SHIP1 Is Present but Strongly Downregulated in T-ALL, and after Restoration Suppresses Leukemia Growth in a T-ALL Xenotransplantation Mouse Model
Abstract
1. Introduction
2. Material and Methods
2.1. Patient Material
2.2. Construction of Plasmids
2.3. Lentiviral Transduction
2.4. Cell Culture
2.5. Jurkat Tet-On Cells
Microarray Analysis
2.6. Gene Expression and Bioinformatic Analysis
2.7. Western Blot Analysis
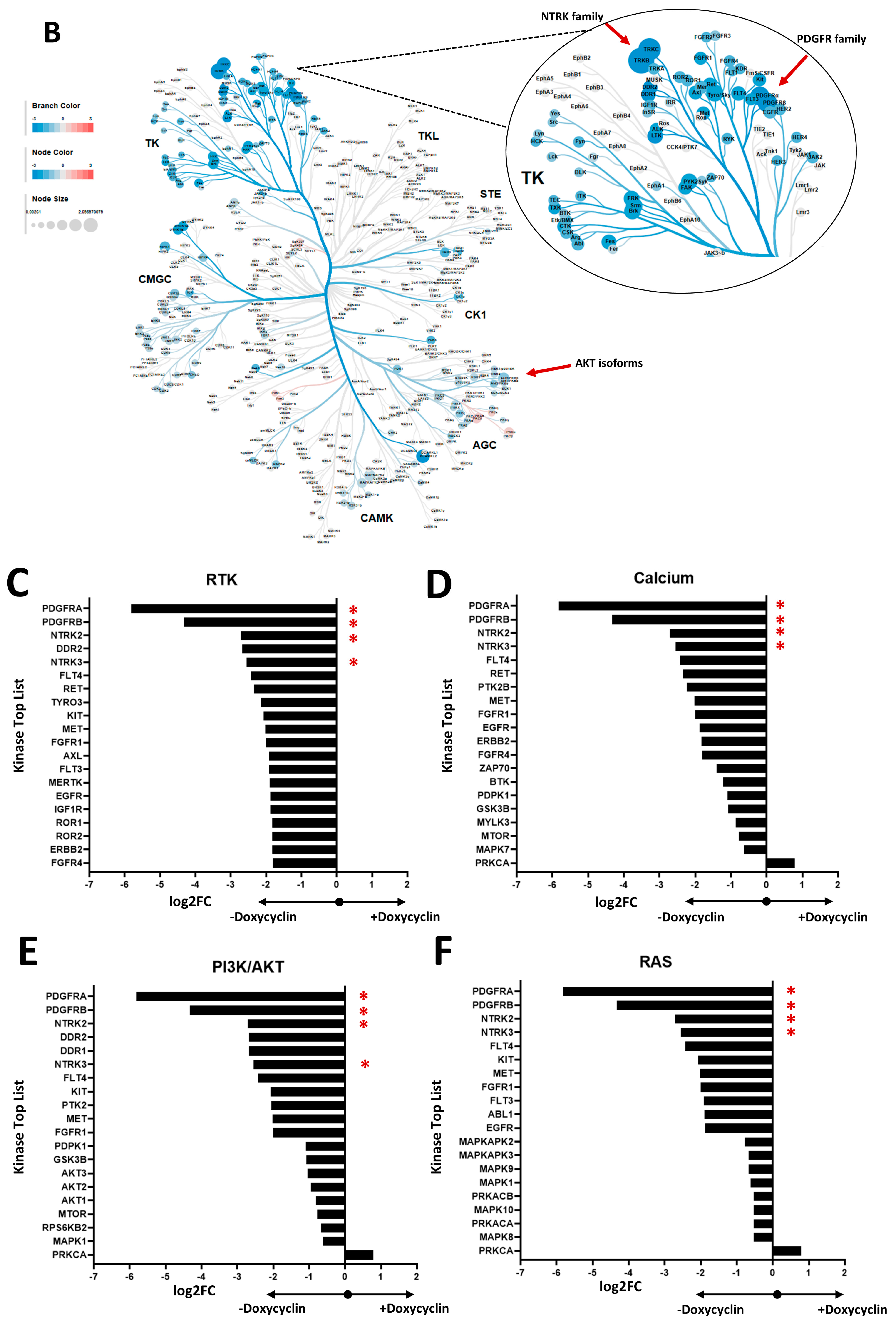
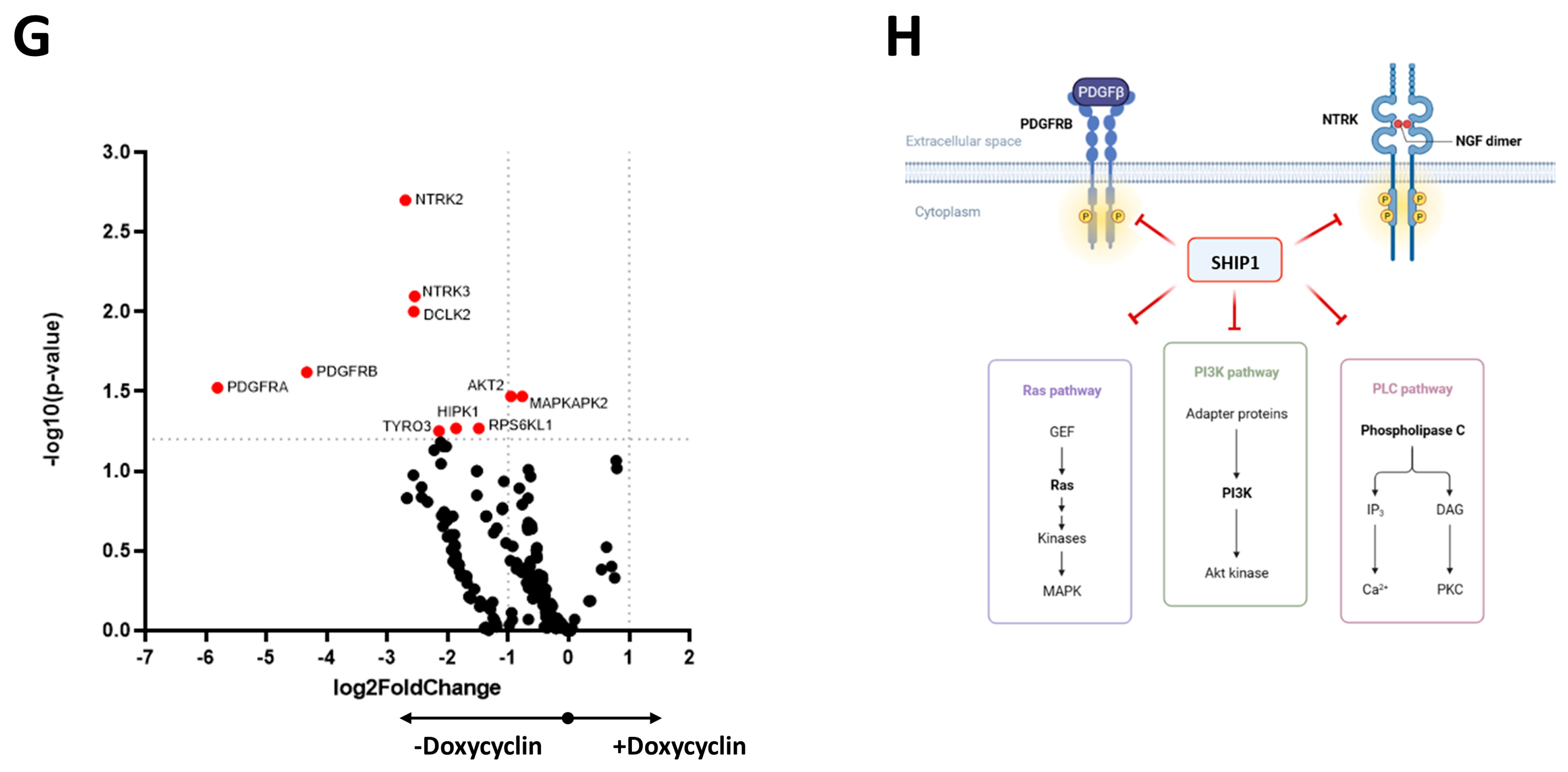
2.8. Functional Kinome Profiling
2.9. RNA Isolation, cDNA Synthesis and Real-Time Quantitative PCR (qPCR)
2.10. Flow Cytometry
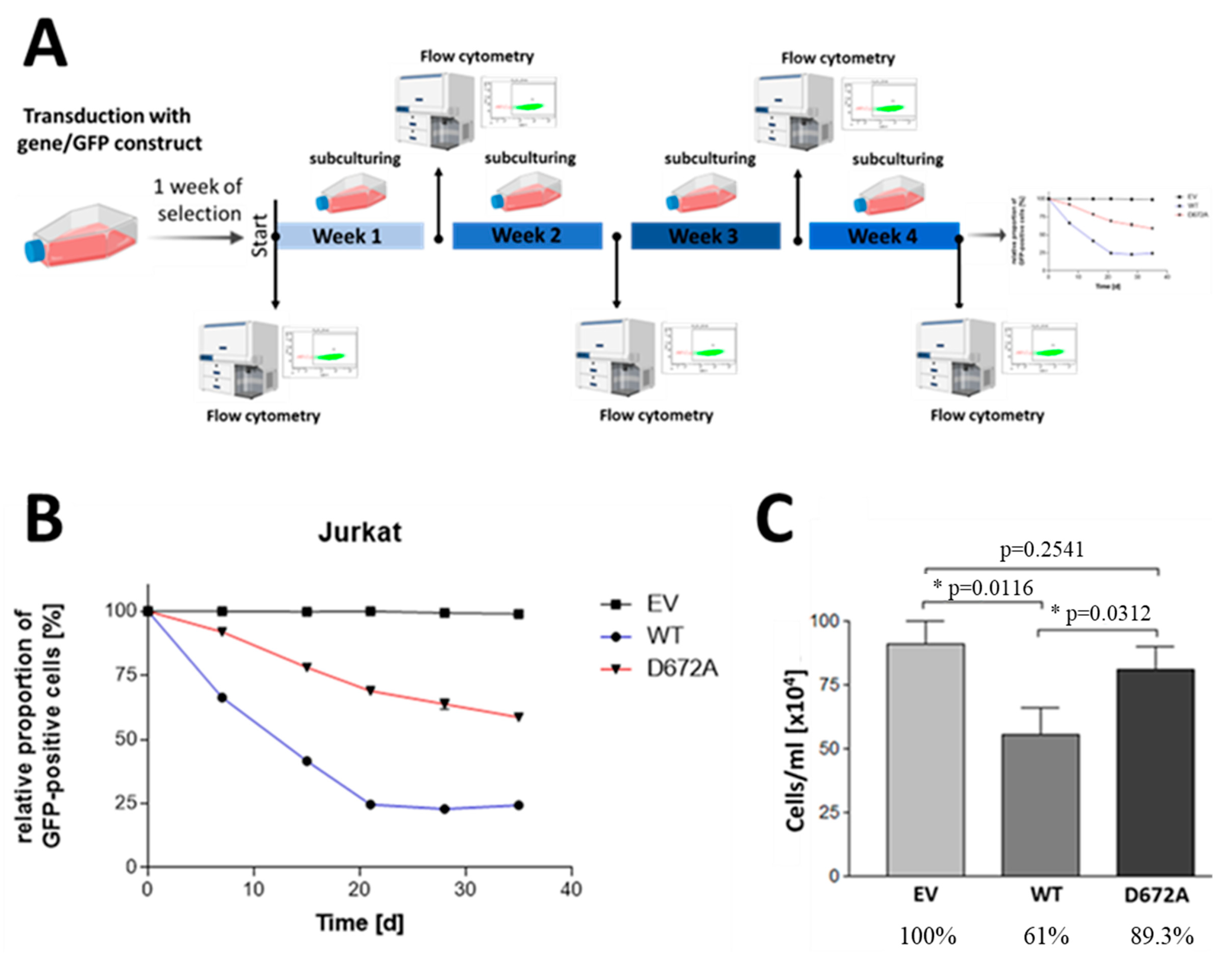
2.11. Determination of Cell Growth
2.12. Live-Cell Imaging
2.13. Animal Experiment
2.14. Statistical Analysis
3. Results
3.1. Expression Analysis of SHIP1 in Leukemia Cells of T-ALL Patients
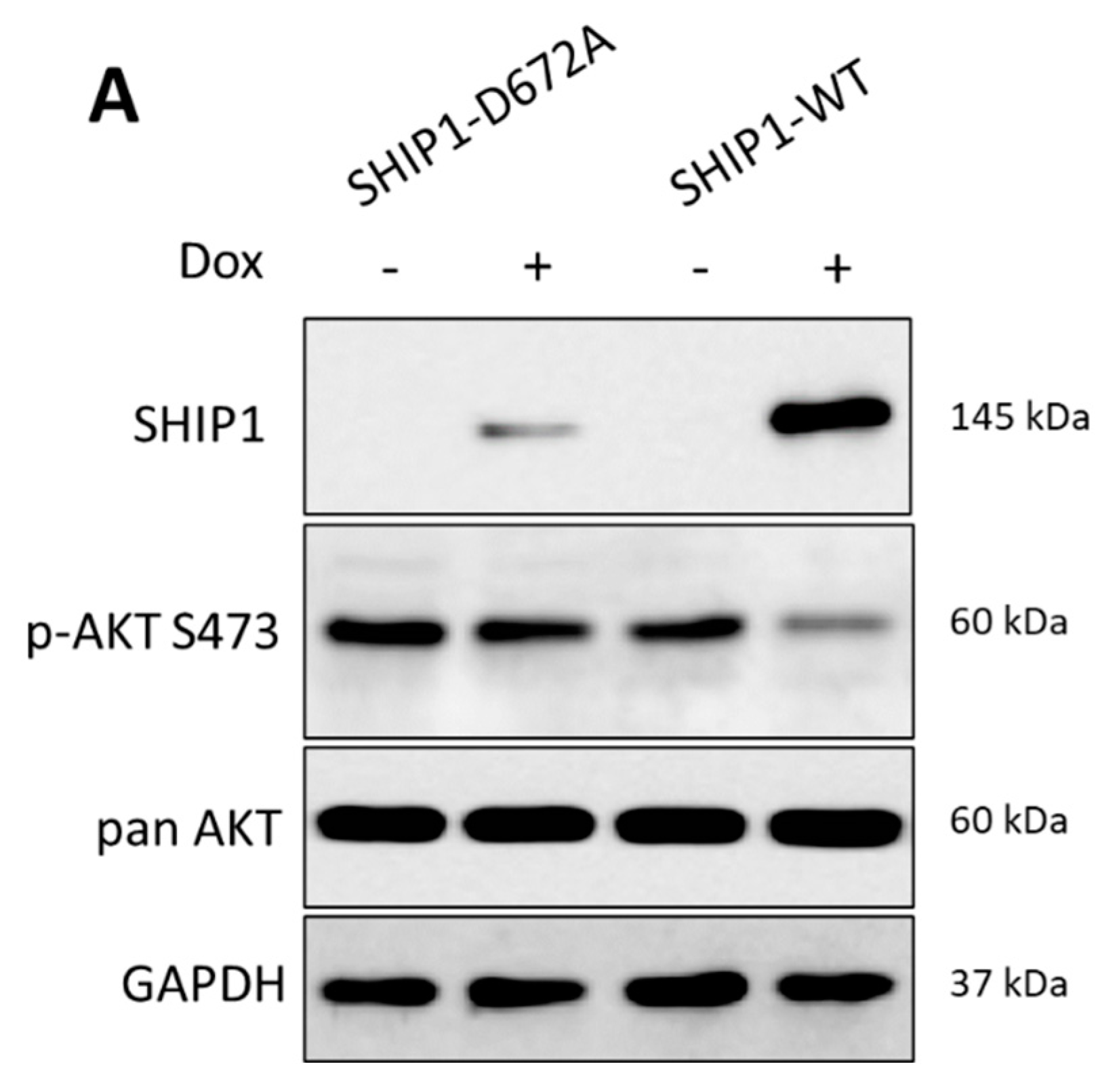
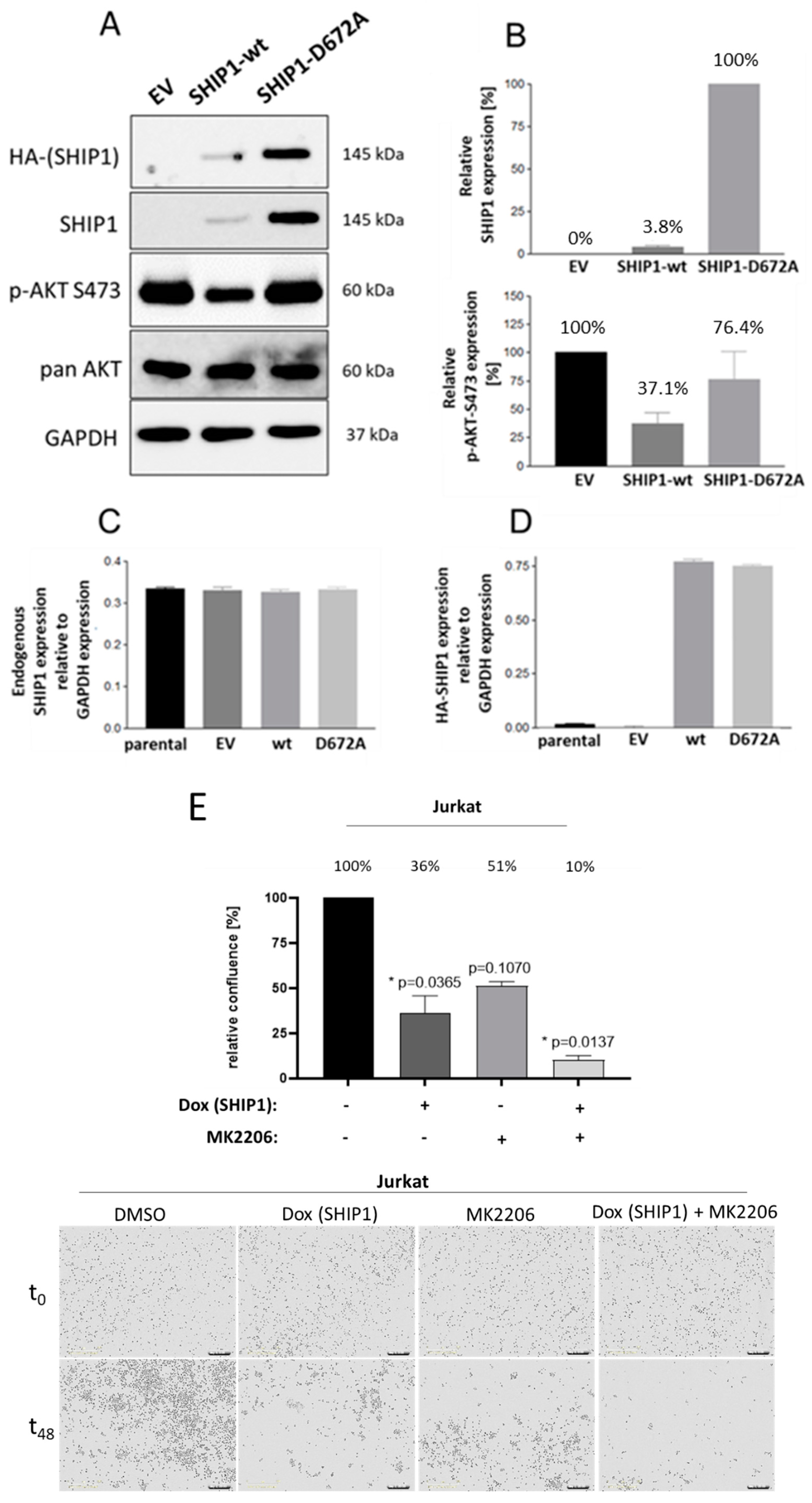
3.2. Investigation of the Reduced SHIP1 Expression in the T-ALL Cell Line Jurkat
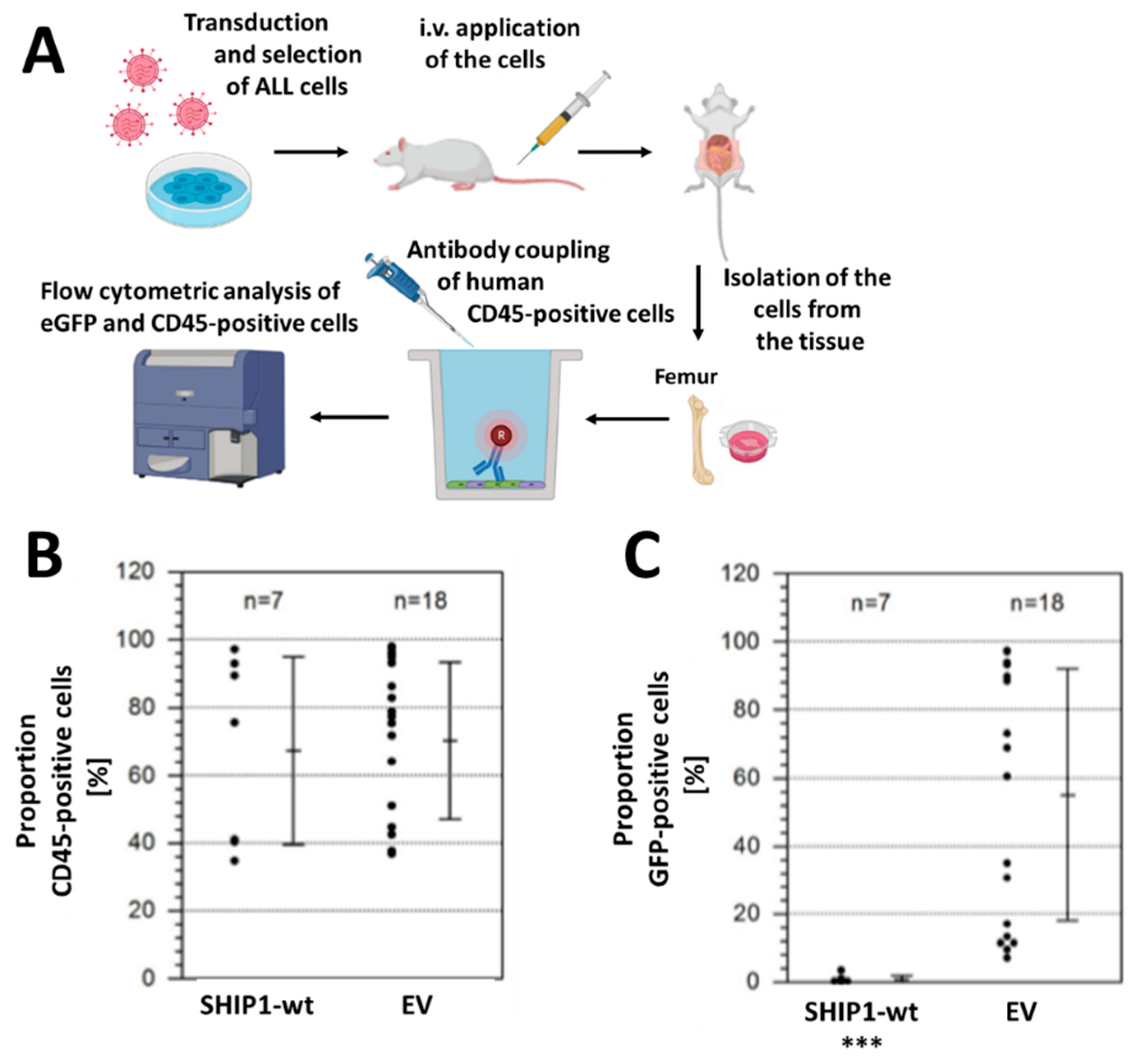
3.3. Xenotransplantation Model of the Consequences of Reconstitution of SHIP1 Expression in T-ALL Cells
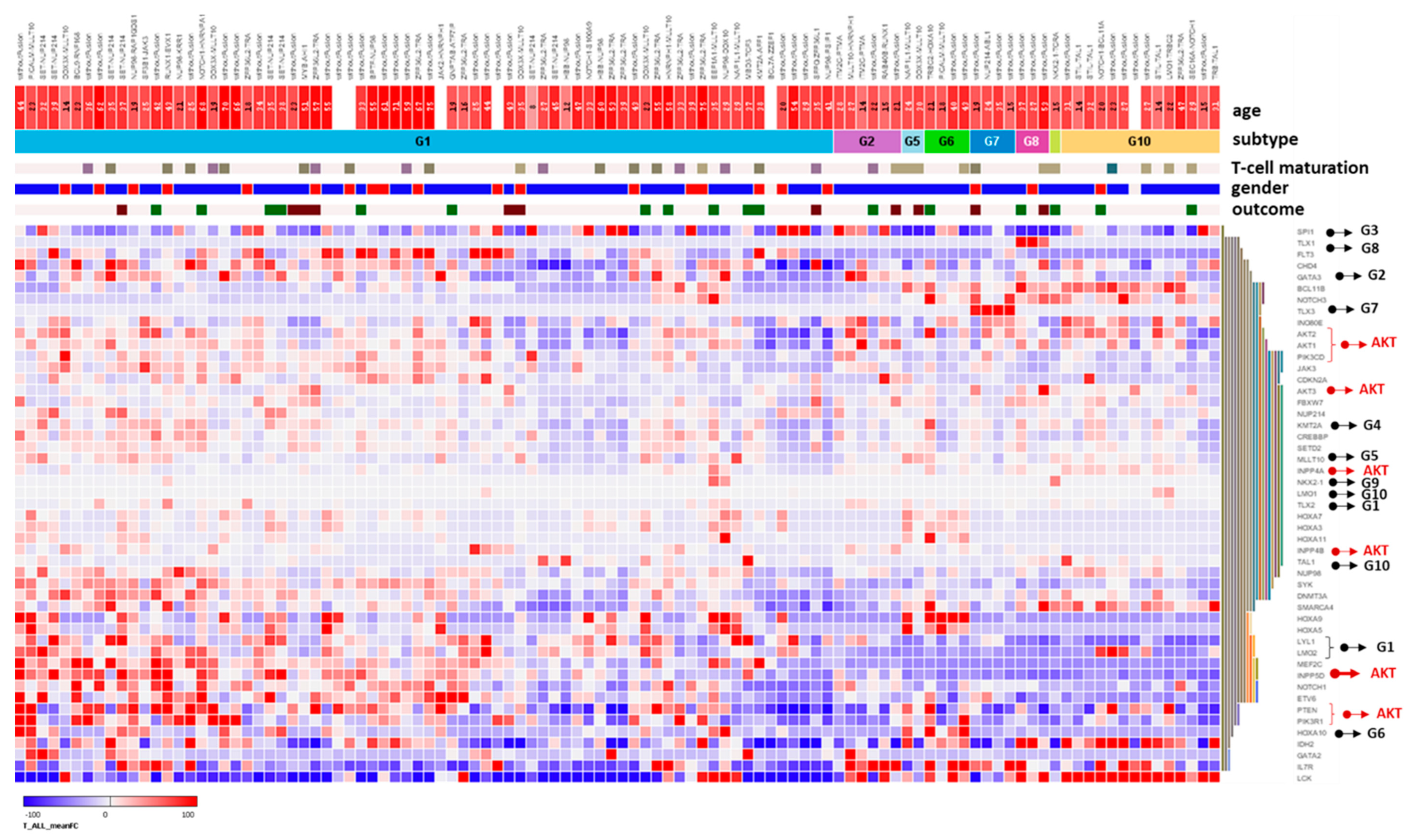
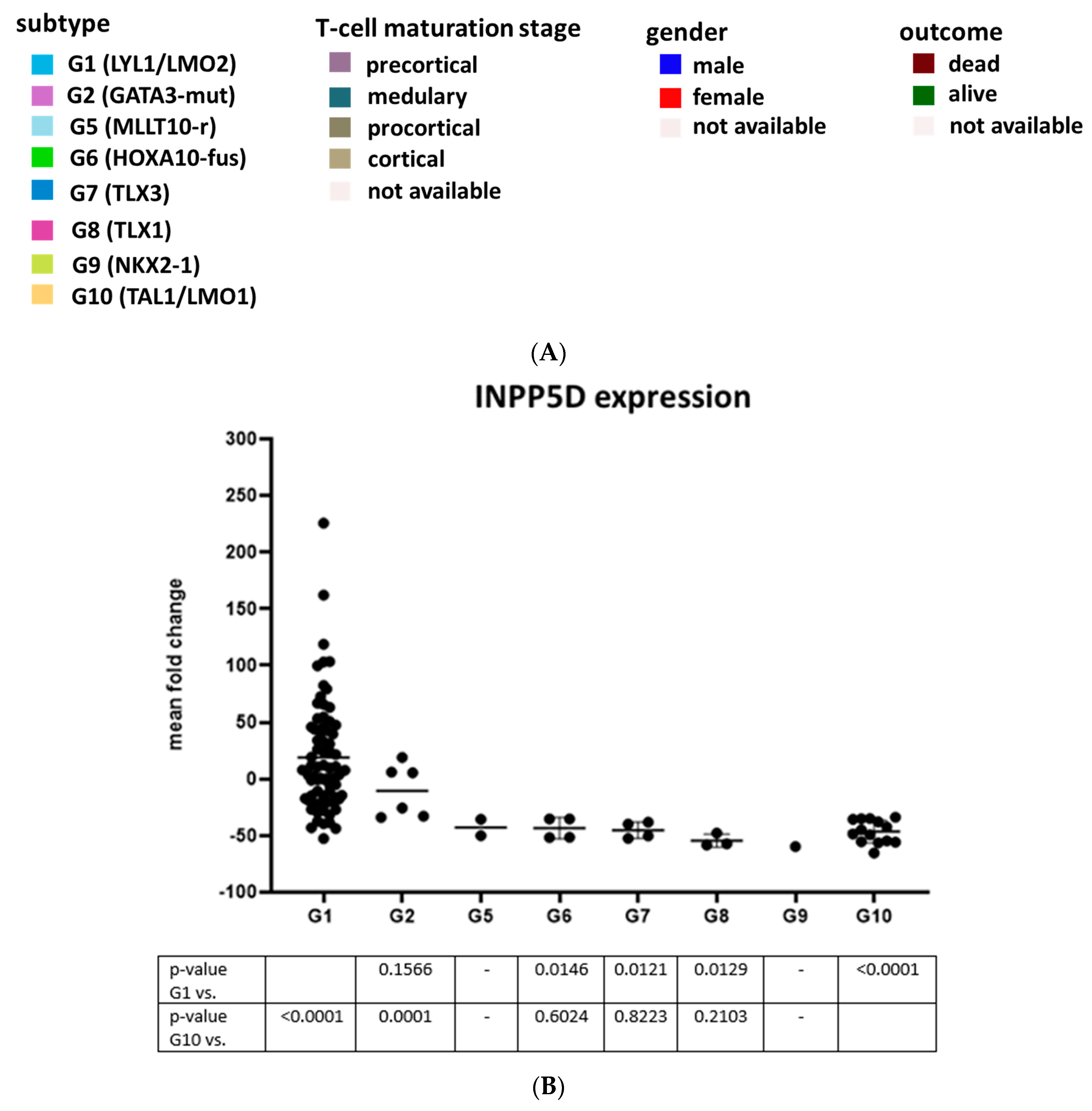
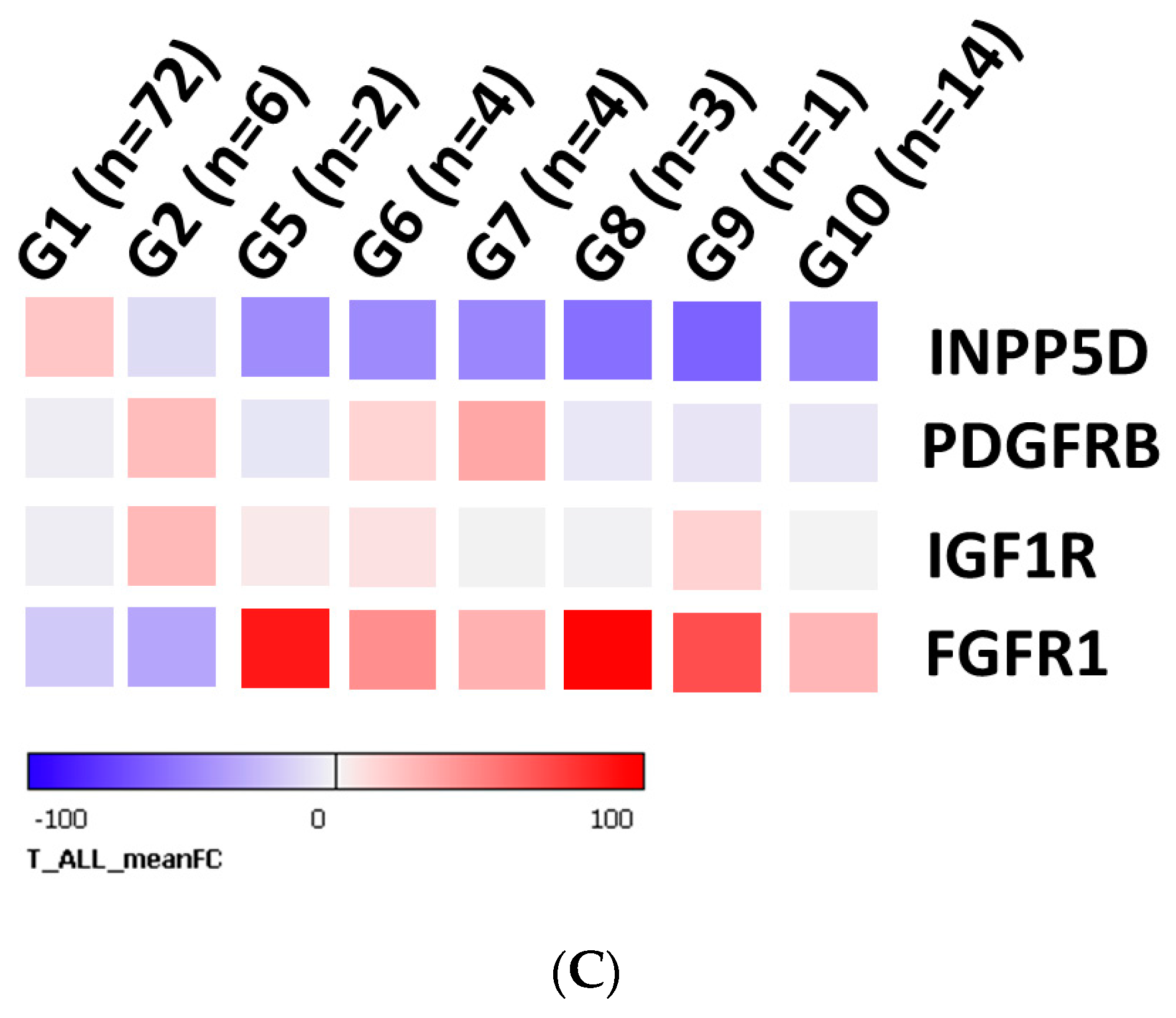
3.4. Gene Expression Analysis of INPP5D and Molecular Classification of INPP5D in T-ALL Subgroups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kaatsch, P.; Grabow, D.; Spix, C. German Childhood Cancer Registry—Annual Report 2018 (1980–2017); Institute of Medical Biostatistics, Epidemiology and Informatics (IMBEI) at the University Medical Center of the Johannes Gutenberg University: Mainz, Germany, 2019. [Google Scholar]
- Pui, C.H.; Robison, L.L.; Look, A.T. Acute lymphoblastic leukaemia. Lancet 2008, 371, 1030–1043. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef]
- Damen, J.E.; Liu, L.; Rosten, P.; Humphries, R.K.; Jefferson, A.B.; Majerus, P.W.; Krystal, G. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc. Natl. Acad. Sci. USA 1996, 93, 1689–1693. [Google Scholar] [CrossRef] [PubMed]
- Drayer, A.L.; Pesesse, X.; De Smedt, F.; Woscholski, R.; Parker, P.; Erneux, C. Cloning and expression of a human placenta inositol 1,3,4,5-tetrakisphosphate and phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase. Biochem. Biophys. Res. Commun. 1996, 225, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Ware, M.D.; Rosten, P.; Damen, J.E.; Liu, L.; Humphries, R.K.; Krystal, G. Cloning and characterization of human SHIP, the 145-kD inositol 5-phosphatase that associates with SHC after cytokine stimulation. Blood 1996, 88, 2833–2840. [Google Scholar] [CrossRef] [PubMed]
