Testicular Germ Cell Tumor Tissue Biomarker Analysis: A Comparison of Human Protein Atlas and Individual Testicular Germ Cell Tumor Component Immunohistochemistry
Abstract
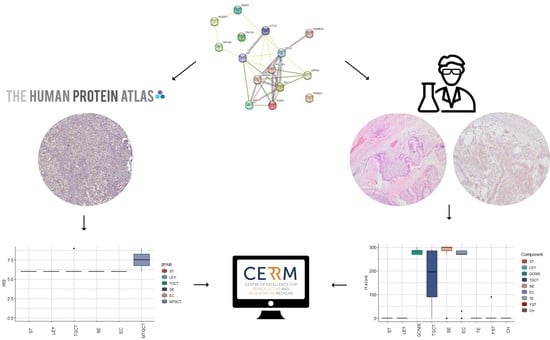
:1. Introduction
2. Materials and Methods
2.1. HPA Analysis
2.2. In-House Analysis
2.2.1. Patients
2.2.2. Ethical Statement
2.2.3. H&E Analysis
2.2.4. IHC
2.2.5. IHC Analysis
3. Results
3.1. Patient Data
3.2. HPA Analysis
3.3. IHC Analysis
3.4. TGCT Patients vs. Non-Neoplastic Patients Testicular Tissue
3.5. Comparison of Results
4. Discussion
4.1. Healthy Controls
4.2. TGCT IHC Analysis
4.3. Study Highlights
- The HPA is a useful online tool for exploration of gene expression on a protein level;
- Due to the TGCT heterogeneity of histological components, bulk protein expression, as shown in the HPA, should be avoided;
- Discrepancies in key TGCT diagnostic biomarkers were detected in the HPA;
- MAGEC2, HOXA9 and 5 mC were confirmed as potential TGCT biomarkers;
- DPPA3, CALCA and TDGF1 were identified as potential novel TGCT biomarkers;
- MGMT was confirmed as a biomarker of healthy testicular tissue;
- RASSF1 and PRSS21 were identified as biomarkers of healthy testicular tissue;
- SALL4, SOX17, RASSF1 and PRSS21 dysregulation in the surrounding testicular tissue with complete preserved spermatogenesis was detected.
4.4. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Znaor, A.; Skakkebaek, N.E.; Rajpert-De Meyts, E.; Kuliš, T.; Laversanne, M.; Gurney, J.; Sarfati, D.; McGlynn, K.A.; Bray, F. Global Patterns in Testicular Cancer Incidence and Mortality in 2020. Int. J. Cancer 2022, 151, 692–698. [Google Scholar] [CrossRef]
- Zhang, T.; Ji, L.; Liu, B.; Guan, W.; Liu, Q.; Gao, Y. Testicular Germ Cell Tumors: A Clinicopathological and Immunohistochemical Analysis of 145 Cases. Int. J. Clin. Exp. Pathol. 2018, 11, 4622–4629. [Google Scholar]
- Znaor, A.; Skakkebæk, N.E.; Rajpert-De Meyts, E.; Laversanne, M.; Kuliš, T.; Gurney, J.; Sarfati, D.; McGlynn, K.A.; Bray, F. Testicular Cancer Incidence Predictions in Europe 2010–2035: A Rising Burden despite Population Ageing. Int. J. Cancer 2020, 147, 820–828. [Google Scholar] [CrossRef]
- Idrees, M.T.; Ulbright, T.M.; Oliva, E.; Young, R.H.; Montironi, R.; Egevad, L.; Berney, D.; Srigley, J.R.; Epstein, J.I.; Tickoo, S.K.; et al. The World Health Organization 2016 Classification of Testicular Non-Germ Cell Tumours: A Review and Update from the International Society of Urological Pathology Testis Consultation Panel. Histopathology 2017, 70, 513–521. [Google Scholar] [CrossRef]
- Fink, C.; Baal, N.; Wilhelm, J.; Sarode, P.; Weigel, R.; Schumacher, V.; Nettersheim, D.; Schorle, H.; Schröck, C.; Bergmann, M.; et al. On the Origin of Germ Cell Neoplasia in Situ: Dedifferentiation of Human Adult Sertoli Cells in Cross Talk with Seminoma Cells in Vitro. Neoplasia 2021, 23, 731–742. [Google Scholar] [CrossRef]
