Early Detection of Pre-Cancerous and Cancerous Cells Using Raman Spectroscopy-Based Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Sample Preparation for Raman Measurements
2.3. Raman Measurements
2.4. Spectral Preprocessing
2.5. Machine Learning Analysis
2.6. Principal Component Analysis
2.7. Linear Discriminant Analysis
2.8. Validation
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Didkowska, J.; Wojciechowska, U.; Michalek, I.M.; Caetano dos Santos, F.L. Cancer incidence and mortality in Poland in 2019. Sci. Rep. 2022, 12, 10875. [Google Scholar] [CrossRef]
- Wardle, J.; Robb, K.; Vernon, S.; Waller, J. Screening for prevention and early diagnosis of cancer. Am. Psychol. 2015, 70, 119. [Google Scholar] [CrossRef] [Green Version]
- Parsa, N. Environmental factors inducing human cancers. Iran. J. Public Health 2012, 41, 1. [Google Scholar]
- Baba, A.I.; Câtoi, C. Comparative Oncology; Publishing House of the Romanian Academy: Bucharest, Romania, 2007. [Google Scholar]
- Yancik, R. Cancer burden in the aged: An epidemiologic and demographic overview. Cancer Interdiscip. Int. J. Am. Cancer Soc. 1997, 80, 1273–1283. [Google Scholar] [CrossRef]
- Berger, N.A.; Savvides, P.; Koroukian, S.M.; Kahana, E.F.; Deimling, G.T.; Rose, J.H.; Bowman, K.F.; Miller, R.H. Cancer in the elderly. Trans. Am. Clin. Climatol. Assoc. 2006, 117, 147. [Google Scholar]
- Hu, Z.; Tang, J.; Wang, Z.; Zhang, K.; Zhang, L.; Sun, Q. Deep learning for image-based cancer detection and diagnosis—A survey. Pattern Recognit. 2018, 83, 134–149. [Google Scholar] [CrossRef]
- Smith, R.A.; Cokkinides, V.; Eyre, H.J. Cancer screening in the United States, 2007: A review of current guidelines, practices, and prospects. CA A Cancer J. Clin. 2007, 57, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Galway, K.; Black, A.; Cantwell, M.M.; Cardwell, C.R.; Mills, M.; Donnelly, M. Psychosocial interventions to improve quality of life and emotional wellbeing for recently diagnosed cancer patients. Cochrane Database Syst. Rev. 2012, 11, CD007064. [Google Scholar]
