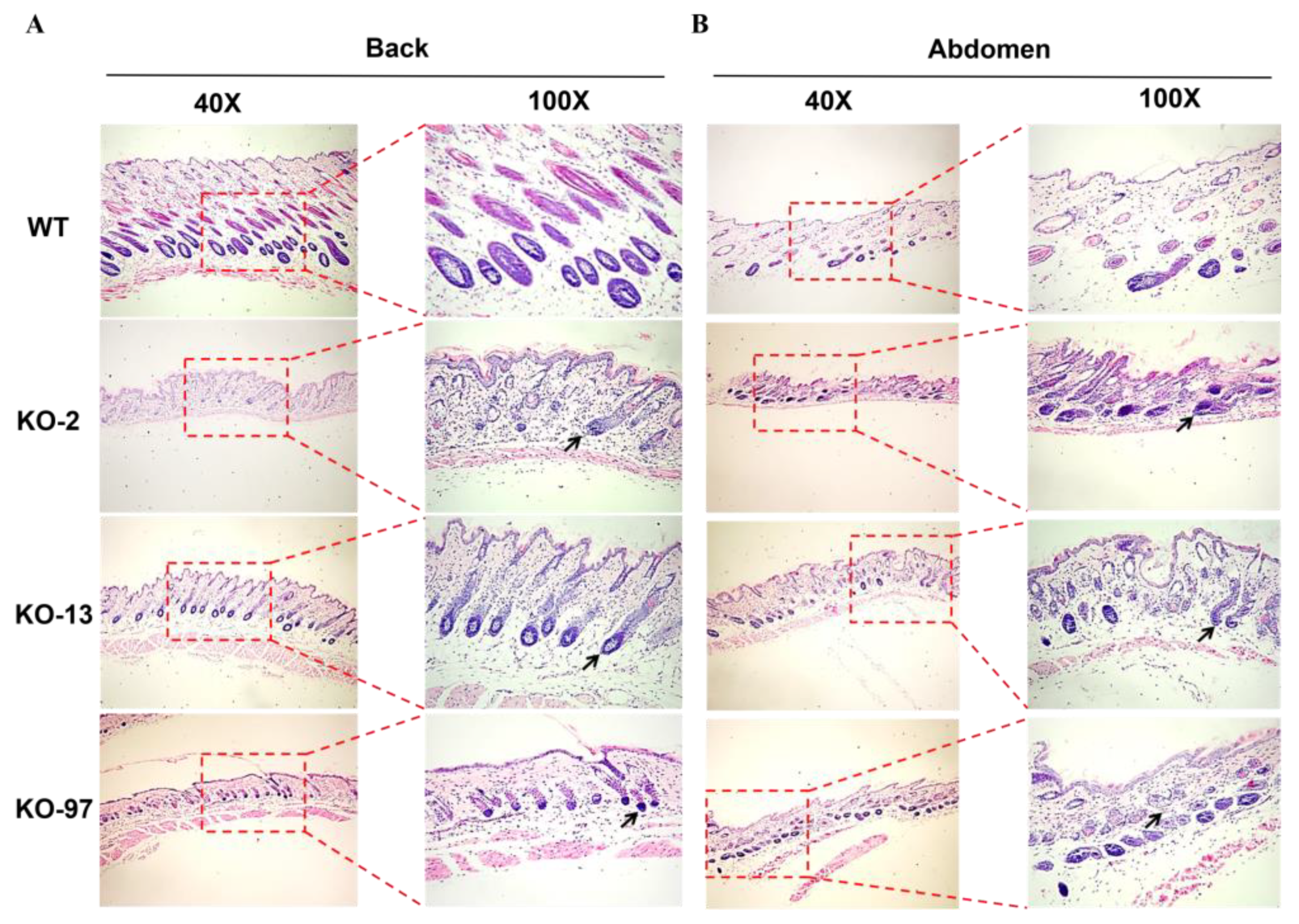
3.2. Phenotype Analysis of Krt71 Knockout Mice
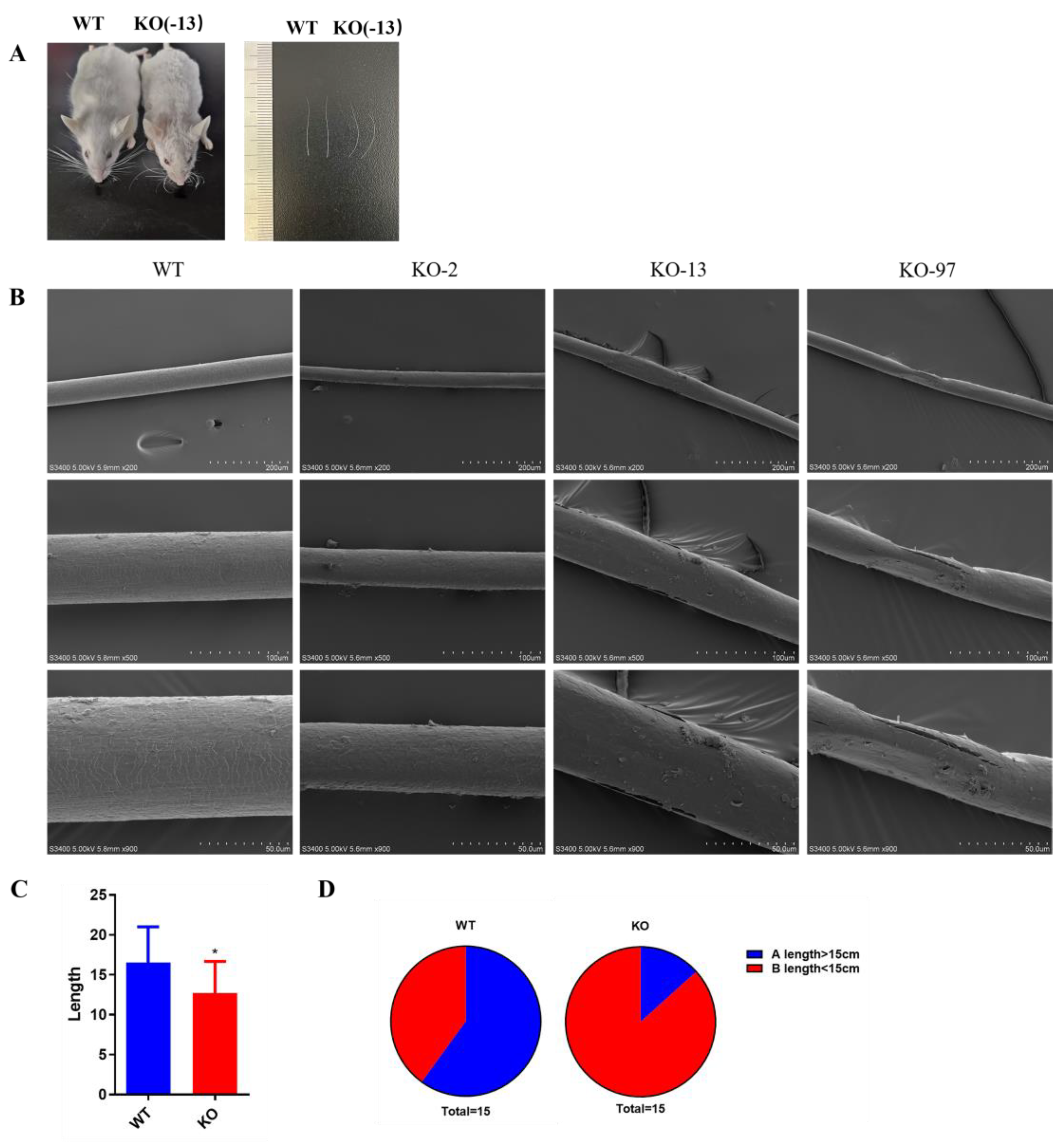
3.3. Structural Changes of Beards in Krt71–KO Mice
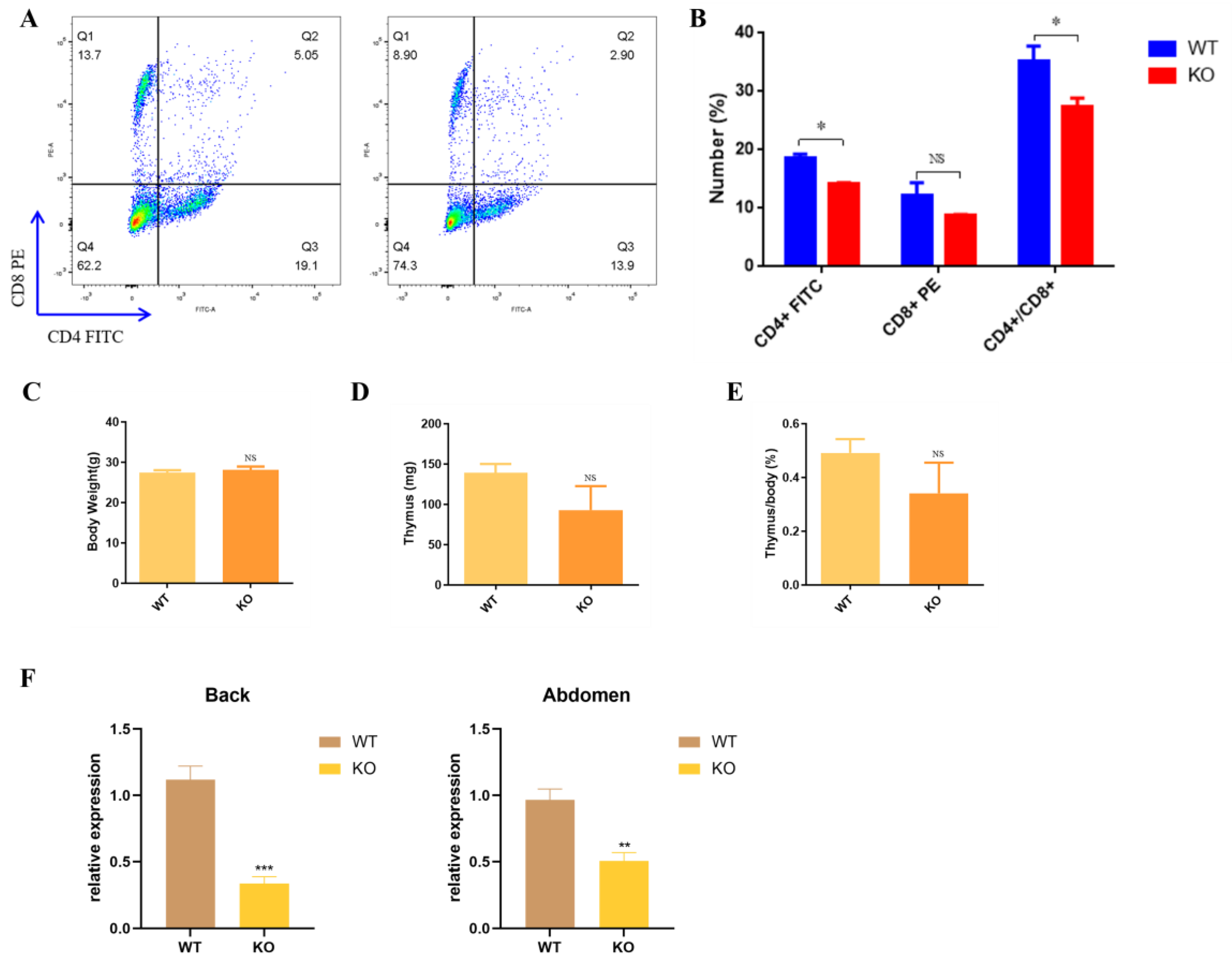
3.4. Nude Appearance of Krt71–KO Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orentreich, N. Disorders of the hair and scalp in childhood. Pediatr. Clin. N. Am. 1971, 18, 953–974. [Google Scholar] [CrossRef] [PubMed]
- Itin, P.H.; Fistarol, S.K. Hair shaft abnormalities–clues to diagnosis and treatment. Dermatology 2005, 211, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Chien, A.J.; Valentine, M.C.; Sybert, V.P. Hereditary woolly hair and keratosis pilaris. J. Am. Acad. Dermatol. 2006, 54, S35–S39. [Google Scholar] [CrossRef] [PubMed]
- Pasternack, S.M.; von Kugelgen, I.; Al Aboud, K.; Lee, Y.A.; Ruschendorf, F.; Voss, K.; Hillmer, A.M.; Molderings, G.J.; Franz, T.; Ramirez, A.; et al. G protein–coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat. Genet. 2008, 40, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Kazantseva, A.; Goltsov, A.; Zinchenko, R.; Grigorenko, A.P.; Abrukova, A.V.; Moliaka, Y.K.; Kirillov, A.G.; Guo, Z.R.; Lyle, S.; Ginter, E.K. Human hair growth deficiency is linked to a genetic defect in the phospholipase gene LIPH. Science 2006, 314, 982–985. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, A.; Farooq, M.; Fujikawa, H.; Inoue, A.; Ohyama, M.; Ehama, R.; Nakanishi, J.; Hagihara, M.; Iwabuchi, T.; Aoki, J.; et al. A Missense Mutation within the Helix Initiation Motif of the Keratin K71 Gene Underlies Autosomal Dominant Woolly Hair/Hypotrichosis. J. Investig. Dermatol. 2012, 132, 2342–2349. [Google Scholar] [CrossRef] [Green Version]