- Burton, J.; Wojewodzic, M.W.; Rounge, T.B.; Haugen, T.B. A Role of the TEX101 Interactome in the Common Aetiology Behind Male Subfertility and Testicular Germ Cell Tumor. Front. Oncol. 2022, 12, 892043. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.; Gillis, A.; Jerónimo, C.; Henrique, R.; Looijenga, L. Human Germ Cell Tumors Are Developmental Cancers: Impact of Epigenetics on Pathobiology and Clinic. Int. J. Mol. Sci. 2019, 20, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berney, D.M.; Cree, I.; Rao, V.; Moch, H.; Srigley, J.R.; Tsuzuki, T.; Amin, M.B.; Comperat, E.M.; Hartmann, A.; Menon, S.; et al. An Introduction to the WHO 5th Edition 2022 Classification of Testicular Tumours. Histopathology 2022, 81, 459–466. [Google Scholar] [CrossRef]
- Baroni, T.; Arato, I.; Mancuso, F.; Calafiore, R.; Luca, G. On the Origin of Testicular Germ Cell Tumors: From Gonocytes to Testicular Cancer. Front. Endocrinol. 2019, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Comiter, C.V.; Kibel, A.S.; Richie, J.P.; Nucci, M.R.; Renshaw, A.A. Prognostic features of teratomas with malignant transformation: A clinicopathological study of 21 cases. J. Urol. 1998, 159, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Oliver, T.R.W.; Chappell, L.; Sanghvi, R.; Deighton, L.; Ansari-Pour, N.; Dentro, S.C.; Young, M.D.; Coorens, T.H.H.; Jung, H.; Butler, T.; et al. Clonal Diversification and Histogenesis of Malignant Germ Cell Tumours. Nat. Commun. 2022, 13, 4272. [Google Scholar] [CrossRef]
- Gopalan, A.; Dhall, D.; Olgac, S.; Fine, S.W.; Korkola, J.E.; Houldsworth, J.; Chaganti, R.S.; Bosl, G.J.; Reuter, V.E.; Tickoo, S.K. Testicular Mixed Germ Cell Tumors: A Morphological and Immunohistochemical Study Using Stem Cell Markers, OCT3/4, SOX2 and GDF3, with Emphasis on Morphologically Difficult-to-Classify Areas. Mod. Pathol. 2009, 22, 1066–1074. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, E.; Antonelli, L.; Albers, P.; Cary, C.; Gillessen Sommer, S.; Heidenreich, A.; Oing, C.; Oldenburg, J.; Pierorazio, P.M.; Stephenson, A.J.; et al. Oncological Follow-up Strategies for Testicular Germ Cell Tumours: A Narrative Review. Eur. Urol. Open Sci. 2022, 44, 142–149. [Google Scholar] [CrossRef]
- Zengerling, F.; Beyersdorff, D.; Busch, J.; Heinzelbecker, J.; Pfister, D.; Ruf, C.; Winter, C.; Albers, P.; Kliesch, S.; Schmidt, S. Prognostic Factors in Patients with Clinical Stage I Nonseminoma—Beyond Lymphovascular Invasion: A Systematic Review. World J. Urol. 2022, 40, 2879–2887. [Google Scholar] [CrossRef]
- Lobo, J.; Gillis, A.J.M.; van den Berg, A.; Looijenga, L.H.J. Prediction of Relapse in Stage I Testicular Germ Cell Tumor Patients on Surveillance: Investigation of Biomarkers. BMC Cancer 2020, 20, 728. [Google Scholar] [CrossRef]
- Pierorazio, P.M.; Cheaib, J.G.; Tema, G.; Patel, H.D.; Gupta, M.; Sharma, R.; Zhang, A.; Bass, E.B. Performance Characteristics of Clinical Staging Modalities for Early Stage Testicular Germ Cell Tumors: A Systematic Review. J. Urol. 2020, 203, 894–901. [Google Scholar] [CrossRef] [PubMed]
- von Eyben, F.E.; Jensen, M.B.; Høyer, S. Frequency and Markers of Precursor Lesions and Implications for the Pathogenesis of Testicular Germ Cell Tumors. Clin. Genitourin. Cancer 2018, 16, e211. [Google Scholar] [CrossRef]