- Salman, A.; Shufan, E.; Zeiri, L.; Huleihel, M. Detection and identification of cancerous murine fibroblasts, transformed by murine sarcoma virus in culture, using Raman spectroscopy and advanced statistical methods. Biochim. Biophys. Acta 2013, 1830, 2720–2727. [Google Scholar] [CrossRef]
- Das Gupta, S.; Finnilä, M.A.J.; Karhula, S.S.; Kauppinen, S.; Joukainen, A.; Kröger, H.; Korhonen, R.K.; Thambyah, A.; Rieppo, L.; Saarakkala, S. Raman microspectroscopic analysis of the tissue-specific composition of the human osteochondral junction in osteoarthritis: A pilot study. Acta Biomater. 2020, 106, 145–155. [Google Scholar] [CrossRef]
- Zhang, F.; Tan, Y.; Ding, J.; Cao, D.; Gong, Y.; Zhang, Y.; Yang, J.; Yin, T. Application and Progress of Raman Spectroscopy in Male Reproductive System. Front. Cell Dev. Biol. 2021, 9, 823546. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, O.; Toner, M.; Flint, S.; Byrne, H.J.; Lyng, F.M. The Potential of Raman Spectroscopy in the Diagnosis of Dysplastic and Malignant Oral Lesions. Cancers 2021, 13, 619. [Google Scholar] [CrossRef] [PubMed]
- Ralbovsky, N.M.; Lednev, I.K. Towards development of a novel universal medical diagnostic method: Raman spectroscopy and machine learning. Chem. Soc. Rev. 2020, 49, 7428–7453. [Google Scholar] [CrossRef]
- Huang, Y.; Swarup, V.P.; Bishnoi, S.W. Rapid Raman imaging of stable, functionalized nanoshells in mammalian cell cultures. Nano Lett. 2009, 9, 2914–2920. [Google Scholar] [CrossRef]
- Salman, A.; Shufan, E.; Zeiri, L.; Huleihel, M. Characterization and detection of Vero cells infected with Herpes Simplex Virus type 1 using Raman spectroscopy and advanced statistical methods. Methods 2014, 68, 364–370. [Google Scholar] [CrossRef]
- Huleihel, M.; Shufan, E.; Zeiri, L.; Salman, A. Detection of Vero Cells Infected with Herpes Simplex Types 1 and 2 and Varicella Zoster Viruses Using Raman Spectroscopy and Advanced Statistical Methods. PLoS ONE 2016, 11, e0153599. [Google Scholar] [CrossRef] [Green Version]
