Single-Cell RNA Sequencing: Opportunities and Challenges for Studies on Corneal Biology in Health and Disease
Abstract
:1. Introduction
2. Collection of Single-Cell Corneal RNA Sequencing Studies
3. Classical and Novel Marker Genes Defining Cell States in the Cornea
3.1. The Limbus and Limbal Epithelium
3.2. The Central Corneal Epithelium
3.3. The Conjunctiva
3.4. The Corneal Stroma
3.5. The Corneal Endothelium
4. Single-Cell RNA Sequencing Applied to Human Corneal Organoids
5. Opportunities and Challenges of Using Single-Cell RNA Sequencing
5.1. Corneal Biology in Health
5.2. Corneal Biology in Disease
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sridhar, M.S. Anatomy of cornea and ocular surface. Indian J. Ophthalmol. 2018, 66, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Spadea, L.; Maraone, G.; Verboschi, F.; Vingolo, E.M.; Tognetto, D. Effect of corneal light scatter on vision: A review of the literature. Int. J. Ophthalmol. 2016, 9, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Reins, R.Y.; McDermott, A.M. Aldehyde dehydrogenase (ALDH) 3A1 expression by the human keratocyte and its repair phenotypes. Exp. Eye Res. 2006, 83, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.E. Bowman’s layer in the cornea– structure and function and regeneration. Exp. Eye Res. 2020, 195, 108033. [Google Scholar] [CrossRef]
- Torricelli, A.A.M.; Singh, V.; Santhiago, M.R.; Wilson, S.E. The Corneal Epithelial Basement Membrane: Structure, Function, and Disease. Investig. Opthalmology Vis. Sci. 2013, 54, 6390–6400. [Google Scholar] [CrossRef]
- Yazdanpanah, G.; Jabbehdari, S.; Djalilian, A.R. Limbal and corneal epithelial homeostasis. Curr. Opin. Ophthalmol. 2017, 28, 348–354. [Google Scholar] [CrossRef]
- Nuzzi, A.; Giuffrida, F.P.; Luccarelli, S.; Nucci, P. Corneal Epithelial Regeneration: Old and New Perspectives. Int. J. Mol. Sci. 2022, 23, 13114. [Google Scholar] [CrossRef]
- Majo, F.; Rochat, A.; Nicolas, M.; Jaoudé, G.A.; Barrandon, Y. Oligopotent stem cells are distributed throughout the mammalian ocular surface. Nature 2008, 456, 250–254. [Google Scholar] [CrossRef]
- Amitai-Lange, A.; Altshuler, A.; Bubley, J.; Dbayat, N.; Tiosano, B.; Shalom-Feuerstein, R. Lineage Tracing of Stem and Progenitor Cells of the Murine Corneal Epithelium. Stem Cells Dayt. Ohio. 2014, 33, 230–239. [Google Scholar] [CrossRef]
- Dorà, N.J.; Hill, R.E.; Collinson, J.M.; West, J.D. Lineage tracing in the adult mouse corneal epithelium supports the limbal epithelial stem cell hypothesis with intermittent periods of stem cell quiescence. Stem Cell Res. 2015, 15, 665–677. [Google Scholar] [CrossRef] [Green Version]
- Di Girolamo, N.; Bobba, S.; Raviraj, V.; Delic, N.C.; Slapetova, I.; Nicovich, P.R.; Halliday, G.M.; Wakefield, D.; Whan, R.; Lyons, J.G. Tracing the Fate of Limbal Epithelial Progenitor Cells in the Murine Cornea. Stem Cells 2014, 33, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, S.P.; Zieske, J.D. ZO1 in Corneal Epithelium: Association to the Zonula Occludens and Adherens Junctions. Exp. Eye Res. 1997, 64, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Dota, A.; Cooper, L.J.; Fullwood, N.J.; Nakamura, T.; Tsuzuki, M.; Mochida, C.; Kinoshita, S. Tight junction-related protein expression and distribution in human corneal epithelium. Exp. Eye Res. 2003, 76, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Walker, H.; Akula, M.; West-Mays, J.A. Corneal development: Role of the periocular mesenchyme and bi-directional signaling. Exp. Eye Res. 2020, 201, 108231. [Google Scholar] [CrossRef] [PubMed]
- Tidu, A.; Schanne-Klein, M.-C.; Borderie, V.M. Development, structure, and bioengineering of the human corneal stroma: A review of collagen-based implants. Exp. Eye Res. 2020, 200, 108256. [Google Scholar] [CrossRef]
- Reneker, L.W.; Silversides, D.W.; Xu, L.; Overbeek, P.A. Formation of corneal endothelium is essential for anterior segment development—A transgenic mouse model of anterior segment dysgenesis. Dev. Camb. Engl. 2000, 127, 533–542. [Google Scholar] [CrossRef]
- Wolosin, J.M.; Budak, M.T.; Akinci, M.M. Ocular surface epithelial and stem cell development. Int. J. Dev. Biol. 2004, 48, 981–991. [Google Scholar] [CrossRef] [Green Version]
- Graw, J. Eye Development. Curr. Top Dev. Biol. 2010, 90, 343–386. [Google Scholar] [CrossRef]
- Dhouailly, D.; Pearton, D.J.; Michon, F. The vertebrate corneal epithelium: From early specification to constant renewal. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2014, 243, 1226–1241. [Google Scholar] [CrossRef]
- Lwigale, P.Y. Corneal Development: Different Cells from a Common Progenitor. Prog. Mol. Biol. Transl. Sci. 2015, 134, 43–59. [Google Scholar] [CrossRef]
- Williams, A.L.; Bohnsack, B.L. Neural crest derivatives in ocular development: Discerning the eye of the storm. Birth Defects Res. Part C Embryo Today Rev. 2015, 105, 87–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miesfeld, J.B.; Brown, N.L. Eye organogenesis: A hierarchical view of ocular development. Curr. Top Dev. Biol. 2019, 132, 351–393. [Google Scholar] [CrossRef] [PubMed]