- Ye, H.; Ulbright, T.M. Difficult Differential Diagnoses in Testicular Pathology. Arch. Pathol. Lab. Med. 2012, 136, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Rajpert-De Meyts, E.; Jørgensen, N.; Petersen, J.H.; Almstrup, K.; Aksglaede, L.; Lauritsen, J.; Rørth, M.; Daugaard, G.; Skakkebæk, N.E. Optimized Detection of Germ Cell Neoplasia in Situ in Contralateral Biopsy Reduces the Risk of Second Testis Cancer. BJU Int. 2022, 130, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Basiri, A.; Movahhed, S.; Parvin, M.; Salimi, M.; Rezaeetalab, G.H. The Histologic Features of Intratubular Germ Cell Neoplasia and Its Correlation with Tumor Behavior. Investig. Clin. Urol. 2016, 57, 191. [Google Scholar] [CrossRef]
- Krasic, J.; Skara, L.; Bojanac, A.K.; Ulamec, M.; Jezek, D.; Kulis, T.; Sincic, N. The Utility of CfDNA in TGCT Patient Management: A Systematic Review. Ther. Adv. Med. Oncol. 2022, 14, 175883592210903. [Google Scholar] [CrossRef]
- Nason, G.J.; Sweet, J.; Landoni, L.; Leao, R.; Anson-Cartwright, L.; Mok, S.; Guzylak, V.; D’Angelo, A.; Fang, Z.Y.; Geist, I.; et al. Discrepancy in Pathology Reports upon Second Review of Radical Orchiectomy Specimens for Testicular Germ Cell Tumors. Can. Urol. Assoc. J. 2020, 14, 411–415. [Google Scholar] [CrossRef]
- Holland, P.; Karmas, E.; Merrimen, J.; Wood, L.A. Accuracy of Germ Cell Tumor Histology and Stage within a Canadian Cancer Registry. Can. Urol. Assoc. J. 2022, 17, 44–48. [Google Scholar] [CrossRef]
- Lobo, J.; Leão, R. Editorial: Diagnostic and Predictive Biomarkers in Testicular Germ Cell Tumors. Front. Oncol. 2022, 12, 1027363. [Google Scholar] [CrossRef]
- Jamin, S.P.; Hikmet, F.; Mathieu, R.; Jégou, B.; Lindskog, C.; Chalmel, F.; Primig, M. Combined RNA/Tissue Profiling Identifies Novel Cancer/Testis Genes. Mol. Oncol. 2021, 15, 3003–3023. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Raos, D.; Oršolić, D.; Mašić, S.; Tomić, M.; Krasić, J.; Tomašković, I.; Gabaj, N.N.; Gelo, N.; Kaštelan, Ž.; Kuliš, T.; et al. CfDNA Methylation in Liquid Biopsies as Potential Testicular Seminoma Biomarker. Epigenomics 2022, 14, 1493–1507. [Google Scholar] [CrossRef]
- Raos, D.; Krasic, J.; Masic, S.; Abramovic, I.; Coric, M.; Kruslin, B.; Katusic Bojanac, A.; Bulic-Jakus, F.; Jezek, D.; Ulamec, M.; et al. In Search of TGCT Biomarkers: A Comprehensive In Silico and Histopathological Analysis. Dis. Markers 2020, 2020, 8892312. [Google Scholar] [CrossRef]
- Krasic, J.; Skara, L.; Ulamec, M.; Katusic Bojanac, A.; Dabelic, S.; Bulic-Jakus, F.; Jezek, D.; Sincic, N. Teratoma Growth Retardation by HDACi Treatment of the Tumor Embryonal Source. Cancers 2020, 12, 3416. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Varella-Garcia, M.; Bunn, P.A.; Di Maria, M.V.; Veve, R.; Bremnes, R.M.; Barón, A.E.; Zeng, C.; Franklin, W.A. Epidermal Growth Factor Receptor in Non–Small-Cell Lung Carcinomas: Correlation Between Gene Copy Number and Protein Expression and Impact on Prognosis. J. Clin. Oncol. 2003, 21, 3798–3807. [Google Scholar] [CrossRef]
- Guo, R.; Ma, L.; Bai, X.; Miao, L.; Li, Z.; Yang, J. A Scoring Method for Immunohistochemical Staining on Ki67. Appl. Immunohistochem. Mol. Morphol. 2021, 29, e20–e28. [Google Scholar] [CrossRef]
- Siegmund, S.E.; Mehra, R.; Acosta, A.M. An Update on Diagnostic Tissue-Based Biomarkers in Testicular Tumors. Hum. Pathol. 2023, 133, 32–55. [Google Scholar] [CrossRef]