- Silveira, L., Jr.; Sathaiah, S.; Zângaro, R.A.; Pacheco, M.T.; Chavantes, M.C.; Pasqualucci, C.A. Near-infrared Raman spectroscopy of human coronary arteries: Histopathological classification based on Mahalanobis distance. J. Clin. Laser Med. Surg. 2003, 21, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Ramos, I.R.M.; Malkin, A.; Lyng, F.M. Current advances in the application of Raman spectroscopy for molecular diagnosis of cervical cancer. BioMed Res. Int. 2015, 2015, 561242. [Google Scholar] [CrossRef] [Green Version]
- Rubina, S.; Krishna, C.M. Raman spectroscopy in cervical cancers: An update. J. Cancer Res. Ther. 2015, 11, 10. [Google Scholar]
- Santos, I.P.; Caspers, P.J.; Bakker Schut, T.C.; van Doorn, R.; Noordhoek Hegt, V.; Koljenović, S.; Puppels, G.J. Raman spectroscopic characterization of melanoma and benign melanocytic lesions suspected of melanoma using high-wavenumber Raman spectroscopy. Anal. Chem. 2016, 88, 7683–7688. [Google Scholar] [CrossRef]
- Lui, H.; Zhao, J.; McLean, D.; Zeng, H. Real-time Raman Spectroscopy for In Vivo Skin Cancer DiagnosisRaman Spectroscopy of Skin Cancer. Cancer Res. 2012, 72, 2491–2500. [Google Scholar] [CrossRef] [Green Version]
- Haka, A.S.; Shafer-Peltier, K.E.; Fitzmaurice, M.; Crowe, J.; Dasari, R.R.; Feld, M.S. Diagnosing breast cancer by using Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 12371–12376. [Google Scholar] [CrossRef]
- Lazaro-Pacheco, D.; Shaaban, A.M.; Rehman, S.; Rehman, I. Raman spectroscopy of breast cancer. Appl. Spectrosc. Rev. 2020, 55, 439–475. [Google Scholar] [CrossRef]
- Hanna, K.; Krzoska, E.; Shaaban, A.M.; Muirhead, D.; Abu-Eid, R.; Speirs, V. Raman spectroscopy: Current applications in breast cancer diagnosis, challenges and future prospects. Br. J. Cancer 2022, 126, 1125–1139. [Google Scholar] [CrossRef]
- Zheng, Q.; Li, J.; Yang, L.; Zheng, B.; Wang, J.; Lv, N.; Luo, J.; Martin, F.L.; Liu, D.; He, J. Raman spectroscopy as a potential diagnostic tool to analyse biochemical alterations in lung cancer. Analyst 2020, 145, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Karnachoriti, M.; Stathopoulos, I.; Kouri, M.; Spyratou, E.; Orfanoudakis, S.; Lykidis, D.; Lambropoulou, Μ.; Danias, N.; Arkadopoulos, N.; Efstathopoulos, E. Biochemical differentiation between cancerous and normal human colorectal tissues by micro-Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 299, 122852. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Gasser, S. Generating Primary Fibroblast Cultures from Mouse Ear and Tail Tissues. J. Vis. Exp. 2016, 107, e53565. [Google Scholar] [CrossRef] [Green Version]
- Berman, J.J.; Albores-Saavedra, J.; Bostwick, D.; DeLellis, R.; Eble, J.; Hamilton, S.R.; Hruban, R.H.; Mutter, G.L.; Page, D.; Rohan, T. Precancer: A conceptual working definition: Results of a Consensus Conference. Cancer Detect. Prev. 2006, 30, 387–394. [Google Scholar] [CrossRef]
- Marim, F.M.; Silveira, T.N.; Lima Jr, D.S.; Zamboni, D.S. A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS ONE 2010, 5, e15263. [Google Scholar] [CrossRef]
- Stone, N.; Kendall, C.; Smith, J.; Crow, P.; Barr, H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004, 126, 141–157. [Google Scholar] [CrossRef]
- Auner, G.W.; Koya, S.K.; Huang, C.; Broadbent, B.; Trexler, M.; Auner, Z.; Elias, A.; Mehne, K.C.; Brusatori, M.A. Applications of Raman spectroscopy in cancer diagnosis. Cancer Metastasis Rev. 2018, 37, 691–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, H.; Baranska, M. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib. Spectrosc. 2007, 43, 13–25. [Google Scholar] [CrossRef]
- Teh, S.K.; Zheng, W.; Ho, K.Y.; Teh, M.; Yeoh, K.G.; Huang, Z. Near-infrared Raman spectroscopy for optical diagnosis in the stomach: Identification of helicobacter-pylori infection and intestinal metaplasia. Int. J. Cancer 2010, 126, 1920–1927. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.; Kendall, C.; Shepherd, N.; Crow, P.; Barr, H. Near-infrared Raman spectroscopy for the classification of epithelial pre-cancers and cancers. J. Raman Spectrosc. 2002, 33, 564–573. [Google Scholar] [CrossRef]
- Chan, J.W.; Taylor, D.S.; Zwerdling, T.; Lane, S.M.; Ihara, K.; Huser, T. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys. J. 2006, 90, 648–656. [Google Scholar] [CrossRef] [Green Version]