- Grocott, T.; Johnson, S.; Bailey, A.P.; Streit, A. Neural crest cells organize the eye via TGF-β and canonical Wnt signalling. Nat. Commun. 2011, 2, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Rheaume, B.A.; Jereen, A.; Bolisetty, M.; Sajid, M.S.; Yang, Y.; Renna, K.; Sun, L.; Robson, P.; Trakhtenberg, E.F. Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun. 2018, 9, 2759. [Google Scholar] [CrossRef] [Green Version]
- Tran, N.M.; Shekhar, K.; Whitney, I.E.; Jacobi, A.; Benhar, I.; Hong, G.; Yan, W.; Adiconis, X.; Arnold, M.E.; Lee, J.M.; et al. Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron 2019, 104, 1039–1055.e12. [Google Scholar] [CrossRef]
- Shiau, F.; Ruzycki, P.A.; Clark, B.S. A single-cell guide to retinal development: Cell fate decisions of multipotent retinal progenitors in scRNA-seq. Dev. Biol. 2021, 478, 41–58. [Google Scholar] [CrossRef]
- van Zyl, T.; Yan, W.; McAdams, A.M.; Monavarfeshani, A.; Hageman, G.S.; Sanes, J.R. Cell atlas of the human ocular anterior segment: Tissue-specific and shared cell types. Proc. Natl. Acad. Sci. USA 2022, 119, e2200914119. [Google Scholar] [CrossRef]
- Gautam, P.; Hamashima, K.; Chen, Y.; Zeng, Y.; Bar Makovoz, B.; Parikh, B.H.; Lee, H.Y.; Lau, K.A.; Su, X.; Wong, R.C.B.; et al. Multi-species single-cell transcriptomic analysis of ocular compartment regulons. Nat. Commun. 2021, 12, 5675. [Google Scholar] [CrossRef]
- Collin, J.; Queen, R.; Zerti, D.; Bojic, S.; Dorgau, B.; Moyse, N.; Molina, M.M.; Yang, C.; Dey, S.; Reynolds, G.; et al. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf. 2021, 21, 279–298. [Google Scholar] [CrossRef]
- Català, P.; Groen, N.; Dehnen, J.A.; Soares, E.; van Velthoven, A.J.H.; Nuijts, R.M.M.A.; Dickman, M.M.; LaPointe, V.L.S. Single cell transcriptomics reveals the heterogeneity of the human cornea to identify novel markers of the limbus and stroma. Sci. Rep. 2021, 11, 21727. [Google Scholar] [CrossRef] [PubMed]
- Ligocki, A.J.; Fury, W.; Gutierrez, C.; Adler, C.; Yang, T.; Ni, M.; Bai, Y.; Wei, Y.; Lehmann, G.L.; Romano, C. Molecular characteristics and spatial distribution of adult human corneal cell subtypes. Sci. Rep. 2021, 11, 16323. [Google Scholar] [CrossRef] [PubMed]
- Maiti, G.; de Barros, M.R.M.; Hu, N.; Dolgalev, I.; Roshan, M.; Foster, J.W.; Tsirigos, A.; Wahlin, K.J.; Chakravarti, S. Single cell RNA-seq of human cornea organoids identifies cell fates of a developing immature cornea. PNAS Nexus 2022, 1, pgac246. [Google Scholar] [CrossRef] [PubMed]
- Dou, S.; Wang, Q.; Bin Zhang, B.; Wei, C.; Wang, H.; Liu, T.; Duan, H.; Jiang, H.; Liu, M.; Qi, X.; et al. Single-cell atlas of keratoconus corneas revealed aberrant transcriptional signatures and implicated mechanical stretch as a trigger for keratoconus pathogenesis. Cell Discov. 2022, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-Q.; Kim, S.; Li, J.-M.; Gao, Q.; Choi, J.; Bian, F.; Hu, J.; Zhang, Y.; Lu, R.; Li, Y.; et al. Single-cell transcriptomics identifies limbal stem cell population and cell types mapping its differentiation trajectory in limbal basal epithelium of human cornea. Ocul. Surf. 2021, 20, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, N.; Wang, J.; Wray, B.; Patel, P.; Yang, W.; Peng, H.; Lavker, R.M. Single-Cell RNA Transcriptome Helps Define the Limbal/Corneal Epithelial Stem/Early Transit Amplifying Cells and How Autophagy Affects This Population. Investig. Opthalmology Vis. Sci. 2019, 60, 3570–3583. [Google Scholar] [CrossRef] [Green Version]
- Sonam, S.; Bangru, S.; Perry, K.J.; Chembazhi, U.V.; Kalsotra, A.; Henry, J.J. Cellular and molecular profiles of larval and adult Xenopus corneal epithelia resolved at the single-cell level. Dev. Biol. 2022, 491, 13–30. [Google Scholar] [CrossRef]
- Lin, J.B.; Shen, X.; Pfeifer, C.W.; Shiau, F.; Santeford, A.; Ruzycki, P.A.; Clark, B.S.; Liu, Q.; Huang, A.J.W.; Apte, R.S. Dry eye disease in mice activates adaptive corneal epithelial regeneration distinct from constitutive renewal in homeostasis. Proc. Natl. Acad. Sci. USA 2023, 120, e2204134120. [Google Scholar] [CrossRef]
- Stumpf, P.S.; Du, X.; Imanishi, H.; Kunisaki, Y.; Semba, Y.; Noble, T.; Smith, R.C.G.; Rose-Zerili, M.; West, J.J.; Oreffo, R.O.C.; et al. Transfer learning efficiently maps bone marrow cell types from mouse to human using single-cell RNA sequencing. Commun. Biol. 2020, 3, 736. [Google Scholar] [CrossRef]
- Bakken, T.E.; van Velthoven, C.T.; Menon, V.; Hodge, R.D.; Yao, Z.; Nguyen, T.N.; Graybuck, L.T.; Horwitz, G.D.; Bertagnolli, D.; Goldy, J.; et al. Single-cell and single-nucleus RNA-seq uncovers shared and distinct axes of variation in dorsal LGN neurons in mice, non-human primates, and humans. eLife 2021, 10, e64875. [Google Scholar] [CrossRef]
- Gao, S.; Wu, Z.; Kannan, J.; Mathews, L.; Feng, X.; Kajigaya, S.; Young, N.S. Comparative Transcriptomic Analysis of the Hematopoietic System between Human and Mouse by Single Cell RNA Sequencing. Cells 2021, 10, 973. [Google Scholar] [CrossRef] [PubMed]
- Chromium Single Cell V(D)J Reagent Kits with Feature Barcoding Technology for Cell Surface Protein, Document Number CG000186 Rev A, 10× Genomics, (25 July 2019). n.d. Available online: https://support.10xgenomics.com/permalink/user-guide-chromium-single-cell-5-reagent-kits-user-guide-v2-chemistry-dual-index-with-feature-barcoding-technology-for-cell-surface-protein-and-immune-receptor-mapping (accessed on 2 June 2023).
