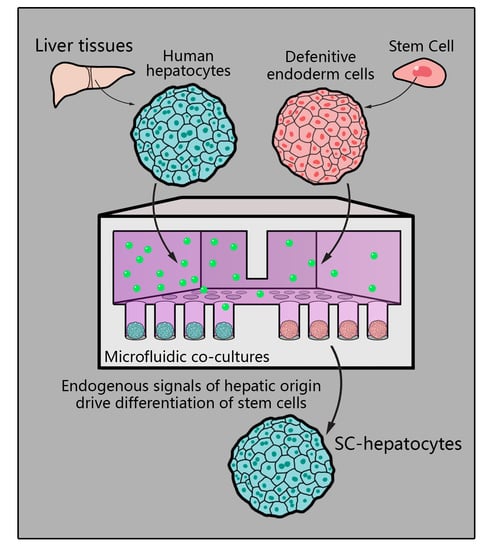
Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microfluidic Co-Culture Device Fabrication
2.1.1. Fabrication of Microfluidic Co-Culture Devices
2.1.2. Mold Fabrications
2.1.3. PDMS Device Fabrication
2.2. Cultures of Human Hepatocytes Isolated from the Liver of Humanized Liver Chimeric Mice
2.3. Hepatic Differentiation of hPSCs under Standard Culture Conditions
2.4. Hepatic Differentiation of hPSCs in Microfluidic 3D Co-Cultures
2.5. Flow Cytometry