- Ulbright, T.M.; Tickoo, S.K.; Berney, D.M.; Srigley, J.R. Best Practices Recommendations in the Application of Immunohistochemistry in Testicular Tumors: Report From the International Society of Urological Pathology Consensus Conference. Am. J. Surg. Pathol. 2014, 38, e50–e59. [Google Scholar] [CrossRef] [Green Version]
- Hart, A.H.; Hartley, L.; Parker, K.; Ibrahim, M.; Looijenga, L.H.J.; Pauchnik, M.; Chow, C.W.; Robb, L. The Pluripotency Homeobox Gene NANOG Is Expressed in Human Germ Cell Tumors. Cancer 2005, 104, 2092–2098. [Google Scholar] [CrossRef]
- Liu, A.; Cheng, L.; Du, J.; Peng, Y.; Allan, R.W.; Wei, L.; Li, J.; Cao, D. Diagnostic Utility of Novel Stem Cell Markers SALL4, OCT4, NANOG, SOX2, UTF1, and TCL1 in Primary Mediastinal Germ Cell Tumors. Am. J. Surg. Pathol. 2010, 34, 697–706. [Google Scholar] [CrossRef]
- Sonne, S.B.; Perrett, R.M.; Nielsen, J.E.; Baxter, M.A.; Kristensen, D.M.; Leffers, H.; Hanley, N.A.; Rajpert-de-Meyts, E. Analysis of SOX2 Expression in Developing Human Testis and Germ Cell Neoplasia. Int. J. Dev. Biol. 2010, 54, 755–760. [Google Scholar] [CrossRef] [Green Version]
- Nonaka, D. Differential Expression of SOX2 and SOX17 in Testicular Germ Cell Tumors. Am. J. Clin. Pathol. 2009, 131, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Li, J.; Guo, C.C.; Allan, R.W.; Humphrey, P.A. SALL4 Is a Novel Diagnostic Marker for Testicular Germ Cell Tumors. Am. J. Surg. Pathol. 2009, 33, 1065–1077. [Google Scholar] [CrossRef]
- Miettinen, M.; Wang, Z.; McCue, P.A.; Sarlomo-Rikala, M.; Rys, J.; Biernat, W.; Lasota, J.; Lee, Y.-S. SALL4 Expression in Germ Cell and Non–Germ Cell Tumors: A Systematic Immunohistochemical Study of 3215 Cases. Am. J. Surg. Pathol. 2014, 38, 410–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, S.K.; Weiss, L.M.; Chu, P.G. D2-40 Immunohistochemistry in the Differential Diagnosis of Seminoma and Embryonal Carcinoma: A Comparative Immunohistochemical Study with KIT (CD117) and CD30. Mod. Pathol. 2007, 20, 320–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lind, G.E.; Skotheim, R.I.; Lothe, R.A. The Epigenome of Testicular Germ Cell Tumors. APMIS 2007, 115, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Skotheim, R.I.; Abeler, V.M.; Nesland, J.M.; Fosså, S.D.; Holm, R.; Wagner, U.; Flørenes, V.A.; Aass, N.; Kallioniemi, O.P.; Lothe, R.A. Candidate Genes for Testicular Cancer Evaluated by In Situ Protein Expression Analyses on Tissue Microarrays. Neoplasia 2003, 5, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, P.K.; Barghorn, A.; Fritzsche, F.R.; Riener, M.-O.; Kristiansen, G.; Knuth, A.; Moch, H. MAGEC2 Is a Sensitive and Novel Marker for Seminoma: A Tissue Microarray Analysis of 325 Testicular Germ Cell Tumors. Mod. Pathol. 2011, 24, 829–835. [Google Scholar] [CrossRef] [Green Version]
- Stoop, H.; Honecker, F.; van de Geijn, G.; Gillis, A.; Cools, M.; de Boer, M.; Bokemeyer, C.; Wolffenbuttel, K.; Drop, S.; de Krijger, R.; et al. Stem Cell Factor as a Novel Diagnostic Marker for Early Malignant Germ Cells. J. Pathol. 2008, 216, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Strohmeyer, T.; Reese, D.; Press, M.; Ackermann, R.; Hartmann, M.; Slamon, D. Expression of the C-Kit Proto-Oncogene and Its Ligand Stem Cell Factor (SCF) in Normal and Malignant Human Testicular Tissue. J. Urol. 1995, 153, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Spiller, C.M.; Gillis, A.J.M.; Burnet, G.; Stoop, H.; Koopman, P.; Bowles, J.; Looijenga, L.H.J. Cripto: Expression, Epigenetic Regulation and Potential Diagnostic Use in Testicular Germ Cell Tumors. Mol. Oncol. 