- Yu, L.; Cao, Y.; Yang, J.Y.H.; Yang, P. Benchmarking clustering algorithms on estimating the number of cell types from single-cell RNA sequencing data. Genome Biol. 2022, 23, 49. [Google Scholar] [CrossRef] [PubMed]
- Duò, A.; Robinson, M.D.; Soneson, C. A systematic performance evaluation of clustering methods for single-cell RNA-seq data. F1000Research 2020, 7, 1141. [Google Scholar] [CrossRef] [PubMed]
- Wiegleb, G.; Reinhardt, S.; Dahl, A.; Posnien, N. Tissue dissociation for single-cell and single-nuclei RNA sequencing for low amounts of input material. Front. Zool. 2022, 19, 27. [Google Scholar] [CrossRef]
- Patterson-Cross, R.B.; Levine, A.J.; Menon, V. Selecting single cell clustering parameter values using subsampling-based robustness metrics. BMC Bioinform. 2021, 22, 39. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [Green Version]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2014, 43, D789–D798. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S.; Shimmura, S.; Kawakita, T.; Miyashita, H.; Den, S.; Shimazaki, J.; Tsubota, K. Cytokeratin 15 Can Be Used to Identify the Limbal Phenotype in Normal and Diseased Ocular Surfaces. Investig. Opthalmology Vis. Sci. 2006, 47, 4780–4786. [Google Scholar] [CrossRef] [Green Version]
- Nasser, W.; Amitai-Lange, A.; Soteriou, D.; Hanna, R.; Tiosano, B.; Fuchs, Y.; Shalom-Feuerstein, R. Corneal-Committed Cells Restore the Stem Cell Pool and Tissue Boundary following Injury. Cell Rep. 2018, 22, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Sartaj, R.; Zhang, C.; Wan, P.; Pasha, Z.; Guaiquil, V.; Liu, A.; Liu, J.; Luo, Y.; Fuchs, E.; Rosenblatt, M.I. Characterization of slow cycling corneal limbal epithelial cells identifies putative stem cell markers. Sci. Rep. 2017, 7, 3793. [Google Scholar] [CrossRef] [Green Version]
- Park, M.; Richardson, A.; Pandzic, E.; Lobo, E.P.; Whan, R.; Watson, S.L.; Lyons, J.G.; Wakefield, D.; Di Girolamo, N. Visualizing the Contribution of Keratin-14+ Limbal Epithelial Precursors in Corneal Wound Healing. Stem Cell Rep. 2018, 12, 14–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tracz, A.F.; Szczylik, C.; Porta, C.; Czarnecka, A.M. Insulin-like growth factor-1 signaling in renal cell carcinoma. BMC Cancer 2016, 16, 453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.-L.; Yang, S.-C.; Lai, C.-Y.; Wang, C.-K.; Chang, C.-F.; Lin, C.-Y.; Chen, W.-J.; Lin, P.-Y.; Wu, H.-C.; Ma, N.; et al. CXCL14 Maintains hESC Self-Renewal through Binding to IGF-1R and Activation of the IGF-1R Pathway. Cells 2020, 9, 1706. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-Y.; Hsieh, H.-L.; Hsiao, L.-D.; Yang, C.-M. PI3-K/Akt/JNK/NF-κB is essential for MMP-9 expression and outgrowth in human limbal epithelial cells on intact amniotic membrane. Stem Cell Res. 2012, 9, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, S.; Oh, D.; Baclagon, E.R.; Zheng, J.J.; Deng, S.X. Wnt Signaling Is Required for the Maintenance of Human Limbal Stem/Progenitor Cells In Vitro. Investig. Opthalmology Vis. Sci. 2019, 60, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Serror, L.; Nir, E.; Dhiraj, D.; Altshuler, A.; Khreish, M.; Tiosano, B.; Hasson, P.; Panman, L.; Luxenburg, C.; et al. SOX2 Regulates P63 and Stem/Progenitor Cell State in the Corneal Epithelium. Stem Cells Dayt. Ohio. 2018, 37, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Biaoxue, R.; Xiguang, C.; Hua, L.; Shuanying, Y. Stathmin-dependent molecular targeting therapy for malignant tumor: The latest 5 years’ discoveries and developments. J. Transl. Med. 2016, 14, 279. [Google Scholar] [CrossRef] [Green Version]
- Feng, F.; Zhao, Z.; Cai, X.; Heng, X.; Ma, X. Cyclin-dependent kinase subunit2 (CKS2) promotes malignant phenotypes and epithelial-mesenchymal transition-like process in glioma by activating TGFβ/SMAD signaling. Cancer Med. 2022, 12, 5889–5907. [Google Scholar] [CrossRef]
- Gerdes, J.; Schwab, U.; Lemke, H.; Stein, H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int. J. Cancer 1983, 31, 13–20. [Google Scholar] [CrossRef]
- Ye, L.; Guo, L.; He, Z.; Wang, X.; Lin, C.; Zhang, X.; Wu, S.; Bao, Y.; Yang, Q.; Song, L.; et al. Upregulation of E2F8 promotes cell proliferation and tumorigenicity in breast cancer by modulating G1/S phase transition. Oncotarget 2016, 7, 23757–23771. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Lu, J.; Fang, Q.; Lu, Y.; Xie, C.; Wu, H.; Yin, Z. UBE2C functions as a potential oncogene by enhancing cell proliferation, migration, invasion, and drug resistance in hepatocellular carcinoma cells. Biosci. Rep. 2019, 39, BSR20182384. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Biswas, S.; Ma, G.; Zhao, P.; Li, B.; Li, J. Canonical NF-κB signaling maintains corneal epithelial integrity and prevents corneal aging via retinoic acid. eLife 2021, 10, e67315. [Google Scholar] [CrossRef]
- D’alessio, A.C.; Fan, Z.P.; Wert, K.J.; Baranov, P.; Cohen, M.A.; Saini, J.S.; Cohick, E.; Charniga, C.; Dadon, D.; Hannett, N.M.; et al. A Systematic Approach to Identify Candidate Transcription Factors that Control Cell Identity. Stem Cell Rep. 2015, 5, 763–775. [Google Scholar] [CrossRef] [Green Version]