2.6. Immunofluorescent Staining of Cells
2.7. ELISA
2.8. RT-PCR Analysis of Hepatic and Stem Cell Gene Expression
2.9. Modeling HGF Secretion and Distribution in the Microfluidic Device
2.10. Statistical Analysis
3. Results
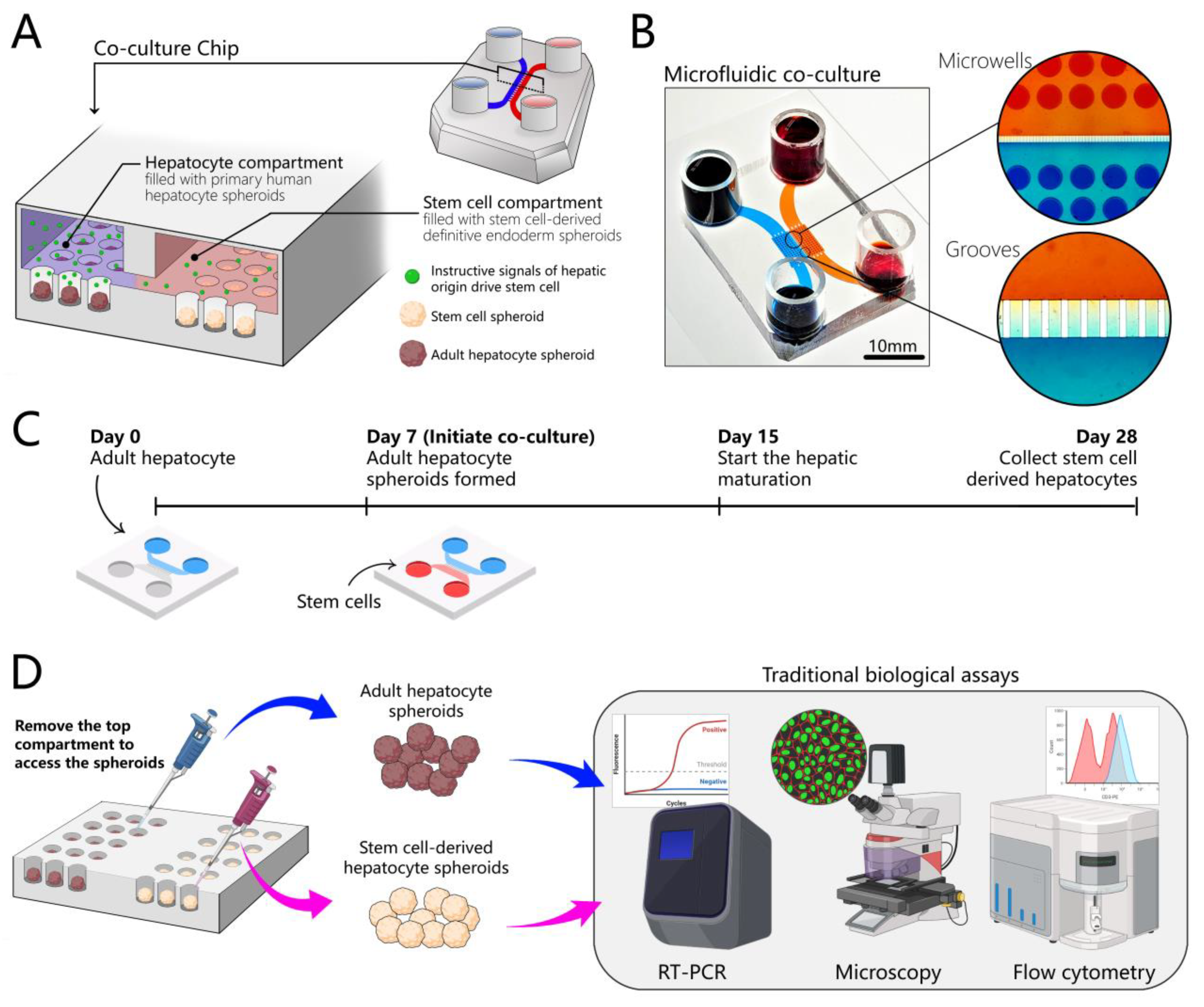