2016, 10, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Spiller, C.M.; Feng, C.-W.; Jackson, A.; Gillis, A.J.M.; Rolland, A.D.; Looijenga, L.H.J.; Koopman, P.; Bowles, J. Endogenous Nodal Signaling Regulates Germ Cell Potency during Mammalian Testis Development. Development 2012, 139, 4123–4132. [Google Scholar] [CrossRef] [Green Version]
- Ezeh, U.I.; Turek, P.J.; Reijo, R.A.; Clark, A.T. Human Embryonic Stem Cell GenesOCT4, NANOG, STELLAR, AndGDF3 Are Expressed in Both Seminoma and Breast Carcinoma. Cancer 2005, 104, 2255–2265. [Google Scholar] [CrossRef]
- Netto, G.J.; Nakai, Y.; Nakayama, M.; Jadallah, S.; Toubaji, A.; Nonomura, N.; Albadine, R.; Hicks, J.L.; Epstein, J.I.; Yegnasubramanian, S.; et al. Global DNA Hypomethylation in Intratubular Germ Cell Neoplasia and Seminoma, but Not in Nonseminomatous Male Germ Cell Tumors. Mod. Pathol. 2008, 21, 1337–1344. [Google Scholar] [CrossRef] [Green Version]
- Nettersheim, D.; Heukamp, L.C.; Fronhoffs, F.; Grewe, M.J.; Haas, N.; Waha, A.; Honecker, F.; Waha, A.; Kristiansen, G.; Schorle, H. Analysis of TET Expression/Activity and 5mC Oxidation during Normal and Malignant Germ Cell Development. PLoS ONE 2013, 8, e82881. [Google Scholar] [CrossRef]
- Bartsch, G.; Jennewein, L.; Harter, P.N.; Antonietti, P.; Blaheta, R.A.; Kvasnicka, H.-M.; Kögel, D.; Haferkamp, A.; Mittelbronn, M.; Mani, J. Autophagy-Associated Proteins BAG3 and P62 in Testicular Cancer. Oncol. Rep. 2016, 35, 1629–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klümper, N.; Syring, I.; Offermann, A.; Adler, D.; Vogel, W.; Müller, S.C.; Ellinger, J.; Strauß, A.; Radzun, H.J.; Ströbel, P.; et al. Differential Expression of Mediator Complex Subunit MED15 in Testicular Germ Cell Tumors. Diagn. Pathol. 2015, 10, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, B.; Haggeney, T.; Fietz, D.; Indumathy, S.; Loveland, K.L.; Hedger, M.; Kliesch, S.; Weidner, W.; Bergmann, M.; Schuppe, H.-C. Specific Immune Cell and Cytokine Characteristics of Human Testicular Germ Cell Neoplasia. Hum. Reprod. 2016, 31, 2192–2202. [Google Scholar] [CrossRef] [Green Version]
- Gayer, F.A.; Fichtner, A.; Legler, T.J.; Reichardt, H.M. A Coculture Model Mimicking the Tumor Microenvironment Unveils Mutual Interactions between Immune Cell Subtypes and the Human Seminoma Cell Line TCam-2. Cells 2022, 11, 885. [Google Scholar] [CrossRef]
- Badia, R.R.; Patel, A.; Chertack, N.; Howard, J.M.; Bagrodia, A.; Bakare, T. Impact of Testicular Cancer Stage on Semen Parameters in Patients before Orchiectomy. Urol. Oncol. Semin. Orig. Investig. 2023, 41, 151.e11–151.e15. [Google Scholar] [CrossRef]
- Skakkebæk, N.E.; Rajpert-De Meyts, E.; Main, K.M. Testicular Dysgenesis Syndrome: An Increasingly Common Developmental Disorder with Environmental Aspects: Opinion. Hum. Reprod. 2001, 16, 972–978. [Google Scholar] [CrossRef] [Green Version]
- Hoei-Hansen, C.E.; Holm, M.; Rajpert-De Meyts, E.; Skakkebaek, N.E. Histological Evidence of Testicular Dysgenesis in Contralateral Biopsies from 218 Patients with Testicular Germ Cell Cancer. J. Pathol. 2003, 200, 370–374. [Google Scholar] [CrossRef]