- Shimomura, Y.; Wajid, M.; Petukhova, L.; Kurban, M.; Christiano, A.M. Autosomal–Dominant Woolly Hair Resulting from Disruption of Keratin 74 (KRT74), a Potential Determinant of Human Hair Texture. Am. J. Hum. Genet. 2010, 86, 632–638. [Google Scholar] [CrossRef] [Green Version]
- Betz, R.C.; Cabral, R.M.; Christiano, A.M.; Sprecher, E. Unveiling the Roots of Monogenic Genodermatoses: Genotrichoses as a Paradigm. J. Investig. Dermatol. 2012, 132, 906–914. [Google Scholar] [CrossRef] [Green Version]
- Romano, M.T.; Tafazzoli, A.; Mattern, M.; Sivalingam, S.; Wolf, S.; Rupp, A.; Thiele, H.; Altmuller, J.; Nurnberg, P.; Ellwanger, J.; et al. Bi–allelic Mutations in LSS, Encoding Lanosterol Synthase, Cause Autosomal–Recessive Hypotrichosis Simplex. Am. J. Hum. Genet. 2018, 103, 777–785. [Google Scholar] [CrossRef] [Green Version]
- Coulombe, P.A.; Omary, M.B. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 2002, 14, 110–122. [Google Scholar] [CrossRef]
- Moll, R.; Divo, M.; Langbein, L. The human keratins: Biology and pathology. Histochem. Cell Biol. 2008, 129, 705–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langbein, L.; Rogers, M.A.; Praetzel, S.; Winter, H.; Schweizer, J. K6irs1, K6irs2, K6irs3, and K6irs4 represent the inner–root–sheath–specific type II epithelial keratins of the human hair follicle. J. Investig. Dermatol. 