- Cheng, W.T.; Liu, M.T.; Liu, H.N.; Lin, S.Y. Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma. Microsc. Res. Tech. 2005, 68, 75–79. [Google Scholar] [CrossRef]
- Gaeta, R.; Franchi, A.; Capanna, R.; Mario, D.A. Contribution of Raman Spectroscopy to diagnosis and grading of chondrogenic tumours. Virchows Arch. 2019, 475, S43–S44. [Google Scholar]
- Ye, J.; Tian, Z.; Wei, H.; Li, Y. Baseline correction method based on improved asymmetrically reweighted penalized least squares for the Raman spectrum. Appl. Opt. 2020, 59, 10933–10943. [Google Scholar] [CrossRef]
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4. [Google Scholar]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification; Wiley: Hoboken, NJ, USA, 2001. [Google Scholar]
- Salman, A.; Lapidot, I.; Pomerantz, A.; Tsror, L.; Shufan, E.; Moreh, R.; Mordechai, S.; Huleihel, M. Identification of fungal phytopathogens using Fourier transform infrared-attenuated total reflection spectroscopy and advanced statistical methods. J. Biomed. Opt. 2012, 17, 017002. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J.H.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: Berlin/Heidelberg, Germany, 2009; Volume 2. [Google Scholar]
- Subasi, A.; Gursoy, M.I. EEG signal classification using PCA, ICA, LDA and support vector machines. Expert Syst. Appl. 2010, 37, 8659–8666. [Google Scholar] [CrossRef]
- Pardee, A.B. Cancer Cells and Normal Cells. Proc. Am. Philos. Soc. 1976, 120, 87–92. [Google Scholar]
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein glycosylation in cancer. Annu. Rev. Pathol. 2015, 10, 473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, A.J.; Puzio-Kuter, A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010, 330, 1340–1344. [Google Scholar] [CrossRef] [Green Version]
- Fadaka, A.; Ajiboye, B.; Ojo, O.; Adewale, O.; Olayide, I.; Emuowhochere, R. Biology of glucose metabolization in cancer cells. J. Oncol. Sci. 2017, 3, 45–51. [Google Scholar] [CrossRef]


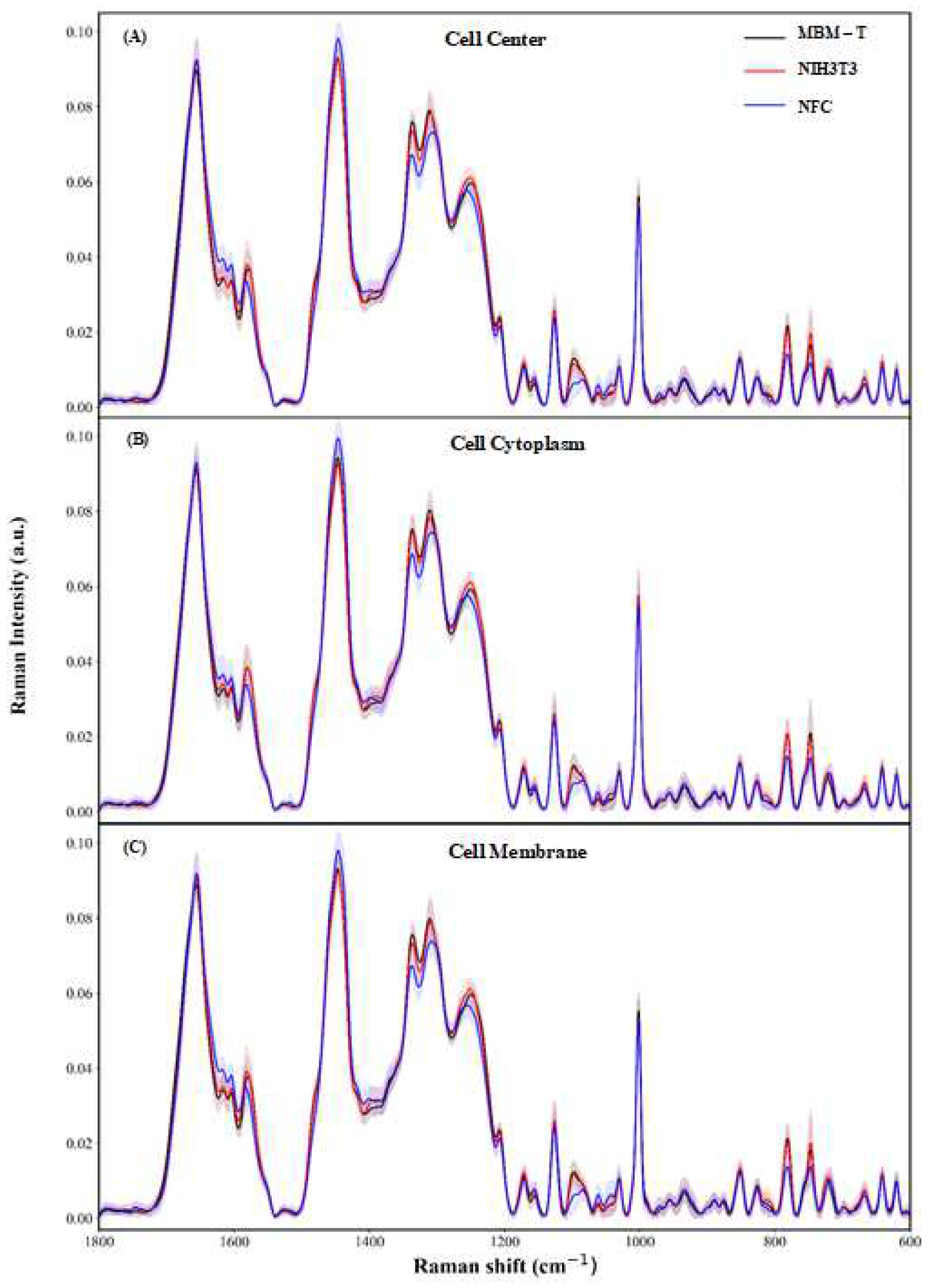

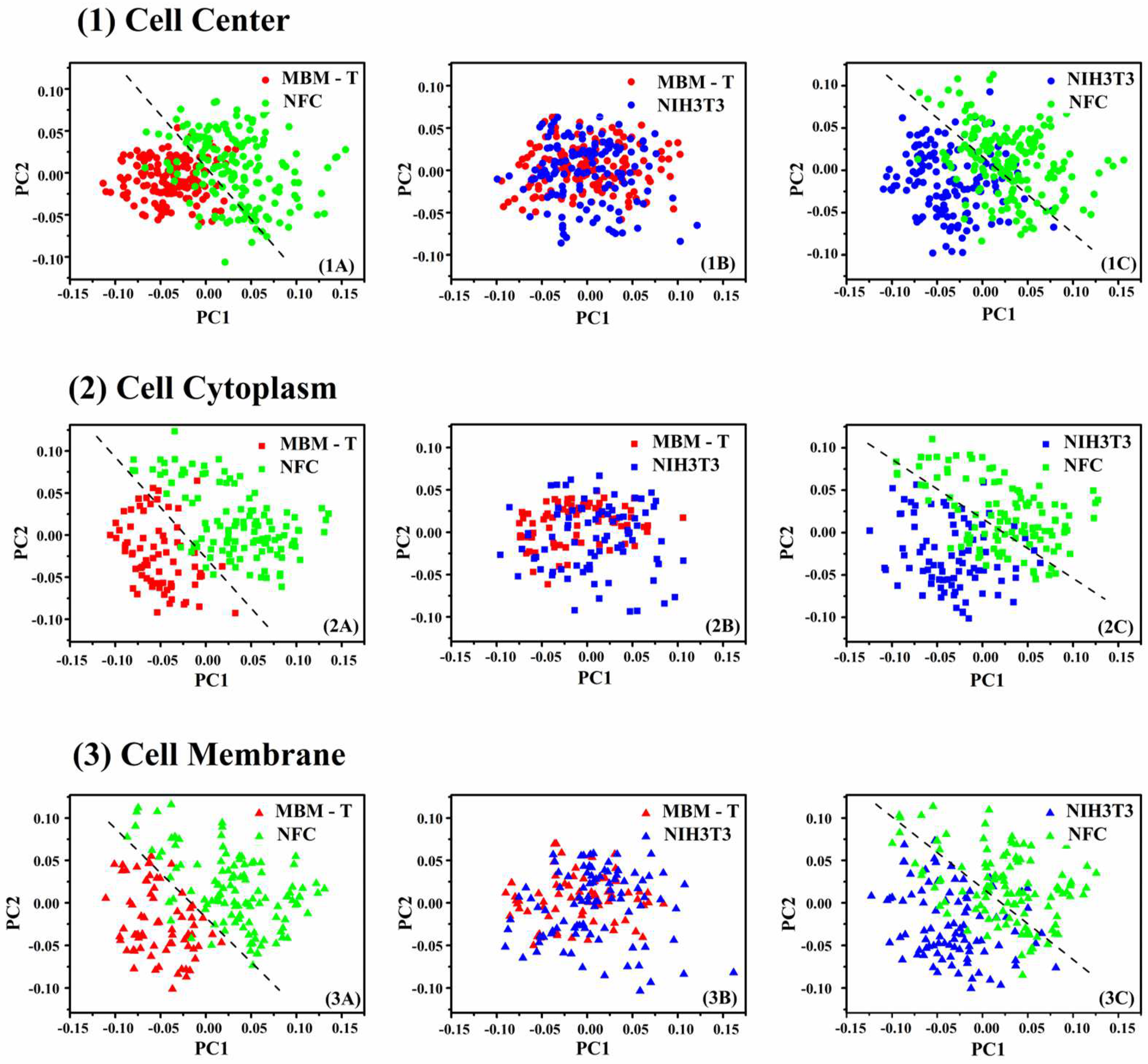
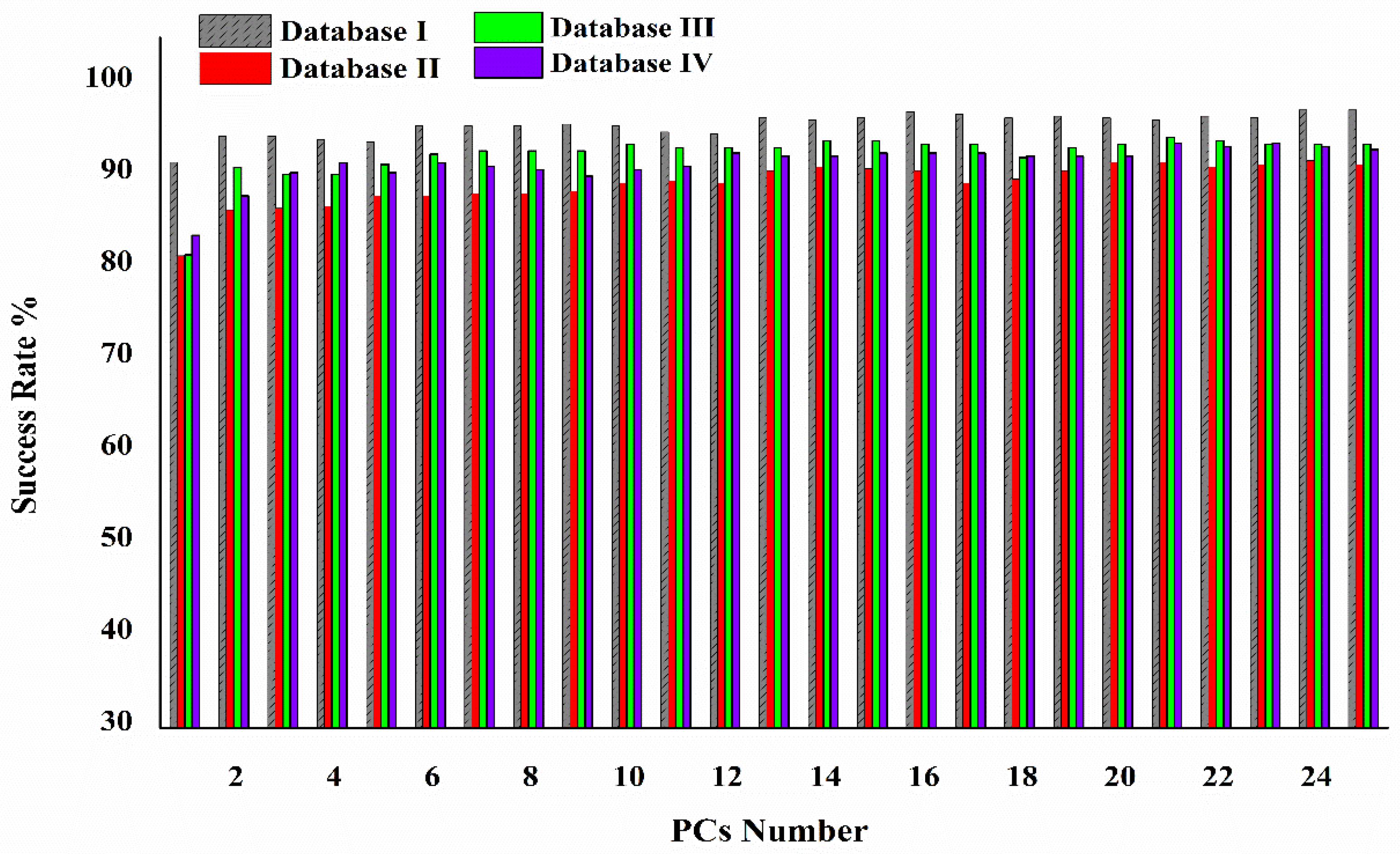
| NFC | NIH/3T3 | MBMT | Total | ||