- Almeida, N.; Chung, M.W.H.; Drudi, E.M.; Engquist, E.N.; Hamrud, E.; Isaacson, A.; Tsang, V.S.K.; Watt, F.M.; Spagnoli, F.M. Employing core regulatory circuits to define cell identity. EMBO J. 2021, 40, e106785. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhu, L.; Liu, J.; Huang, H.; Guo, H.; Wang, L.; Li, L.; Gu, S.; Tan, J.; Zhong, J.; et al. Loss of FOXC1 contributes to the corneal epithelial fate switch and pathogenesis. Signal Transduct. Target. Ther. 2021, 6, 5. [Google Scholar] [CrossRef]
- Li, M.; Huang, H.; Li, L.; He, C.; Zhu, L.; Guo, H.; Wang, L.; Liu, J.; Wu, S.; Liu, J.; et al. Core transcription regulatory circuitry orchestrates corneal epithelial homeostasis. Nat. Commun. 2021, 12, 420. [Google Scholar] [CrossRef]
- Barbaro, V.; Testa, A.; Di Iorio, E.; Mavilio, F.; Pellegrini, G.; De Luca, M. C/EBPδ regulates cell cycle and self-renewal of human limbal stem cells. J. Cell Biol. 2007, 177, 1037–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merjava, S.; Neuwirth, A.; Tanzerova, M.; Jirsova, K. The spectrum of cytokeratins expressed in the adult human cornea, limbus and perilimbal conjunctiva. Histol. Histopathol. 2011, 26, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Miranda, A.; Nakatsu, M.N.; Zarei-Ghanavati, S.; Nguyen, C.V.; Deng, S.X. Keratin 13 is a more specific marker of conjunctival epithelium than keratin 19. Mol. Vis. 2011, 17, 1652–1661. [Google Scholar] [PubMed]
- de Paiva, C.S.; Chen, Z.; Corrales, R.M.; Pflugfelder, S.C.; Li, D. ABCG2 Transporter Identifies a Population of Clonogenic Human Limbal Epithelial Cells. Stem Cells 2005, 23, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Mariappan, I.; Maddileti, S.; Savy, S.; Tiwari, S.; Gaddipati, S.; Fatima, A.; Sangwan, V.S.; Balasubramanian, D.; Vemuganti, G.K. In vitro culture and expansion of human limbal epithelial cells. Nat. Protoc. 2010, 5, 1470–1479. [Google Scholar] [CrossRef]
- Kramerov, A.A.; Saghizadeh, M.; Maguen, E.; Rabinowitz, Y.S.; Ljubimov, A.V. Persistence of reduced expression of putative stem cell markers and slow wound healing in cultured diabetic limbal epithelial cells. Mol. Vis. 2015, 21, 1357–1367. [Google Scholar]
- Ksander, B.R.; Kolovou, P.E.; Wilson, B.J.; Saab, K.R.; Guo, Q.; Ma, J.; McGuire, S.P.; Gregory, M.S.; Vincent, W.J.B.; Perez, V.L.; et al. ABCB5 is a limbal stem cell gene required for corneal development and repair. Nature 2014, 511, 353–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaharuddin, B.; Osei-Bempong, C.; Ahmad, S.; Rooney, P.; Ali, S.; Oldershaw, R.; Meeson, A. Human limbal mesenchymal stem cells express ABCB5 and can grow on amniotic membrane. Regen. Med. 2016, 11, 273–286. [Google Scholar] [CrossRef] [Green Version]
- Mathan, J.J.; Ismail, S.; McGhee, J.J.; McGhee, C.N.J.; Sherwin, T. Sphere-forming cells from peripheral cornea demonstrate the ability to repopulate the ocular surface. Stem Cell Res. Ther. 2016, 7, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.K.; Lee, G.-H.; Lee, B.; Maeng, Y.-S. Establishment of Novel Limbus-Derived, Highly Proliferative ABCG2+/ABCB5+ Limbal Epithelial Stem Cell Cultures. Stem Cells Int. 2017, 2017, 7678637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, K.B.; Collins, C.A.; Nascimento, E.; Tan, D.W.; Frye, M.; Itami, S.; Watt, F.M. Lrig1 Expression Defines a Distinct Multipotent Stem Cell Population in Mammalian Epidermis. Cell Stem Cell 2009, 4, 427–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bath, C.; Muttuvelu, D.; Emmersen, J.; Vorum, H.; Hjortdal, J.; Zachar, V. Transcriptional Dissection of Human Limbal Niche Compartments by Massive Parallel Sequencing. PLoS ONE 2013, 8, e64244. [Google Scholar] [CrossRef]
- Parfitt, G.J.; Kavianpour, B.; Wu, K.L.; Xie, Y.; Brown, D.J.; Jester, J.V. Immunofluorescence Tomography of Mouse Ocular Surface Epithelial Stem Cells and Their Niche Microenvironment. Investig. Opthalmology Vis. Sci. 2015, 56, 7338–7344. [Google Scholar] [CrossRef] [Green Version]
- Farrelly, O.; Suzuki-Horiuchi, Y.; Brewster, M.; Kuri, P.; Huang, S.; Rice, G.; Bae, H.; Xu, J.; Dentchev, T.; Lee, V.; et al. Two-photon live imaging of single corneal stem cells reveals compartmentalized organization of the limbal niche. Cell Stem Cell 2021, 28, 1233–1247.e4. [Google Scholar] [CrossRef]
- Krenning, L.; Sonneveld, S.; Tanenbaum, M.E. Time-resolved single-cell sequencing identifies multiple waves of mRNA decay during the mitosis-to-G1 phase transition. eLife 2022, 11, e71356. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, M.; Lin, S.; Jian, R.; Li, X.; Chan, J.; Dong, G.; Fang, H.; Robinson, A.E.; Snyder, M.P.; et al. A Quantitative Proteome Map of the Human Body. Cell 2020, 183, 269–283.e19. [Google Scholar] [CrossRef] [PubMed]