- Markulin, D.; Vojta, A.; Samaržija, I.; Gamulin, M.; Bečeheli, I.; Jukić, I.; Maglov, Č.; Zoldoš, V.; Fučić, A. Association Between RASSF1A Promoter Methylation and Testicular Germ Cell Tumor: A Meta-Analysis and a Cohort Study. Cancer Genom. Proteom. 2017, 14, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Eildermann, K.; Aeckerle, N.; Debowski, K.; Godmann, M.; Christiansen, H.; Heistermann, M.; Schweyer, S.; Bergmann, M.; Kliesch, S.; Gromoll, J.; et al. Developmental Expression of the Pluripotency Factor Sal-Like Protein 4 in the Monkey, Human and Mouse Testis: Restriction to Premeiotic Germ Cells. Cells Tissues Organs. 2012, 196, 206–220. [Google Scholar] [CrossRef]
- Chan, A.-L.; La, H.M.; Legrand, J.M.D.; Mäkelä, J.-A.; Eichenlaub, M.; De Seram, M.; Ramialison, M.; Hobbs, R.M. Germline Stem Cell Activity Is Sustained by SALL4-Dependent Silencing of Distinct Tumor Suppressor Genes. Stem. Cell Rep. 2017, 9, 956–971. [Google Scholar] [CrossRef] [Green Version]
- Morgia, G. New Advances in Clinical Biomarkers in Testis Cancer. Front. Biosci. 2010, E2, 456–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Liu, Q.; Lin, L.; Zheng, X. Reductions in Calcitonin Gene-Related Peptide May Be Associated with the Impairment of the Contralateral Testis in Unilateral Cryptorchidism. Exp. Ther. Med. 2015, 9, 1797–1800. [Google Scholar] [CrossRef] [PubMed]
- Orsatti, A.; Sirolli, M.; Ambrosi, F.; Franceschini, T.; Giunchi, F.; Franchini, E.; Grillini, M.; Massari, F.; Mollica, V.; Bianchi, F.M.; et al. SOX2 and PRAME in the “Reprogramming” of Seminoma Cells. Pathol. Res. Pract. 2022, 237, 154044. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.; Stoop, H.; Gillis, A.J.M.; Looijenga, L.H.J.; Oosterhuis, W. Interobserver Agreement in Vascular Invasion Scoring and the Added Value of Immunohistochemistry for Vascular Markers to Predict Disease Relapse in Stage I Testicular Nonseminomas. Am. J. Surg. Pathol. 2019, 43, 1711–1719. [Google Scholar] [CrossRef]
- Killian, J.K.; Dorssers, L.C.J.; Trabert, B.; Gillis, A.J.M.; Cook, M.B.; Wang, Y.; Waterfall, J.J.; Stevenson, H.; Smith, W.I.; Noyes, N.; et al. Imprints and DPPA3 Are Bypassed during Pluripotency- and Differentiation-Coupled Methylation Reprogramming in Testicular Germ Cell Tumors. Genome Res. 2016, 26, 1490–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, A.L.; Moreira-Barbosa, C.; Lobo, J.; Vilela-Salgueiro, B.; Cantante, M.; Guimarães, R.; Lopes, P.; Braga, I.; Oliveira, J.; Antunes, L.; et al. DNA Methylation Profiling as a Tool for Testicular Germ Cell Tumors Subtyping. Epigenomics 2018, 10, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, C.M.D.S.; van Helvoort Lengert, A.; Cárcano, F.M.; Silva, E.C.A.; Brait, M.; Lopes, L.F.; Vidal, D.O. MGMT and CALCA Promoter Methylation Are Associated with Poor Prognosis in Testicular Germ Cell Tumor Patients. Oncotarget 2017, 8, 50608–50617. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.; Jaiswal, D.; Singh, K.; Trivedi, S.; Agrawal, N.K.; Gupta, G.; Rajender, S.; Singh, K. Azoospermic Infertility Is Associated with Altered Expression of DNA Repair Genes. DNA Repair 2019, 75, 39–47. [Google Scholar] [CrossRef]