|---|---|---|---|---|---|
| No. of measurements | Cell center | 169 | 132 | 143 | 444 |
| Cytoplasm | 115 | 86 | 73 | 274 | |
| Rich in membrane | 116 | 91 | 72 | 279 | |
| Total No. of measurements | 400 | 309 | 288 | 997 | |
| No. of cells | 169 | 136 | 152 | 457 | |
| Peak Position/cm−1 | Major Assignments |
|---|---|
| 1745 | vw υ(C=O) lipid |
| 1657 | vs Amide I |
| 1618 | s υ(C=C) Tyr, Trp |
| 1602 | s υ(C=C) Phe, Tyr |
| 1578 | s DNA: A, G, C=C, δ(N–H) and υ(C–N) amide II |
| 1528 | vw–C=C–carotenoid |
| 1443 | vs δ(CH2) lipids, υ(C–H) proteins (collagen) |
| 1398 | s δ(CH2), C=O symmetric stretching |
| 1337 | s CH3CH2 γ of collagen and polynucleotide chain (DNA bases) |
| 1311 | s CH3CH2 τ of lipids, collagen, Trp |
| 1250 | s Amide III |
| 1209 | m υ(C–C6H5), Trp, Phe, Tyr |
| 1175 | w υ(C–H) Tyr, Phe, Cyt, G |
| 1156 | w υ (C–C, C–N) proteins, carotenoids |
| 1128 | m υ (C–N, C–C) Skeletal |
| 1096 | w symmetric υ(PO2−) DNA BK, υ(C–N) |
| 1088 | w υ(C–C) skeletal, υ(C–N) proteins, υ(C–C) lipid |
| 1064 | vw Proline (assignment to collagen) |
| 1045 | vw Collagen |
| 1032 | w υ(C–H) Phe, ν(C–C) skeletal, υ(C–N) proteins |
| 1001 | s υ(C–C), Symmetric ring breathing mode of Phe |
| 971 | vw Cyt (DNA and RNA) |
| 957 | w υ(C–C) hydroxyapatite, carotenoid, cholesterol, υ(PO4−3), (CH3) proteins (α-helix) |
| 934 | w υ(C–C) skeletal, proline, valine, protein BK (α-helix conformation), glycogen |
| 889 | w BK, proteins C–C skeletal |
| 874 | w υ(C–C) Hydroxyproline, Trp |
| 850 | w Ring breathing mode of Tyr, υ(C–C) proline ring |
| 826 | w υ(O–P–O) DNA, proteins, phosphodiester, proline, hydroxyproline, Tyr |
| 811 | vw phosphodiester, υ(O–P–O) RNA |
| 781 | m υ(O–P–O) DNA, RNA, T, Cyt, U, phosphodiester, BK |
| 750 | m Symmetric breathing of Trp, υ(C–S) Cys, DNA: T |
| 721 | w υ(C–S, C–C) protein, CH2 rocking, C–N (membrane phospholipid head) |
| 668 | w υ(C–S) Cys |
| 642 | w υ(C–C) τ Tyr, υ(C–S) protein, Phe |
| 619 | w υ(C–C) τ Phe, protein |
| Predicted | |||
|---|---|---|---|
| Positive | Negative | ||
| TRUE | Positive | True Positive (TP) | False Negative (FN) |