- Vattulainen, M.; Ilmarinen, T.; Viheriälä, T.; Jokinen, V.; Skottman, H. Corneal epithelial differentiation of human pluripotent stem cells generates ABCB5+ and ∆Np63α+ cells with limbal cell characteristics and high wound healing capacity. Stem Cell Res. Ther. 2021, 12, 609. [Google Scholar] [CrossRef] [PubMed]
- Stasi, K.; Goings, D.; Huang, J.; Herman, L.; Pinto, F.; Addis, R.C.; Klein, D.; Massaro-Giordano, G.; Gearhart, J.D. Optimal Isolation and Xeno-Free Culture Conditions for Limbal Stem Cell Function. Investig. Opthalmology Vis. Sci. 2014, 55, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.-Y.; Wang, P.-R.; Luo, L.-J.; Chen, S.-T. Stabilization of collagen nanofibers with l-lysine improves the ability of carbodiimide cross-linked amniotic membranes to preserve limbal epithelial progenitor cells. Int. J. Nanomed. 2014, 9, 5117–5130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djalilian, A.; Namavari, A.; Ito, A.; Balali, S.; Afshar, A.; Lavker, R.; Yue, B.J.T. Down-regulation of Notch signaling during corneal epithelial proliferation. Mol. Vis. 2008, 14, 1041–1049. [Google Scholar]
- Nakamura, T.; Ohtsuka, T.; Sekiyama, E.; Cooper, L.J.; Kokubu, H.; Fullwood, N.J.; Barrandon, Y.; Kageyama, R.; Kinoshita, S. Hes1 Regulates Corneal Development and the Function of Corneal Epithelial Stem/Progenitor Cells. Stem Cells Dayt. Ohio. 2008, 26, 1265–1274. [Google Scholar] [CrossRef]
- Laux-Fenton, W.T.; Donaldson, P.J.; Kistler, J.; Green, C.R. Connexin Expression Patterns in the Rat Cornea: Molecular evidence for communication compartments. Cornea 2003, 22, 457–464. [Google Scholar] [CrossRef]
- Maimouni, S.; Issa, N.; Cheng, S.; Ouaari, C.; Cheema, A.; Kumar, D.; Byers, S. Tumor suppressor RARRES1- A novel regulator of fatty acid metabolism in epithelial cells. PLoS ONE 2018, 13, e0208756. [Google Scholar] [CrossRef] [Green Version]
- Choreño-Parra, J.A.; Thirunavukkarasu, S.; Zúñiga, J.; Khader, S.A. The protective and pathogenic roles of CXCL17 in human health and disease: Potential in respiratory medicine. Cytokine Growth Factor Rev. 2020, 53, 53–62. [Google Scholar] [CrossRef]
- Turner, H.C.; Budak, M.T.; Akinci, M.A.M.; Wolosin, J.M. Comparative Analysis of Human Conjunctival and Corneal Epithelial Gene Expression with Oligonucleotide Microarrays. Investig. Opthalmology Vis. Sci. 2007, 48, 2050–2061. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Bjork, P.; Källberg, E.; Olsson, A.; Riva, M.; Mörgelin, M.; Liberg, D.; Ivars, F.; Leanderson, T. Vesicular Location and Transport of S100A8 and S100A9 Proteins in Monocytoid Cells. PLoS ONE 2015, 10, e0145217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, E.C.; Liu, C.-Y.; Chikama, T.-I.; Hayashi, Y.; Kao, C.W.-C.; Birk, D.E.; Funderburgh, J.L.; Jester, J.V.; Kao, W.W.-Y. Keratocan, a Cornea-specific Keratan Sulfate Proteoglycan, Is Regulatedby Lumican. J. Biol. Chem. 2005, 280, 25541–25547. [Google Scholar] [CrossRef] [Green Version]
- Nassiri, F.; Cusimano, M.D.; Scheithauer, B.W.; Rotondo, F.; Fazio, A.; Yousef, G.M.; Syro, L.V.; Kovacs, K.; Lloyd, R.V. Endoglin (CD105): A review of its role in angiogenesis and tumor diagnosis, progression and therapy. Anticancer Res. 2011, 31, 2283–2290. [Google Scholar] [PubMed]
- Takagi, Y.; Fukumitsu, R.; Yoshida, K.; Miyamoto, S. Endoglin (CD105) is a more appropriate marker than CD31 for detecting microvessels in carotid artery plaques. Surg. Neurol. Int. 2013, 4, 132. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zafrullah, N.; Devaux, F.; Hemmavanh, C.; Adams, S.; Ziebarth, N.M.; Koch, M.; Birk, D.E.; Espana, E.M. Collagen XII Is a Regulator of Corneal Stroma Structure and Function. Investig. Opthalmology Vis. Sci. 2020, 61, 61. [Google Scholar] [CrossRef]
- Chng, Z.; Peh, G.S.L.; Herath, W.B.; Cheng, T.Y.D.; Ang, H.-P.; Toh, K.-P.; Robson, P.; Mehta, J.S.; Colman, A. High Throughput Gene Expression Analysis Identifies Reliable Expression Markers of Human Corneal Endothelial Cells. PLoS ONE 2013, 8, e67546. [Google Scholar] [CrossRef] [Green Version]
- Nakai, H.; Tsuchiya, Y.; Koike, N.; Asano, T.; Ueno, M.; Umemura, Y.; Sasawaki, Y.; Ono, R.; Hamuro, J.; Sotozono, C.; et al. Comprehensive Analysis Identified the Circadian Clock and Global Circadian Gene Expression in Human Corneal Endothelial Cells. Investig. Opthalmology Vis. Sci. 2022, 63, 16. [Google Scholar] [CrossRef]
- Hinz, B.; Phan, S.H.; Thannickal, V.J.; Galli, A.; Bochaton-Piallat, M.-L.; Gabbiani, G. The Myofibroblast: One Function, Multiple Origins. Am. J. Pathol. 2007, 170, 1807–1816. [Google Scholar] [CrossRef]
- Rockey, D.C.; Weymouth, N.; Shi, Z. Smooth Muscle α Actin (Acta2) and Myofibroblast Function during Hepatic Wound Healing. PLoS ONE 2013, 8, e77166. [Google Scholar] [CrossRef] [Green Version]