- Odai, H.; Sasaki, K.; Iwamatsu, A.; Nakamoto, T.; Ueno, H.; Yamagata, T.; Mitani, K.; Yazaki, Y.; Hirai, H. Purification and molecular cloning of SH2- and SH3-containing inositol polyphosphate-5-phosphatase, which is involved in the signaling pathway of granulocyte-macrophage colony-stimulating factor, erythropoietin, and Bcr-Abl. Blood 1997, 89, 2745–2756. [Google Scholar] [CrossRef]
- Silva, A.; Yunes, J.A.; Cardoso, B.A.; Martins, L.R.; Jotta, P.Y.; Abecasis, M.; Nowill, A.E.; Leslie, N.R.; Cardoso, A.A.; Barata, J.T. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J. Clin. Investig. 2008, 118, 3762–3774. [Google Scholar] [CrossRef]
- Morishita, N.; Tsukahara, H.; Chayama, K.; Ishida, T.; Washio, K.; Miyamura, T.; Yamashita, N.; Oda, M.; Morishima, T. Activation of Akt is associated with poor prognosis and chemotherapeutic resistance in pediatric B-precursor acute lymphoblastic leukemia. Pediatr. Blood Cancer 2012, 59, 83–89. [Google Scholar] [CrossRef]
- Martelli, A.M.; Chiarini, F.; Evangelisti, C.; Cappellini, A.; Buontempo, F.; Bressanin, D.; Fini, M.; McCubrey, J.A. Two hits are better than one: Targeting both phosphatidylinositol 3-kinase and mammalian target of rapamycin as a therapeutic strategy for acute leukemia treatment. Oncotarget 2012, 3, 371–394. [Google Scholar] [CrossRef]
- Horn, S.; Endl, E.; Fehse, B.; Weck, M.M.; Mayr, G.W.; Jücker, M. Restoration of SHIP activity in a human leukemia cell line downregulates constitutively activated phosphatidylinositol 3-kinase/Akt/GSK-3beta signaling and leads to an increased transit time through the G1 phase of the cell cycle. Leukemia 2004, 18, 1839–1849. [Google Scholar] [CrossRef]
- Blackburn, J.S.; Liu, S.; Wilder, J.L.; Dobrinski, K.P.; Lobbardi, R.; Moore, F.E.; Martinez, S.A.; Chen, E.Y.; Lee, C.; Langenau, D.M. Clonal evolution enhances leukemia propagating cell frequency in T-cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell 2014, 25, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Fransecky, L.; Mochmann, L.H.; Baldus, C.D. Outlook on PI3K/AKT/mTOR inhibition in acute leukemia. Mol. Cell. Ther. 2015, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Girardi, T.; Vicente, C.; Cools, J.; De Keersmaecker, K. The genetics and molecular biology of T-ALL. Blood 2017, 129, 1113–1123. [Google Scholar] [CrossRef]
- Palomero, T.; Sulis, M.L.; Cortina, M.; Real, P.J.; Barnes, K.; Ciofani, M.; Caparros, E.; Buteau, J.; Brown, K.; Perkins, S.L.; et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 2007, 13, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Bhamra, G.; Bamford, S.; Dawson, E.; Kok, C.; Clements, J.; Menzies, A.; Teague, J.W.; Futreal, P.A.; Stratton, M.R. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr. Protoc. Hum. Genet. 2008. [Google Scholar] [CrossRef]
- Luo, J.M.; Liu, Z.L.; Hao, H.L.; Wang, F.X.; Dong, Z.R.; Ohno, R. Mutation analysis of SHIP gene in acute leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2004, 12, 420–426. [Google Scholar]