| Negative | False Positive (FP) | True Negative (TN) | |
| (a) NFC versus (NIH/3T3 + MBM-T) | ||||||||
| Database | No. of NFC | No. of Abnormal | No. of PCs | Acc (%) | SE (%) | SP (%) | PPV (%) | NPV (%) |
| I | 169 | 288 | 2 | 93.1 ± 3.2 | 97 | 96.9 | 94.8 | 98.2 |
| II | 169 | 275 | 4 | 88.4 ± 3.6 | 87 | 94.2 | 90.2 | 92.2 |
| III | 115 | 159 | 6 | 92.2 ± 3.1 | 93 | 95 | 93 | 95 |
| IV | 116 | 163 | 4 | 92.5 ± 2.9 | 89.7 | 96.3 | 94.5 | 92.9 |
| (b) NIH/3T3 versus MBM-T | ||||||||
| Database | No. of NIH/3T3 | No. of MBM-T | No. of PCs | Acc (%) | SE (%) | SP (%) | PPV (%) | NPV (%) |
| I | 136 | 152 | 13 | 81.7 ± 3.3 | 77.9 | 85.5 | 82.8 | 81.3 |
| II | 132 | 143 | 20 | 80.2 ± 2.9 | 78 | 83.2 | 81.1 | 80.4 |
| III | 86 | 73 | 10 | 79.9 ± 2.5 | 74.4 | 86.3 | 86.5 | 74.1 |
| IV | 91 | 72 | 10 | 80.2 ± 2.1 | 82.5 | 80.6 | 81.4 | 79.5 |
| (c) NFC versus NIH/3T3 | ||||||||
| Database | No. of NFC | No. of NIH/3T3 | No. of PCs | Acc (%) | SE (%) | SP (%) | PPV (%) | NPV (%) |
| I | 169 | 136 | 3 | 93.7 ± 3.6 | 98.2 | 92.6 | 94.3 | 97.7 |
| II | 169 | 132 | 5 | 87.4 ± 3.4 | 92.9 | 84.8 | 88.7 | 90.3 |
| III | 115 | 86 | 5 | 93.0 ± 3.2 | 94.8 | 90.7 | 93.2 | 92.9 |
| IV | 116 | 91 | 5 | 89.3 ± 3.1 | 90.1 | 88.2 | 92.1 | 87.2 |
| (d) NFC versus MBM-T | ||||||||
| Database | No. of NFC | No. of MBM-T | No. of PCs | Acc (%) | SE (%) | SP (%) | PPV (%) | NPV (%) |
| I | 169 | 152 | 2 | 96.5 ± 3.5 | 98.8 | 96.1 | 96.5 | 98.6 |
| II | 169 | 143 | 6 | 90.6 ± 2.9 | 92.9 | 94.4 | 95.2 | 91.8 |
| III | 115 | 73 | 5 | 94.1 ± 3.1 | 95.7 | 91.8 | 94.8 | 93.1 |
| IV | 116 | 72 | 5 | 93.6 ± 3.3 | 94 | 93.1 | 95.6 | 90.5 |
| (a) NFC versus (NIH/3T3 and MBM-T) (Normal–Abnormal) | ||||||||
| Region | No. of NFC | No. of (NIH/3T3 + MBM-T) | No. of PCs | Acc (%) | SE (%) | SP (%) | PPV (%) | NPV (%) |
| 1195–600 | 169 | 288 | 4 | 94.5 | 95.3 | 94.1 | 90.4 | 97.1 |
| 1380–1196 | 10 | 94.4 | 93.5 | 95.1 | 91.9 | 96.1 | ||
| 1520–1381 | 21 | 93 | 92.9 | 93.1 | 88.7 | 95.7 | ||
| 1728–1521 | 10 | 92.8 | 92.9 | 92.7 | 88.2 | 95.7 | ||
| (b) NIH/3T3 versus MBM-T (Cancerous–Precancerous) | ||||||||