- Mikhailova, A.; Ilmarinen, T.; Uusitalo, H.; Skottman, H. Small-Molecule Induction Promotes Corneal Epithelial Cell Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2014, 2, 219–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, R.; Ishikawa, Y.; Sasamoto, Y.; Katori, R.; Nomura, N.; Ichikawa, T.; Araki, S.; Soma, T.; Kawasaki, S.; Sekiguchi, K.; et al. Co-ordinated ocular development from human iPS cells and recovery of corneal function. Nature 2016, 531, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Kamarudin, T.A.; Bojic, S.; Collin, J.; Yu, M.; Alharthi, S.; Buck, H.; Shortt, A.; Armstrong, L.; Figueiredo, F.C.; Lako, M. Differences in the Activity of Endogenous Bone Morphogenetic Protein Signaling Impact on the Ability of Induced Pluripotent Stem Cells to Differentiate to Corneal Epithelial-Like Cells. Stem Cells Dayt. Ohio. 2017, 36, 337–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, Y.; Hayashi, R.; Shibata, S.; Quantock, A.J.; Nishida, K. Ocular surface ectoderm instigated by WNT inhibition and BMP4. Stem Cell Res. 2020, 46, 101868. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.W.; Wahlin, K.; Adams, S.M.; Birk, D.E.; Zack, D.J.; Chakravarti, S. Cornea organoids from human induced pluripotent stem cells. Sci. Rep. 2017, 7, 41286. [Google Scholar] [CrossRef]
- Isla-Magrané, H.; Veiga, A.; García-Arumí, J.; Duarri, A. Multiocular organoids from human induced pluripotent stem cells displayed retinal, corneal, and retinal pigment epithelium lineages. Stem Cell Res. Ther. 2021, 12, 581. [Google Scholar] [CrossRef]
- Susaimanickam, P.J.; Maddileti, S.; Kumar, V.; Boyinpally, S.R.; Naik, R.R.; Naik, M.N.; Reddy, G.B.; Sangwan, V.S.; Mariappan, I. Generating minicorneal organoids from human induced pluripotent stem cells. Development 2017, 144, 2338–2351. [Google Scholar] [CrossRef] [Green Version]
- Govindarajan, B.; Gipson, I.K. Membrane-tethered mucins have multiple functions on the ocular surface. Exp. Eye Res. 2010, 90, 655–663. [Google Scholar] [CrossRef] [Green Version]
- Shangguan, Y.; Li, C.; Lin, H.; Ou, M.; Tang, D.; Dai, Y.; Yan, Q. Application of single-cell RNA sequencing in embryonic development. Genomics 2020, 112, 4547–4551. [Google Scholar] [CrossRef]
- Molè, M.A.; Coorens, T.H.H.; Shahbazi, M.N.; Weberling, A.; Weatherbee, B.A.T.; Gantner, C.W.; Sancho-Serra, C.; Richardson, L.; Drinkwater, A.; Syed, N.; et al. A single cell characterisation of human embryogenesis identifies pluripotency transitions and putative anterior hypoblast centre. Nat. Commun. 2021, 12, 3679. [Google Scholar] [CrossRef]
- Cao, S.; Feng, H.; Yi, H.; Pan, M.; Lin, L.; Zhang, Y.S.; Feng, Z.; Liang, W.; Cai, B.; Li, Q.; et al. Single-cell RNA sequencing reveals the developmental program underlying proximal–distal patterning of the human lung at the embryonic stage. Cell Res. 2023, 33, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Grueterich, M.; Espana, E.M.; Tseng, S.C. Ex vivo expansion of limbal epithelial stem cells: Amniotic membrane serving as a stem cell niche. Surv. Ophthalmol. 2003, 48, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.R.; Hamley, I.W.; Connon, C.J. Ex vivo expansion of limbal stem cells is affected by substrate properties. Stem Cell Res. 2012, 8, 403–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puri, S.; Moreno, I.Y.; Sun, M.; Verma, S.; Lin, X.; Gesteira, T.F.; Coulson-Thomas, V.J. Hyaluronan supports the limbal stem cell phenotype during ex vivo culture. Stem Cell Res. Ther. 2022, 13, 384. [Google Scholar] [CrossRef] [PubMed]
- Sareen, D.; Saghizadeh, M.; Ornelas, L.; Winkler, M.A.; Narwani, K.; Sahabian, A.; Funari, V.A.; Tang, J.; Spurka, L.; Punj, V.; et al. Differentiation of Human Limbal-Derived Induced Pluripotent Stem Cells Into Limbal-Like Epithelium. Stem Cells Transl. Med. 2014, 3, 1002–1012. [Google Scholar] [CrossRef] [Green Version]
- Aberdam, E.; Petit, I.; Sangari, L.; Aberdam, D. Induced pluripotent stem cell-derived limbal epithelial cells (LiPSC) as a cellular alternative for in vitro ocular toxicity testing. PLoS ONE 2017, 12, e0179913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunha, D.L.; Eintracht, J.; Harding, P.; Zhou, J.H.; Moosajee, M. Restoration of functional PAX6 in aniridia patient iPSC-derived ocular tissue models using repurposed nonsense suppression drugs. bioRxiv 2022. bioRxiv:2022.10.12.511600. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, J.; Lewy, T.; Rice, C.M.; Elemento, O.; Rendeiro, A.F.; Mason, C.E. Spatial omics technologies at multimodal and single cell/subcellular level. Genome Biol. 2022, 23, 256. [Google Scholar] [CrossRef]
- Mantri, M.; Scuderi, G.J.; Abedini-Nassab, R.; Wang, M.F.Z.; McKellar, D.; Shi, H.; Grodner, B.; Butcher, J.T.; De Vlaminck, I. Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis. Nat. Commun. 2021, 12, 1771. [Google Scholar] [CrossRef]
- Kuppe, C.; Flores, R.O.R.; Li, Z.; Hayat, S.; Levinson, R.T.; Liao, X.; Hannani, M.T.; Tanevski, J.; Wünnemann, F.; Nagai, J.S.; et al. Spatial multi-omic map of human myocardial infarction. Nature 2022, 608, 766–777. [Google Scholar] [CrossRef]