- Zhang, S.J.; Shi, J.Y.; Zhu, Y.M.; Shi, Z.Z.; Yan-Sheng Gu, B.W.; Bai, X.T.; Shen, Z.X.; Li, J.Y. The investigation of mutation and single nucleotide polymorphism of receptor tyrosine kinases and downstream scaffold molecules in acute myeloid leukemia. Leuk. Lymphoma 2006, 47, 2610–2616. [Google Scholar] [CrossRef]
- Gilby, D.C.; Goodeve, A.C.; Winship, P.R.; Valk, P.J.; Delwel, R.; Reilly, J.T. Gene structure, expression profiling and mutation analysis of the tumour suppressor SHIP1 in Caucasian acute myeloid leukaemia. Leukemia 2007, 21, 2390–2393. [Google Scholar] [CrossRef]
- Brauer, H.; Strauss, J.; Wegner, W.; Müller-Tidow, C.; Horstmann, M.; Jücker, M. Leukemia-associated mutations in SHIP1 inhibit its enzymatic activity, interaction with the GM-CSF receptor and Grb2, and its ability to inactivate PI3K/AKT signaling. Cell Signal. 2012, 24, 2095–2101. [Google Scholar] [CrossRef]
- Ehm, P.; Nelson, N.; Giehler, S.; Schaks, M.; Bettin, B.; Kirchmair, J.; Jücker, M. Reduced expression and activity of patient-derived SHIP1 phosphatase domain mutants. Cell Signal. 2023, 101, 110485. [Google Scholar] [CrossRef]
- Hamilton, M.J.; Ho, V.W.; Kuroda, E.; Ruschmann, J.; Antignano, F.; Lam, V.; Krystal, G. Role of SHIP in cancer. Exp. Hematol. 2011, 39, 2–13. [Google Scholar] [CrossRef]
- Helgason, C.D.; Damen, J.E.; Rosten, P.; Grewal, R.; Sorensen, P.; Chappel, S.M.; Borowski, A.; Jirik, F.; Krystal, G.; Humphries, R.K. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998, 12, 1610–1620. [Google Scholar] [CrossRef]
- Liu, Q.; Sasaki, T.; Kozieradzki, I.; Wakeham, A.; Itie, A.; Dumont, D.J.; Penninger, J.M. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev. 1999, 13, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.; Verma, S.; Byrne, C.H.; Shrikhande, G.; Winkler, T.; Algate, P.A.; Rohrschneider, L.R.; Griffin, J.D. BCR/ABL directly inhibits expression of SHIP, an SH2-containing polyinositol-5-phosphatase involved in the regulation of hematopoiesis. Mol. Cell. Biol. 1999, 19, 7473–7480. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.M.; Otero, D.; Kao, E.; Miletic, A.V.; Hother, C.; Ralfkiaer, E.; Rickert, R.C.; Gronbaek, K.; David, M. Onco-miR-155 targets SHIP1 to promote TNFα-dependent growth of B cell lymphomas. EMBO Mol. Med. 2009, 1, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Miletic, A.V.; Anzelon-Mills, A.N.; Mills, D.M.; Omori, S.A.; Pedersen, I.M.; Shin, D.M.; Ravetch, J.V.; Bolland, S.; Morse, H.C., 3rd; Rickert, R.C. Coordinate suppression of B cell lymphoma by PTEN and SHIP phosphatases. J. Exp. Med. 2010, 207, 2407–2420. [Google Scholar] [CrossRef] [PubMed]
- Lo, T.C.; Barnhill, L.M.; Kim, Y.; Nakae, E.A.; Yu, A.L.; Diccianni, M.B. Inactivation of SHIP1 in T-cell acute lymphoblastic leukemia due to mutation and extensive alternative splicing. Leuk. Res. 2009, 33, 1562–1566. [Google Scholar] [CrossRef]