2003, 120, 512–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikkawa, Y.; Oyama, A.; Ishii, R.; Miura, I.; Amano, T.; Ishii, Y.; Yoshikawa, Y.; Masuya, H.; Wakana, S.; Shiroishi, T.; et al. A small deletion hotspot in the type II keratin gene mK6irs1/Krt2–6g on mouse chromosome 15, a candidate for causing the wavy hair of the caracul (Ca) mutation. Genetics 2003, 165, 721–733. [Google Scholar] [CrossRef]
- Runkel, F.; Klaften, M.; Koch, K.; Bohnert, V.; Bussow, H.; Fuchs, H.; Franz, T.; de Angelis, M.H. Morphologic and molecular characterization of two novel Krt71 (Krt2–6g) mutations: Krt71(rco12) and Krt71(rco13). Mamm. Genome 2006, 17, 1172–1182. [Google Scholar] [CrossRef]
- Cadieu, E.; Neff, M.W.; Quignon, P.; Walsh, K.; Chase, K.; Parker, H.G.; VonHoldt, B.M.; Rhue, A.; Boyko, A.; Byers, A.; et al. Coat Variation in the Domestic Dog Is Governed by Variants in Three Genes. Science 2009, 326, 150–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuramoto, T.; Hirano, R.; Kuwamura, M.; Serikawa, T. Identification of the Rat Rex Mutation as a 7–bp Deletion at Splicing Acceptor Site of the Krt71 Gene. J. Vet. Med. Sci. 2010, 72, 909–912. [Google Scholar] [CrossRef] [Green Version]
- Gandolfi, B.; Outerbridge, C.A.; Beresford, L.G.; Myers, J.A.; Pimentel, M.; Alhaddad, H.; Grahn, J.C.; Grahn, R.A.; Lyons, L.A. The naked truth: Sphynx and Devon Rex cat breed mutations in KRT71. Mamm. Genome 2010, 21, 509–515. [Google Scholar] [CrossRef] [Green Version]
- Sui, T.T.; Yuan, L.; Liu, H.; Chen, M.; Deng, J.C.; Wang, Y.; Li, Z.J.; Lai, L.X. CRISPR/Cas9–mediated mutation of PHEX in rabbit recapitulates human X–linked hypophosphatemia (XLH). Hum. Mol. Genet. 2016, 25, 2661–2671. [Google Scholar] [CrossRef] [Green Version]