- Tran, H.T.N.; Ang, K.S.; Chevrier, M.; Zhang, X.; Lee, N.Y.S.; Goh, M.; Chen, J. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020, 21, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunha, D.L.; Arno, G.; Corton, M.; Moosajee, M. The Spectrum of PAX6 Mutations and Genotype-Phenotype Correlations in the Eye. Genes 2019, 10, 1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polisetti, N.; Schlunck, G.; Reinhard, T. PAX6 Expression Patterns in the Adult Human Limbal Stem Cell Niche. Cells 2023, 12, 400. [Google Scholar] [CrossRef] [PubMed]
- Tone, S.O.; Kocaba, V.; Böhm, M.; Wylegala, A.; White, T.L.; Jurkunas, U.V. Fuchs endothelial corneal dystrophy: The vicious cycle of Fuchs pathogenesis. Prog. Retin. Eye Res. 2021, 80, 100863. [Google Scholar] [CrossRef]
- Wang, Q.; Dou, S.; Zhang, B.; Jiang, H.; Qi, X.; Duan, H.; Wang, X.; Dong, C.; Xie, L.; Cao, Y.; et al. Heterogeneity of human corneal endothelium implicates lncRNA NEAT1 in Fuchs endothelial corneal dystrophy. Mol. Ther.—Nucleic Acids 2022, 27, 880–893. [Google Scholar] [CrossRef]
- Moshirfar, M.; Somani, A.N.; Vaidyanathan, U.; Patel, B.C. Fuchs Endothelial Dystrophy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Wilson, S.E.; Liu, J.J.; Mohan, R.R. Stromal-epithelial interactions in the cornea. Prog. Retin. Eye Res. 1999, 18, 293–309. [Google Scholar] [CrossRef]
- Gabison, E.E.; Huet, E.; Baudouin, C.; Menashi, S. Direct epithelial–stromal interaction in corneal wound healing: Role of EMMPRIN/CD147 in MMPs induction and beyond. Prog. Retin. Eye Res. 2009, 28, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Maeda, N.; Nakagawa, T.; Higashiura, R.; Fuchihata, M.; Koh, S.; Nishida, K. Evaluation of corneal epithelial and stromal thickness in keratoconus using spectral-domain optical coherence tomography. Jpn. J. Ophthalmol. 2014, 58, 389–395. [Google Scholar] [CrossRef]
- Bykhovskaya, Y.; Rabinowitz, Y.S. Update on the genetics of keratoconus. Exp. Eye Res. 2020, 202, 108398. [Google Scholar] [CrossRef] [PubMed]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squair, J.W.; Gautier, M.; Kathe, C.; Anderson, M.A.; James, N.D.; Hutson, T.H.; Hudelle, R.; Qaiser, T.; Matson, K.J.E.; Barraud, Q.; et al. Confronting false discoveries in single-cell differential expression. Nat. Commun. 2021, 12, 5692. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [Green Version]
- Van de Sande, B.; Lee, J.S.; Mutasa-Gottgens, E.; Naughton, B.; Bacon, W.; Manning, J.; Wang, Y.; Pollard, J.; Mendez, M.; Hill, J.; et al. Applications of single-cell RNA sequencing in drug discovery and development. Nat. Rev. Drug Discov. 2023, 22, 496–520. [Google Scholar] [CrossRef]
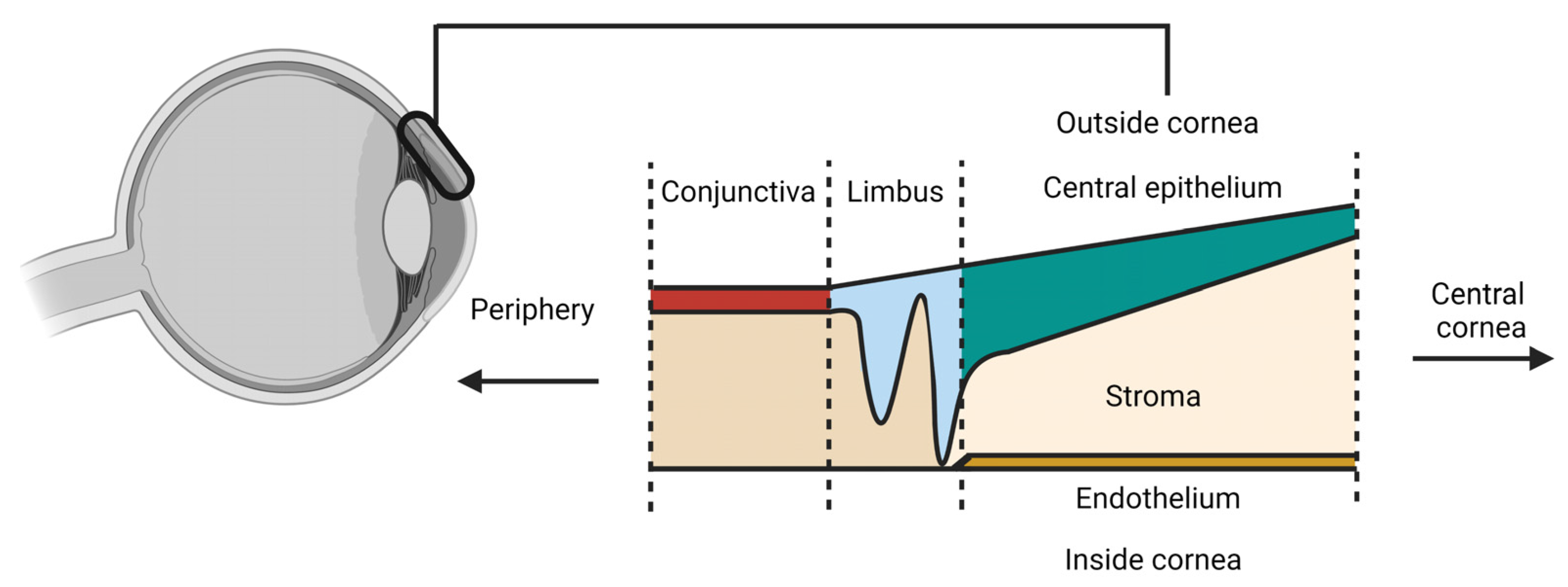
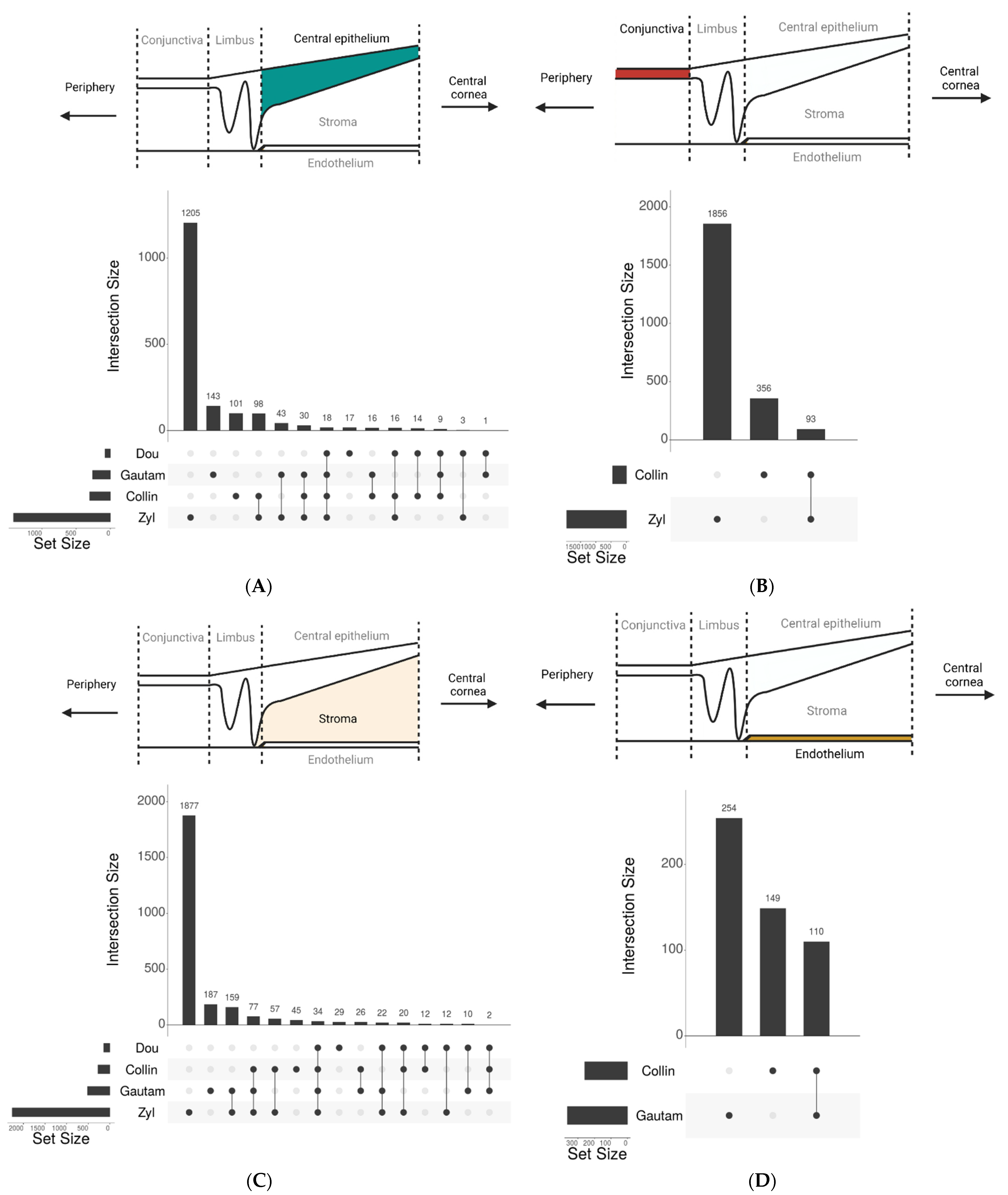
| Study and Year of Publication | Tissues | Total Number of Cells | Total Number of Corneal Cell States | Raw Data Publicly Available | Number of Donors | Dissociation Method |
|---|---|---|---|---|---|---|
| Zyl (2022) * [28] | Ocular anterior segment | 191,992 | 28 | Yes | 6 | Separate dissection |
| Gautam (2021) [29] | Complete eye | Approx. 50,000 | 7 | Yes | 3 | Separate dissection |
| Collin (2021) [30] | Complete cornea | 213,430 | 21 | Yes | 6 | Bulk enzymatic |
| Català (2021) [31] | Complete cornea | 19,472 | 15 | Yes | 8 | Separate dissection |
| Ligocki (2021) [32] | Complete cornea | 16,234 | 16 | No | 6 | Bulk enzymatic |
| Maiti (2022) [33] | Central and peripheral cornea | 53,438 | 12 | Yes | 3 | Separate dissection |
| Dou (2022) [34] | Central cornea | 39,214 | 6 | No | 4 | Bulk enzymatic |
| Li (2021) [35] | Limbus | 16,360 | 12 | Yes | 2 | Bulk enzymatic |
| Layer | Study | Names of Corneal Cell States |
|---|---|---|
| Limbus | Català (2021) [31] | Cells in the limbal stem cell niche, corneal basal limbal epithelial cells, terminally differentiated migratory limbal epithelial cells, transiently amplifying cells, LSC in the limbal stem cell niche or in the peripheral cornea, wing superficial limbal epithelial cells |
| Collin (2021) [30] | qLSC, LNCPs, LPCs, limbal fibroblasts, limbal stroma cells, limbal stromal keratocytes, limbal superficial epithelium, limbal suprabasal cells | |
| Li (2021) [35] | Progenitor cells, transiently amplifying cells | |
| Ligocki (2021) [32] | Early limbal progenitor cells, late limbal progenitor cells, transiently amplifying cells | |
| Maiti (2022) [33] | Limbal progenitor cells, limbal epithelial cells, corneal epithelial stem cells | |
| Zyl (2022) * [28] | Basal limbal epithelial cells, wing limbal epithelial cells, superficial limbal epithelial cells, transiently amplifying cells | |
| Corneal epithelium | Català (2021) [31] | Basal corneal epithelium, terminally differentiated central corneal epithelium, wing superficial epithelial cells |
| Collin (2021) [30] | Basal corneal epithelium, central cornea suprabasal cells, suprabasal corneal epithelium | |
| Dou (2022) [34] | Corneal epithelial cells | |
| Gautam (2021) [29] | ELF3-high corneal epithelial cells, TGFBI-high corneal epithelial cells | |
| Li (2021) [35] | Differentiated cells | |
| Ligocki (2021) [32] | Basal epithelial cells, central superficial mature epithelial cells, transitional epithelial cells, transiently amplifying cells | |