- Freeburn, R.W.; Wright, K.L.; Burgess, S.J.; Astoul, E.; Cantrell, D.A.; Ward, S.G. Evidence that SHIP-1 contributes to phosphatidylinositol 3,4,5-trisphosphate metabolism in T lymphocytes and can regulate novel phosphoinositide 3-kinase effectors. J. Immunol. 2002, 169, 5441–5450. [Google Scholar] [CrossRef]
- Harms, D.O.; Janka-Schaub, G.E. Co-operative study group for childhood acute lymphoblastic leukemia (COALL): Long-term follow-up of trials 82, 85, 89 and 92. Leukemia 2000, 14, 2234–2239. [Google Scholar] [CrossRef]
- Escherich, G.; Horstmann, M.A.; Zimmermann, M.; Janka-Schaub, G.E. Cooperative study group for childhood acute lymphoblastic leukaemia (COALL): Long-term results of trials 82, 85, 89, 92 and 97. Leukemia 2010, 24, 298–308. [Google Scholar] [CrossRef]
- Ehm, P.; Nalaskowski, M.M.; Wundenberg, T.; Jücker, M. The tumor suppressor SHIP1 colocalizes in nucleolar cavities with p53 and components of PML nuclear bodies. Nucleus 2015, 6, 154–164. [Google Scholar] [CrossRef]
- Wang, W.; Malcolm, B.A. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques 1999, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Metzner, A.; Precht, C.; Fehse, B.; Fiedler, W.; Stocking, C.; Günther, A.; Mayr, G.W.; Jücker, M. Reduced proliferation of CD34(+) cells from patients with acute myeloid leukemia after gene transfer of, I.N.P.P.5.D. Gene Ther. 2009, 16, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Schneider, U.; Schwenk, H.U.; Bornkamm, G. Characterization of EBV-genome negative, “null”; “T” cell lines derived from children with acute lymphoblastic leukemia leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer 1977, 19, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Siekmann, I.K.; Dierck, K.; Prall, S.; Klokow, M.; Strauss, J.; Buhs, S.; Wrzeszcz, A.; Bockmayr, M.; Beck, F.; Trochimiuk, M.; et al. Combined inhibition of receptor tyrosine and p21-activated kinases as a therapeutic strategy in childhood ALL. Blood Adv. 2018, 9, 2554–2567. [Google Scholar] [CrossRef]
- Dai, Y.T.; Zhang, F.; Fang, H.; Li, J.F.; Lu, G.; Jiang, L.; Chen, B.; Mao, D.D.; Liu, Y.F.; Wang, J.; et al. Transcriptome-wide subtyping of pediatric and adult T cell acute lymphoblastic leukemia in an international study of 707 cases. PNAS 2022, 119, e2120787119. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Ehm, P.A.H.; Lange, F.; Hentschel, C.; Jepsen, A.; Glück, M.; Nelson, N.; Bettin, B.; de Bruyn Kops, C.; Kirchmair, J.; Nalaskowski, M.; et al. Analysis of the FLVR motif of SHIP1 and its importance for the protein stability of SH2 containing signaling proteins. Cell Signal. 2019, 63, 109380. [Google Scholar] [CrossRef]