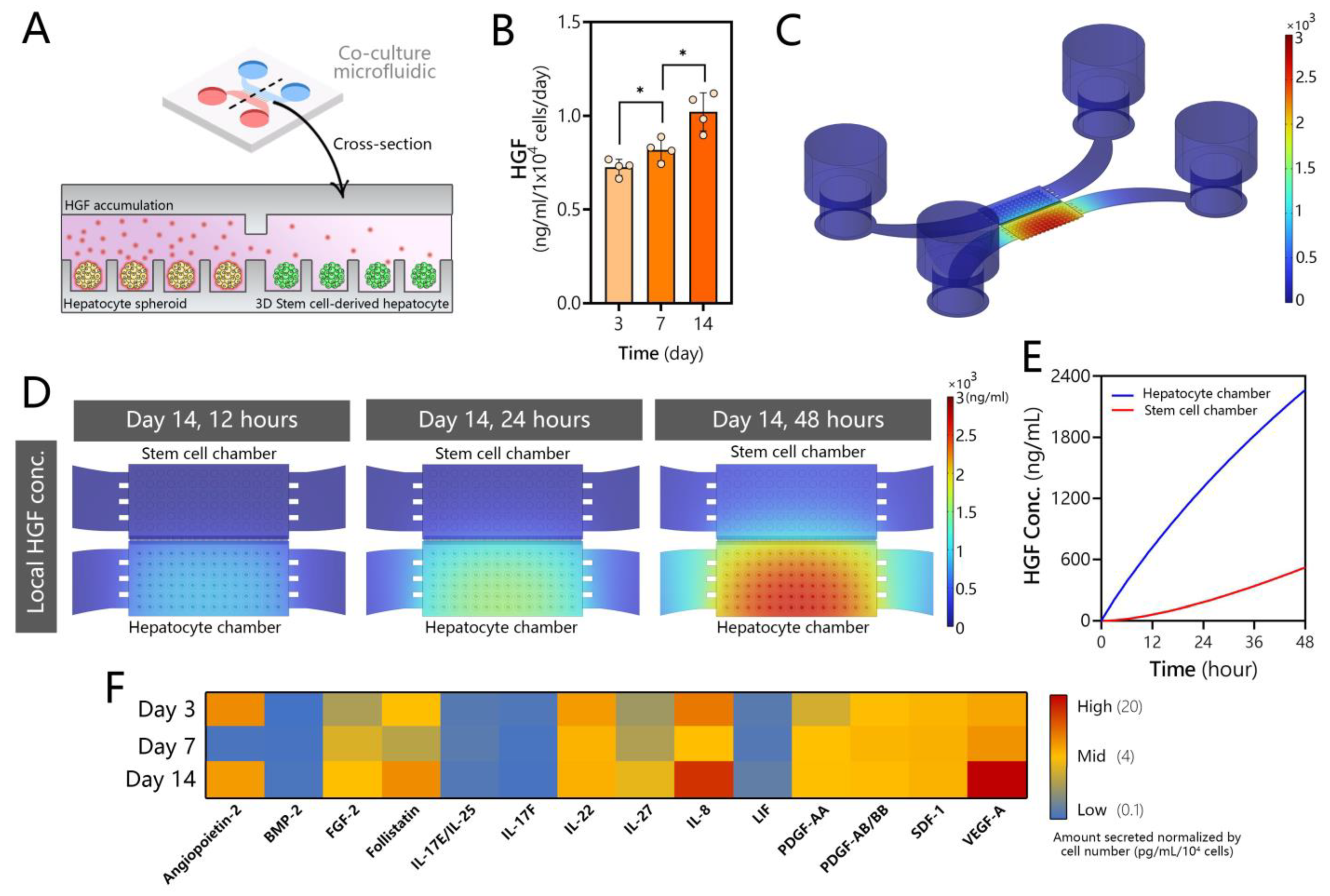
3.1. Design of a Microfluidic Device for 3D Co-Cultures of Hepatocytes and Stem Cells
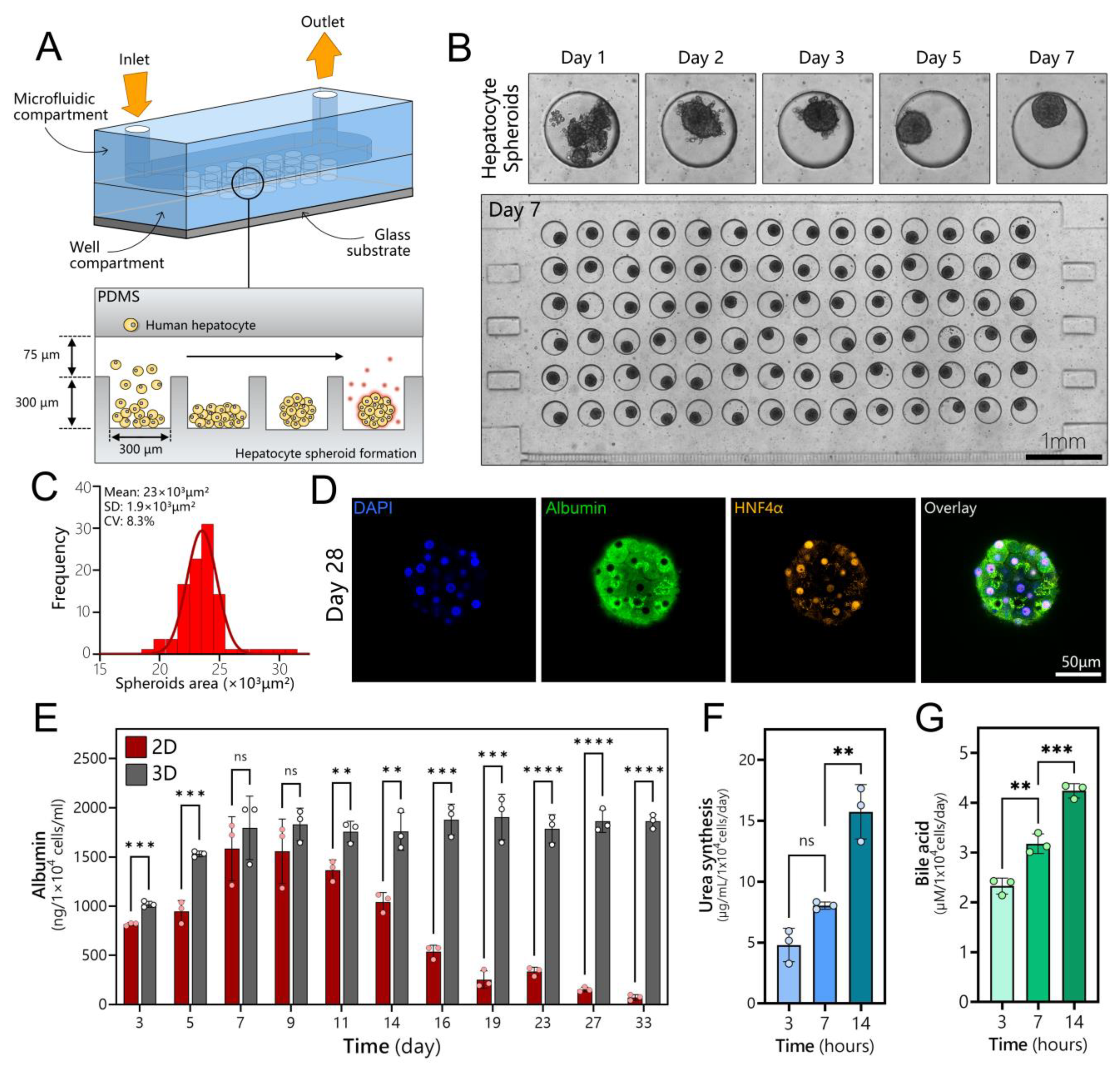
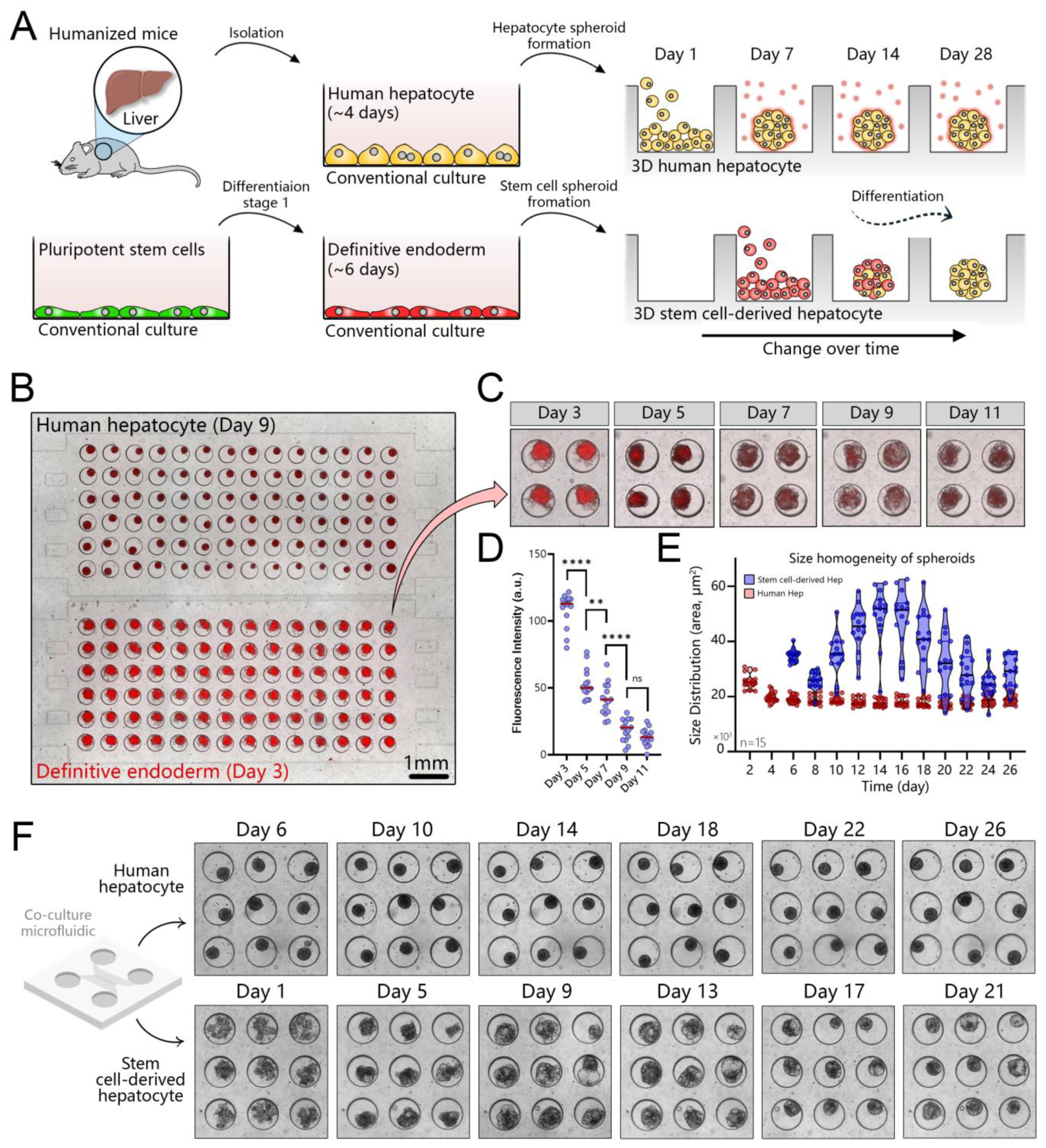