| Maiti (2022) [33] | Corneal epithelial stem cells, differentiated corneal epithelium, differentiated superficial corneal epithelium | |
| Zyl (2022) * [28] | Basal corneal epithelium, superficial-most squamous epithelium, wing superficial cells | |
| Conjunctiva | Collin (2021) [30] | Basal conjunctival epithelium, superficial conjunctival epithelium |
| Gautam (2021) [29] | Conjunctival cells | |
| Li (2021) [35] | Conjunctiva | |
| Ligocki (2021) [32] | Conjunctival epithelial cells | |
| Maiti (2022) [33] | Conjunctival epithelial cells | |
| Zyl (2022) * [28] | Basal conjunctival epithelium, mucin-producing goblet cells, superficial conjunctival epithelium, wing conjunctival epithelium, conjunctival melanocytes | |
| Stroma | Català (2021) [31] | Activated stromal keratocytes, general stromal keratocytes, transitioning keratocytes in the stroma to myofibroblasts |
| Collin (2021) [30] | CSSCs, central stromal keratocytes | |
| Dou (2022) [34] | Corneal stroma cells | |
| Ligocki (2021) [32] | Stromal cells | |
| Maiti (2022) [33] | Corneal stromal cells | |
| Zyl (2022) * [28] | Corneal stromal fibroblasts, corneal stromal keratocytes | |
| Endothelium | Català (2021) [31] | Corneal endothelium stationary cells, corneal endothelium migratory cells |
| Collin (2021) [30] | Corneal endothelium, FCECs | |
| Ligocki (2021) [32] | Corneal endothelial cells | |
| Maiti (2022) [33] | Corneal endothelium | |
| Zyl (2022) * [28] | Endothelial lining, pericytes, vascular endothelium |
| Markers Identified in Single-Cell Studies | Cell States and Studies | Associated Disorder | Affected Cornea Layer | Gene OMIM Number |
|---|---|---|---|---|
| GJB2 | Wing superficial limbal epithelial cells (Català), basal corneal epithelium (Català), basal corneal epithelium (Collin) | Keratitis–Ichthyosis–Deafness (KID) Syndrome | Epithelium, stroma | 121011 |
| TP63 | Limbal progenitor cells (Collin), LNCP (Collin), limbal suprabasal cells (Collin), limbal epithelial stem cells (Català), epithelial stem cells (Maiti) | Ectrodactyly–Ectodermal Dysplasia–Cleft Syndrome | Epithelium | 603273 |
| PAX6 | LNCP (Collin), corneal epithelium (Zyl) | Aniridia | Epithelium | 607108 |
| Axenfeld–Rieger Syndrome | Neural-crest derived structures, stroma, endothelium | 601542 | ||
| PITX2 | Corneal endothelium stationary cells (Català), corneal endothelium migratory cells (Català) | 601090 | ||
| FOXC1 | Limbal stem cells (Li) | 601542 | ||
| TGFBI | TGFBI-hi corneal epithelial cells (Gautam) | Epithelial basement membrane dystrophy | Epithelium | 601692 |
| KRT12 | Basal limbal epithelial cells (Zyl), limbal epithelial basal cells (Català), terminally differentiated migratory limbal epithelial cells (Català), basal and wing cells (Zyl),central superficial mature epithelial cells (Ligocki), corneal epithelium (Dou and Zyl), differentiated cells (Li), differentiated corneal epithelium (Maiti), transitional epithelial cells (Ligocki), wing superficial central epithelium (Català) | Meesmann corneal dystrophy | 601687 | |
| KRT3 | 148043 | |||
| DCN | Corneal stromal cell subsets (Maiti), stromal cells (Ligocki and Dou) | Congenital stromal corneal dystrophy | Stroma | 125255 |
| Posterior amorphous corneal dystrophy | ||||
| LUM | Activated stromal keratocytes (Català), corneal stromal fibroblasts (Zyl), CSSCs (Collin), central stromal keratocytes (Collin), general stromal keratocytes (Català), stromal cells (Ligocki and Dou), transitioning keratocytes stromal myofibroblasts (Català), corneal stromal cell subsets (Maiti), corneal endothelium (Maiti) | 600616 | ||
| KERA | Cornea plana | Stroma, Descemet membrane | 603288 | |
| COL8A2 | Corneal endothelium (Maiti) | Fuchs endothelial and posterior polymorphous corneal dystrophy | Endothelium, Descemet membrane | 120252 |
| SLC4A11 | Corneal endothelium stationary and migratory cells (Català), corneal endothelial cells (Ligocki) | Congenital hereditary and Fuchs endothelial corneal dystrophy | Endothelium | 610206 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arts, J.A.; Laberthonnière, C.; Lima Cunha, D.; Zhou, H. Single-Cell RNA Sequencing: Opportunities and Challenges for Studies on Corneal Biology in Health and Disease. Cells 2023, 12, 1808. https://doi.org/10.3390/cells12131808
Arts JA, Laberthonnière C, Lima Cunha D, Zhou H. Single-Cell RNA Sequencing: Opportunities and Challenges for Studies on Corneal Biology in Health and Disease. Cells. 2023; 12(13):1808. https://doi.org/10.3390/cells12131808
Chicago/Turabian StyleArts, Julian A., Camille Laberthonnière, Dulce Lima Cunha, and Huiqing Zhou. 2023. "Single-Cell RNA Sequencing: Opportunities and Challenges for Studies on Corneal Biology in Health and Disease" Cells 12, no. 13: 1808. https://doi.org/10.3390/cells12131808
APA StyleArts, J. A., Laberthonnière, C., Lima Cunha, D., & Zhou, H. (2023). Single-Cell RNA Sequencing: Opportunities and Challenges for Studies on Corneal Biology in Health and Disease. Cells, 12(13), 1808. https://doi.org/10.3390/cells12131808