- Bußmann, L.; Hoffer, K.; von Bargen, C.M.; Droste, C.; Lange, T.; Kemmling, J.; Schröder-Schwarz, J.; Vu, A.T.; Akingunsade, L.; Nollau, P.; et al. Analyzing tyrosine kinase activity in head and neck cancer by functional kinomics: Identification of hyperactivated Src family kinases as prognostic markers and potential targets. Int. J. Cancer 2021, 149, 1166–1180, Erratum in Int. J. Cancer 2022, 150, E6. [Google Scholar] [CrossRef]
- Hinz, N.; Baranowsky, A.; Horn, M.; Kriegs, M.; Sibbertsen, F.; Smit, D.J.; Clezardin, P.; Lange, T.; Schinke, T.; Jücker, M. Knockdown of AKT3 Activates HER2 and DDR Kinases in Bone-Seeking Breast Cancer Cells, Promotes Metastasis In Vivo and Attenuates the TGFβ/CTGF Axis. Cells 2021, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biol. 2010, 8, e1000412. [Google Scholar] [CrossRef] [PubMed]
- Metz, K.S.; Deoudes, E.M.; Berginski, M.E.; Jimenez-Ruiz, I.; Aksoy, B.A.; Hammerbacher, J.; Gomez, S.M.; Phanstiel, D.H. Coral: Clear and Customizable Visualization of Human Kinome Data. Cell Syst. 2018, 7, 347–350.e1. [Google Scholar] [CrossRef]
- Damen, J.E.; Ware, M.D.; Kalesnikoff, J.; Hughes, M.R.; Krystal, G. SHIP’s C-terminus is essential for its hydrolysis of PIP3 and inhibition of mast cell degranulation. Blood 2001, 97, 1343–1351. [Google Scholar] [CrossRef]
- Soria, J.C.; Lee, H.Y.; Lee, J.I.; Wang, L.; Issa, J.P.; Kemp, B.L.; Liu, D.D.; Kurie, J.M.; Mao, L.; Khuri, F.R. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin. Cancer Res. 2002, 8, 1178–1184. [Google Scholar]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Jotta, P.Y.; Ganazza, M.A.; Silva, A.; Viana, M.B.; da Silva, M.J.; Zambaldi, L.J.; Barata, J.T.; Brandalise, S.R.; Yunes, J.A. Negative prognostic impact of PTEN mutation in pediatric T-cell acute lymphoblastic leukemia. Leukemia 2010, 24, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Diccianni, M.B.; Barnhill, L.M.; Lo, T.; Yu, A.L. SHIP1 and PTEN Co-Inactivation and Akt Pathway Targeting in ALL. Blood 2009, 114, 2386. [Google Scholar] [CrossRef]
- Correia, N.C.; Gírio, A.; Antunes, I.; Martins, L.R.; Barata, J.T. The multiple layers of non-genetic regulation of PTEN tumour suppressor activity. Eur. J. Cancer 2014, 50, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.M.; Soares, M.V.; Ribeiro, P.; Caldas, J.; Póvoa, V.; Martins, L.R.; Melão, A.; Serra-Caetano, A.; de Sousa, A.B.; Lacerda, J.F.; et al. Adult B-cell acute lymphoblastic leukemia cells display decreased PTEN activity and constitutive hyperactivation of PI3K/Akt pathway despite high PTEN protein levels. Haematologica 2014, 99, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Arevalo, M.A.; Rodríguez-Tebar, A. Activation of casein kinase II and inhibition of phosphatase and tensin homologue deleted on chromosome 10 phosphatase by nerve growth factor/p75NTR inhibit glycogen synthase kinase-3beta and stimulate axonal growth. Mol. Biol. Cell 2006, 17, 3369–3377. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, A.; Alimonti, A.; Pandolfi, P.P. PTEN level in tumor suppression: How much is too little? Cancer Res. 2011, 71, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Shojaee, S.; Chan, L.N.; Buchner, M.; Cazzaniga, V.; Cosgun, K.N.; Geng, H.; Qiu, Y.H.; von Minden, M.D.; Ernst, T.; Hochhaus, A.; et al. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat. Med. 2016, 22, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Lasky, J.L.; Chang, C.J.; Mosessian, S.; Lewis, X.; Xiao, Y.; Yeh, J.E.; Chen, J.Y.; Iruela-Arispe, M.L.; Varella-Garcia, M.; et al. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature 2008, 453, 529–533. [Google Scholar] [CrossRef]
- Täger, M.; Horn, S.; Latuske, E.; Ehm, P.; Schaks, M.; Nalaskowski, M.; Fehse, B.; Fiedler, W.; Stocking, C.; Wellbrock, J.; et al. SHIP1, but not an AML-derived SHIP1 mutant, suppresses myeloid leukemia growth in a xenotransplantation mouse model. Gene Ther. 2017, 24, 749–753. [Google Scholar] [CrossRef]
- Bose, S.; Deininger, M.; Gora-Tybor, J.; Goldman, J.M.; Melo, J.V. The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: Biologic significance and implications for the assessment of minimal residual disease. Blood 1998, 92, 3362–3367. [Google Scholar] [CrossRef]
- Ehm, P.; Bettin, B.; Jücker, M. Activated Src kinases downstream of BCR-ABL and Flt3 induces proteasomal degradation of SHIP1 by phosphorylation of tyrosine 1021. Biochim. Biophys. Acta Mol. Cell Res. 2023, 1870, 119467. [Google Scholar] [CrossRef]
- Glück, M.; Dally, L.; Jücker, M.; Ehm, P. JAK2-V617F is a negative regulation factor of SHIP1 protein and thus influences the AKT signaling pathway in patients with Myeloproliferative neoplasm (MPN). Int. J. Biochem. Cell Biol. 2022, 149, 106229. [Google Scholar] [CrossRef]
- Cribbs, A.P.; Kennedy, A.; Gregory, B.; Brennan, F.M. Simplified production and concentration of lentiviral vectors to achieve high transduction in primary human T cells. BMC Biotechnol. 2013, 13, 98. [Google Scholar] [CrossRef]
- Selvaraj, N.; Kedage, V.; Hollenhorst, P.C. Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell Commun. Signal. 2015, 13, 12. [Google Scholar] [CrossRef]
- De Val, S.; Anderson, J.P.; Heidt, A.B.; Khiem, D.; Xu, S.M.; Black, B.L. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev. Biol. 2004, 275, 424–434. [Google Scholar] [CrossRef]
- Hansen, D.V.; Hanson, J.E.; Sheng, M. Microglia in Alzheimer’s disease. J. Cell Biol. 2018, 217, 459–472. [Google Scholar] [CrossRef]
- Canté-Barrett, K.; Meijer, M.T.; Hagelaar, R.; Yang, W.; Yu, J.; Smits, W.K.; Nulle, M.E.; Jansen, J.P.; Pieters, R.; Yang, J.J.; et al. MEF2C opposes Notch in lymphoid lineage decision and drives leukemia in the thymus. JCI Insight 2022, 7, e150363. [Google Scholar] [CrossRef] [PubMed]