3.2. Long-Term Maintenance of Functional Human Hepatocytes
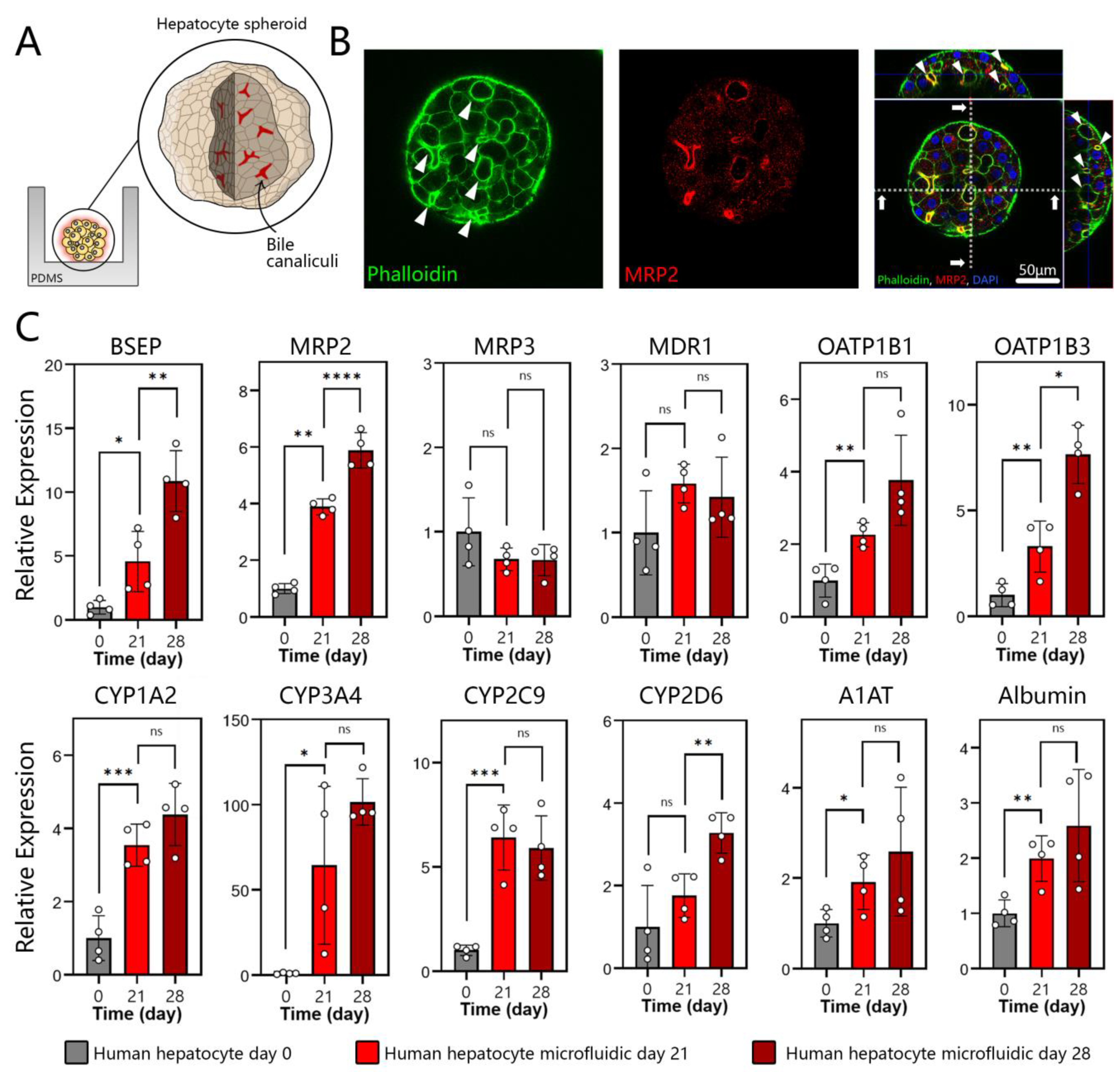
3.3. Polarization, Biliary Excretion, and Gene Expression in Microfluidic Spheroid Cultures of Adult Hepatocytes
3.4. Assessing Production of HGF and Other Secreted