- Bowles, J.; Teasdale, R.P.; James, K.; Koopman, P. Dppa3 Is a Marker of Pluripotency and Has a Human Homologue That Is Expressed in Germ Cell Tumours. Cytogenet. Genome Res. 2003, 101, 261–265. [Google Scholar] [CrossRef]
- Spiller, C.M.; Bowles, J.; Koopman, P. Nodal/Cripto Signaling in Fetal Male Germ Cell Development: Implications for Testicular Germ Cell Tumors. Int. J. Dev. Biol. 2013, 57, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Ranjitha, V.; Khemani, R.; Rao, B.V.; Fonseca, D.; Murthy, S.; Giridhar, A.; Jayakarthik, Y.; Sharma, R.; Raju, K.V.N.; Rao, T.; et al. The Core Four—A Panel of Immunohistochemistry Markers to Diagnose and Subtype Testicular Germ Cell Tumors. Urol. Ann. 2022, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Bonatelli, M.; Silva, E.C.A.; Cárcano, F.M.; Zaia, M.G.; Lopes, L.F.; Scapulatempo-Neto, C.; Pinheiro, C. The Warburg Effect Is Associated With Tumor Aggressiveness in Testicular Germ Cell Tumors. Front. Endocrinol. 2019, 10, 417. [Google Scholar] [CrossRef] [PubMed]
- Barrachina, F.; de la Iglesia, A.; Jodar, M.; Soler-Ventura, A.; Mallofré, C.; Rodriguez-Carunchio, L.; Goudarzi, A.; Corral, J.M.; Ballescà, J.L.; Castillo, J.; et al. Histone H4 Acetylation Is Dysregulated in Active Seminiferous Tubules Adjacent to Testicular Tumours. Hum. Reprod. 2022, 37, 1712–1726. [Google Scholar] [CrossRef]
- Bozoky, B.; Szekely, L.; Ernberg, I.; Savchenko, A. AtlasGrabber: A Software Facilitating the High Throughput Analysis of the Human Protein Atlas Online Database. BMC Bioinform. 2022, 23, 546. [Google Scholar] [CrossRef]







| HPA Data | ||
|---|---|---|
| Controls | Testis | 3 |
| Diagnosis | TGCT | 17 |
| SE | 9 | |
| EC | 5 | |
| MTGCT | 3 | |
| Clinicopathological Data | ||
|---|---|---|
| TGCT patients | 38 | |
| Age (median, range) | 29.5 (18–45) | |
| TNM | T1 | 21 |
| T2 | 13 | |
| T3 | 4 | |
| Stage | I | 33 |
| II | 2 | |
| III | 3 | |
| TGCT diagnosis | EC | 10 |
| TE | 2 | |
| MTGCT | 26 | |
| Components within MTGCT | EC | 19 |
| TE | 15 | |
| YST | 12 | |
| SE | 18 | |
| CH | 1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krasic, J.; Skara Abramovic, L.; Himelreich Peric, M.; Vanjorek, V.; Gangur, M.; Zovko, D.; Malnar, M.; Masic, S.; Demirovic, A.; Juric, B.; et al. Testicular Germ Cell Tumor Tissue Biomarker Analysis: A Comparison of Human Protein Atlas and Individual Testicular Germ Cell Tumor Component Immunohistochemistry. Cells 2023, 12, 1841. https://doi.org/10.3390/cells12141841
Krasic J, Skara Abramovic L, Himelreich Peric M, Vanjorek V, Gangur M, Zovko D, Malnar M, Masic S, Demirovic A, Juric B, et al. Testicular Germ Cell Tumor Tissue Biomarker Analysis: A Comparison of Human Protein Atlas and Individual Testicular Germ Cell Tumor Component Immunohistochemistry. Cells. 2023; 12(14):1841. https://doi.org/10.3390/cells12141841
Chicago/Turabian StyleKrasic, Jure, Lucija Skara Abramovic, Marta Himelreich Peric, Vedran Vanjorek, Marko Gangur, Dragana Zovko, Marina Malnar, Silvija Masic, Alma Demirovic, Bernardica Juric, and et al. 2023. "Testicular Germ Cell Tumor Tissue Biomarker Analysis: A Comparison of Human Protein Atlas and Individual Testicular Germ Cell Tumor Component Immunohistochemistry" Cells 12, no. 14: 1841. https://doi.org/10.3390/cells12141841