- Kong, N.R.; Davis, M.; Chai, L.; Winoto, A.; Tjian, R. MEF2C and EBF1 Co-regulate B Cell-Specific Transcription. PLoS Genet. 2016, 12, e1005845. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Guo, H.; Wang, D.; Tu, R.; Qing, G.; Liu, H. Genome-Wide Analysis Identifies Rag1 and Rag2 as Novel Notch1 Transcriptional Targets in Thymocytes. Front. Cell Dev. Biol. 2021, 9, 703338. [Google Scholar] [CrossRef]
- Smeets, M.F.; Chan, A.C.; Dagger, S.; Bradley, C.K.; Wei, A.; Izon, D.J. Fli-1 overexpression in hematopoietic progenitors deregulates T cell development and induces pre-T cell lymphoblastic leukaemia/lymphoma. PLoS ONE 2013, 8, e62346. [Google Scholar] [CrossRef] [PubMed]
- Lakhanpal, G.K.; Vecchiarelli-Federico, L.M.; Li, Y.J.; Cui, J.W.; Bailey, M.L.; Spaner, D.E.; Dumont, D.J.; Barber, D.L.; Ben-David, Y. The inositol phosphatase SHIP-1 is negatively regulated by Fli-1 and its loss accelerates leukemogenesis. Blood 2010, 116, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sanda, T.; Look, A.T.; Novina, C.D.; von Boehmer, H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J. Exp. Med. 2011, 208, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.R.; Sanda, T.; Lawton, L.N.; Li, X.; Kreslavsky, T.; Novina, C.D.; Brand, M.; Gutierrez, A.; Kelliher, M.A.; Jamieson, C.H.; et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J. Exp. Med. 2013, 210, 1545–1557. [Google Scholar] [CrossRef]
- Mavrakis, K.J.; Van Der Meulen, J.; Wolfe, A.L.; Liu, X.; Mets, E.; Taghon, T.; Khan, A.A.; Setty, M.; Rondou, P.; Vandenberghe, P.; et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat. Genet. 2011, 43, 673–678. [Google Scholar] [CrossRef]
- Espinosa, L.; Cathelin, S.; D’Altri, T.; Trimarchi, T.; Statnikov, A.; Guiu, J.; Rodilla, V.; Inglés-Esteve, J.; Nomdedeu, J.; Bellosillo, B.; et al. The Notch/Hes1 pathway sustains NF-κB activation through CYLD repression in T cell leukemia. Cancer Cell 2010, 18, 268–281. [Google Scholar] [CrossRef]
- Rai, D.; Karanti, S.; Jung, I.; Dahia, P.L.; Aguiar, R.C. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet. Cytogenet. 2008, 181, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Gerloff, D.; Grundler, R.; Wurm, A.A.; Bräuer-Hartmann, D.; Katzerke, C.; Hartmann, J.U.; Madan, V.; Müller-Tidow, C.; Duyster, J.; Tenen, D.G.; et al. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia 2015, 29, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Sulis, M.L.; Williams, O.; Palomero, T.; Tosello, V.; Pallikuppam, S.; Real, P.J.; Barnes, K.; Zuurbier, L.; Meijerink, J.P.; Ferrando, A.A. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood 2008, 112, 733–740. [Google Scholar] [CrossRef]
- Boomer, J.S.; Green, J.M. An enigmatic tail of CD28 signaling. Cold Spring Harb. Perspect. Biol. 2010, 2, a002436. [Google Scholar] [CrossRef] [PubMed]
- Sade, H.; Krishna, S.; Sarin, A. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J. Biol. Chem. 2004, 279, 2937–2944. [Google Scholar] [CrossRef] [PubMed]