- Sui, T.T.; Lau, Y.S.; Liu, D.; Liu, T.J.; Xu, L.; Gao, Y.D.; Lai, L.X.; Li, Z.J.; Han, R.Z. A novel rabbit model of Duchenne muscular dystrophy generated by CRISPR/Cas9. Dis. Model. Mech. 2018, 11, dmm032201. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Yang, D.S.; Ruan, J.X.; Zhang, J.F.; Chen, Y.E.; Xu, J. Production of immunodeficient rabbits by multiplex embryo transfer and multiplex gene targeting. Sci. Rep. 2017, 7, 12202. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.C.; Chen, M.; Liu, Z.Q.; Song, Y.N.; Sui, T.T.; Lai, L.X.; Li, Z.J. The disrupted balance between hair follicles and sebaceous glands in Hoxc13–ablated rabbits. FASEB J. 2019, 33, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Combalia, A.; Brugues, A.; Garcia-Veigas, F.J.; Ferrando, J. Scanning Electron Microscopy and X-ray Microanalysis of Reconstructive Hair Fibers. Int. J. Trichology 2017, 9, 54–57. [Google Scholar] [PubMed]
- Barakat, T.S.; Halbritter, F.; Zhang, M.; Rendeiro, A.F.; Perenthaler, E.; Bock, C.; Chambers, I. Functional Dissection of the Enhancer Repertoire in Human Embryonic Stem Cells. Cell Stem Cell 2018, 23, 276–288.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, X.L.; Liu, Y.F.; Zhang, J.B.; Xu, Q.Q.; Liu, C.K.; Fang, M.Y. Characteristics and Expression Profile of KRT71 Screened by Suppression Subtractive Hybridization cDNA Library in Curly Fleece Chinese Tan Sheep. DNA Cell Biol. 2017, 36, 552–564. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Meng, M.; Wang, H. A Novel Prognostic Biomarker LPAR6 in Hepatocellular Carcinoma via Associating with Immune Infiltrates. J. Clin. Transl. Hepatol. 2022, 10, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Hoenerhoff, M.; Yang, D.S.; Yang, Y.; Deng, C.; Wen, L.A.; Ma, L.Y.; Pallas, B.; Zhao, C.Z.; Koike, Y.; et al. Development of the Nude Rabbit Model. Stem Cell Rep. 2021, 16, 656–665. [Google Scholar] [CrossRef]
- Kur-Piotrowska, A.; Kopcewicz, M.; Kozak, L.P.; Sachadyn, P.; Grabowska, A.; Gawronska-Kozak, B. Neotenic phenomenon in gene expression in the skin of Foxn1–deficient (nude) mice—A projection for regenerative skin wound healing. BMC Genom. 2017, 18, 56. [Google Scholar] [CrossRef] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Yao, H.; Wang, H.; Sui, T. Development of Woolly Hair and Hairlessness in a CRISPR−Engineered Mutant Mouse Model with KRT71 Mutations. Cells 2023, 12, 1781. https://doi.org/10.3390/cells12131781
Zhang T, Yao H, Wang H, Sui T. Development of Woolly Hair and Hairlessness in a CRISPR−Engineered Mutant Mouse Model with KRT71 Mutations. Cells. 2023; 12(13):1781. https://doi.org/10.3390/cells12131781
Chicago/Turabian StyleZhang, Tao, Hongwu Yao, Hejun Wang, and Tingting Sui. 2023. "Development of Woolly Hair and Hairlessness in a CRISPR−Engineered Mutant Mouse Model with KRT71 Mutations" Cells 12, no. 13: 1781. https://doi.org/10.3390/cells12131781
APA StyleZhang, T., Yao, H., Wang, H., & Sui, T. (2023). Development of Woolly Hair and Hairlessness in a CRISPR−Engineered Mutant Mouse Model with KRT71 Mutations. Cells, 12(13), 1781. https://doi.org/10.3390/cells12131781