| No. of NFC | No. of NIH/3T3 | No. of PCs | Acc (%) | SE (%) | SP (%) | PPV (%) | NPV (%) | |
| 1195–600 | 136 | 152 | 32 | 72.2 | 67.6 | 76.3 | 71.9 | 72.5 |
| 1380–1196 | 12 | 72.9 | 64.7 | 80.3 | 74.6 | 71.8 | ||
| 1520–1381 | 8 | 67.4 | 61 | 73 | 66.9 | 67.7 | ||
| 1728–1521 | 11 | 71.9 | 69.1 | 74.3 | 70.7 | 72.9 | ||
| (c) NFC versus MBM-T (Normal–Cancerous) | ||||||||
| No. of NFC | No. of MBM-T | No. of PCs | Acc (%) | SE (%) | SP (%) | PPV (%) | NPV (%) | |
| 1195–600 | 169 | 152 | 11 | 97.8 | 97.6 | 98 | 98.2 | 97.4 |
| 1380–1196 | 8 | 96 | 97.6 | 94.1 | 94.8 | 97.3 | ||
| 1520–1381 | 10 | 95 | 95.9 | 94.1 | 94.7 | 95.3 | ||
| 1728–1521 | 6 | 95.3 | 96.4 | 94.1 | 94.8 | 96 | ||
| (d) NFC versus NIH/3T3 (Normal–Precancerous) | ||||||||
| No. of NIH/3T3 | No. of MBM-T | No. of PCs | Acc (%) | SE (%) | SP (%) | PPV (%) | NPV (%) | |
| 1195–600 | 169 | 152 | 4 | 93.8 | 98.2 | 88.2 | 91.2 | 97.6 |
| 1380–1196 | 10 | 93.4 | 95.9 | 90.4 | 92.6 | 94.6 | ||
| 1520–1381 | 47 | 93.4 | 96.4 | 89.7 | 92.1 | 95.3 | ||
| 1728–1521 | 8 | 90.2 | 95.9 | 83.1 | 87.6 | 94.2 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharaha, U.; Hania, D.; Lapidot, I.; Salman, A.; Huleihel, M. Early Detection of Pre-Cancerous and Cancerous Cells Using Raman Spectroscopy-Based Machine Learning. Cells 2023, 12, 1909. https://doi.org/10.3390/cells12141909
Sharaha U, Hania D, Lapidot I, Salman A, Huleihel M. Early Detection of Pre-Cancerous and Cancerous Cells Using Raman Spectroscopy-Based Machine Learning. Cells. 2023; 12(14):1909. https://doi.org/10.3390/cells12141909
Chicago/Turabian StyleSharaha, Uraib, Daniel Hania, Itshak Lapidot, Ahmad Salman, and Mahmoud Huleihel. 2023. "Early Detection of Pre-Cancerous and Cancerous Cells Using Raman Spectroscopy-Based Machine Learning" Cells 12, no. 14: 1909. https://doi.org/10.3390/cells12141909
APA StyleSharaha, U., Hania, D., Lapidot, I., Salman, A., & Huleihel, M. (2023). Early Detection of Pre-Cancerous and Cancerous Cells Using Raman Spectroscopy-Based Machine Learning. Cells, 12(14), 1909. https://doi.org/10.3390/cells12141909