- Stenton, G.R.; Mackenzie, L.F.; Tam, P.; Cross, J.L.; Harwig, C.; Raymond, J.; Toews, J.; Wu, J.; Ogden, N.; MacRury, T.; et al. Characterization of AQX-1125, a small-molecule SHIP1 activator: Part 1. Effects on inflammatory cell activation and chemotaxis in vitro and pharmacokinetic characterization in vivo. Br. J. Pharmacol. 2013, 168, 1506–1518. [Google Scholar] [CrossRef] [PubMed]
- De Coninck, S.; De Smedt, R.; Lintermans, B.; Goossens, S.; Van Vlierberghe, P. PDGFR-β As a New Therapeutic Target in T-Cell Acute Lymphoblastic Leukemia. Blood 2022, 140, 11513–11514. [Google Scholar] [CrossRef]
- Wiedmann, M.; Wang, X.; Tang, X.; Han, M.; Li, M.; Mao, Z. PI3K/Akt-dependent regulation of the transcription factor myocyte enhancer factor-2 in insulin-like growth factor-1- and membrane depolarization-mediated survival of cerebellar granule neurons. J. Neurosci. Res. 2005, 81, 226–234. [Google Scholar] [CrossRef]
- Ehm, P.; Grottke, A.; Bettin, B.; Jücker, M. Investigation of the function of the PI3-Kinase/AKT signaling pathway for leukemogenesis and therapy of acute childhood lymphoblastic leukemia (ALL). Cell Signal. 2022, 93, 110301. [Google Scholar] [CrossRef]
- Pauls, S.D.; Marshall, A.J. Regulation of immune cell signaling by SHIP1: A phosphatase, scaffold protein, and potential therapeutic target. Eur. J. Immunol. 2017, 47, 932–945. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehm, P.; Rietow, R.; Wegner, W.; Bußmann, L.; Kriegs, M.; Dierck, K.; Horn, S.; Streichert, T.; Horstmann, M.; Jücker, M. SHIP1 Is Present but Strongly Downregulated in T-ALL, and after Restoration Suppresses Leukemia Growth in a T-ALL Xenotransplantation Mouse Model. Cells 2023, 12, 1798. https://doi.org/10.3390/cells12131798
Ehm P, Rietow R, Wegner W, Bußmann L, Kriegs M, Dierck K, Horn S, Streichert T, Horstmann M, Jücker M. SHIP1 Is Present but Strongly Downregulated in T-ALL, and after Restoration Suppresses Leukemia Growth in a T-ALL Xenotransplantation Mouse Model. Cells. 2023; 12(13):1798. https://doi.org/10.3390/cells12131798
Chicago/Turabian StyleEhm, Patrick, Ruth Rietow, Wiebke Wegner, Lara Bußmann, Malte Kriegs, Kevin Dierck, Stefan Horn, Thomas Streichert, Martin Horstmann, and Manfred Jücker. 2023. "SHIP1 Is Present but Strongly Downregulated in T-ALL, and after Restoration Suppresses Leukemia Growth in a T-ALL Xenotransplantation Mouse Model" Cells 12, no. 13: 1798. https://doi.org/10.3390/cells12131798
APA StyleEhm, P., Rietow, R., Wegner, W., Bußmann, L., Kriegs, M., Dierck, K., Horn, S., Streichert, T., Horstmann, M., & Jücker, M. (2023). SHIP1 Is Present but Strongly Downregulated in T-ALL, and after Restoration Suppresses Leukemia Growth in a T-ALL Xenotransplantation Mouse Model. Cells, 12(13), 1798. https://doi.org/10.3390/cells12131798