Signals in Microfluidic Hepatocyte Cultures
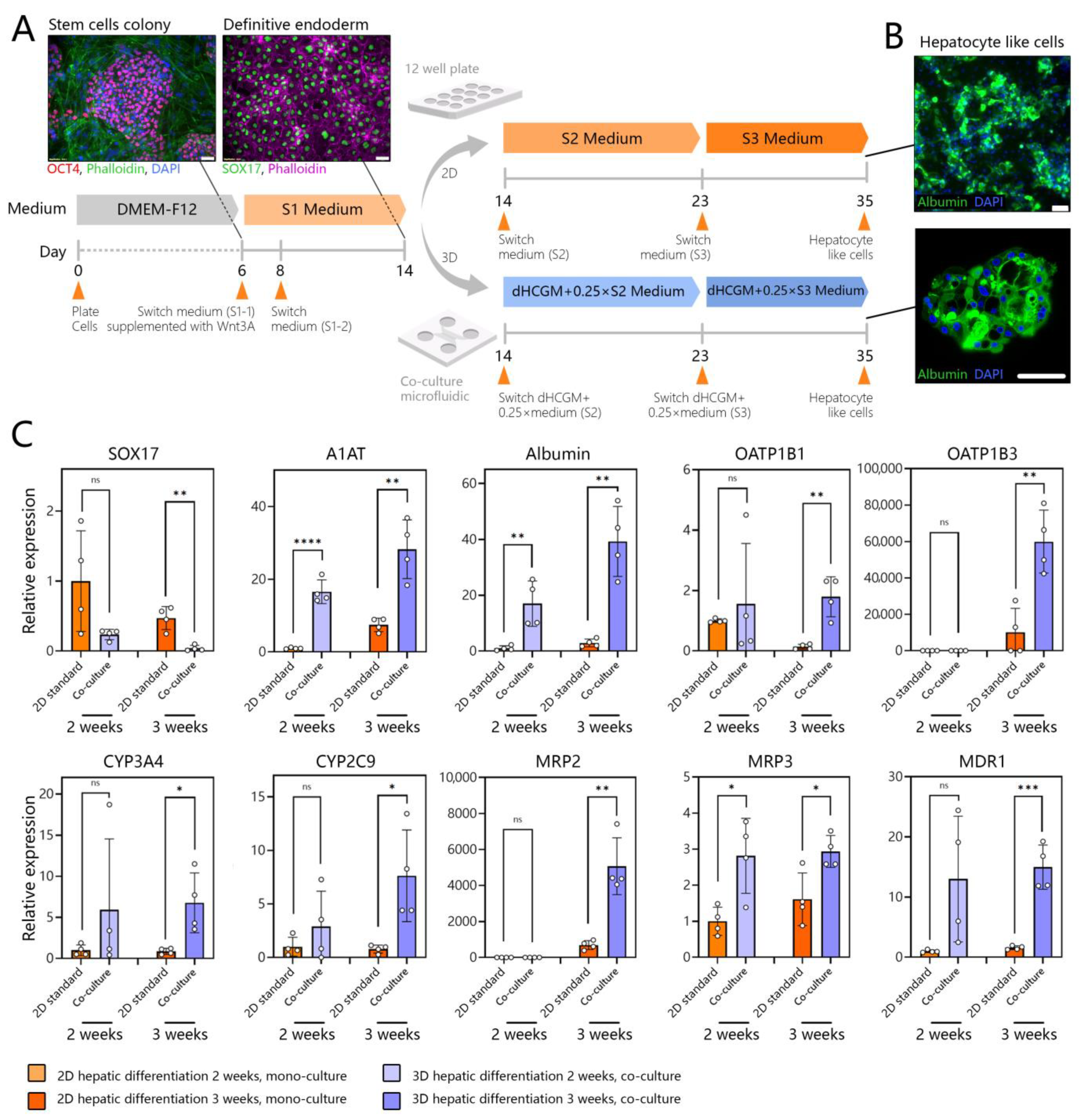
3.5. Creating Co-Cultures to Guide Hepatic Differentiation of Stem Cells
3.6. Assessing Hepatic Differentiation of DE Stem Cells in Co-Cultures with Adult Hepatocytes
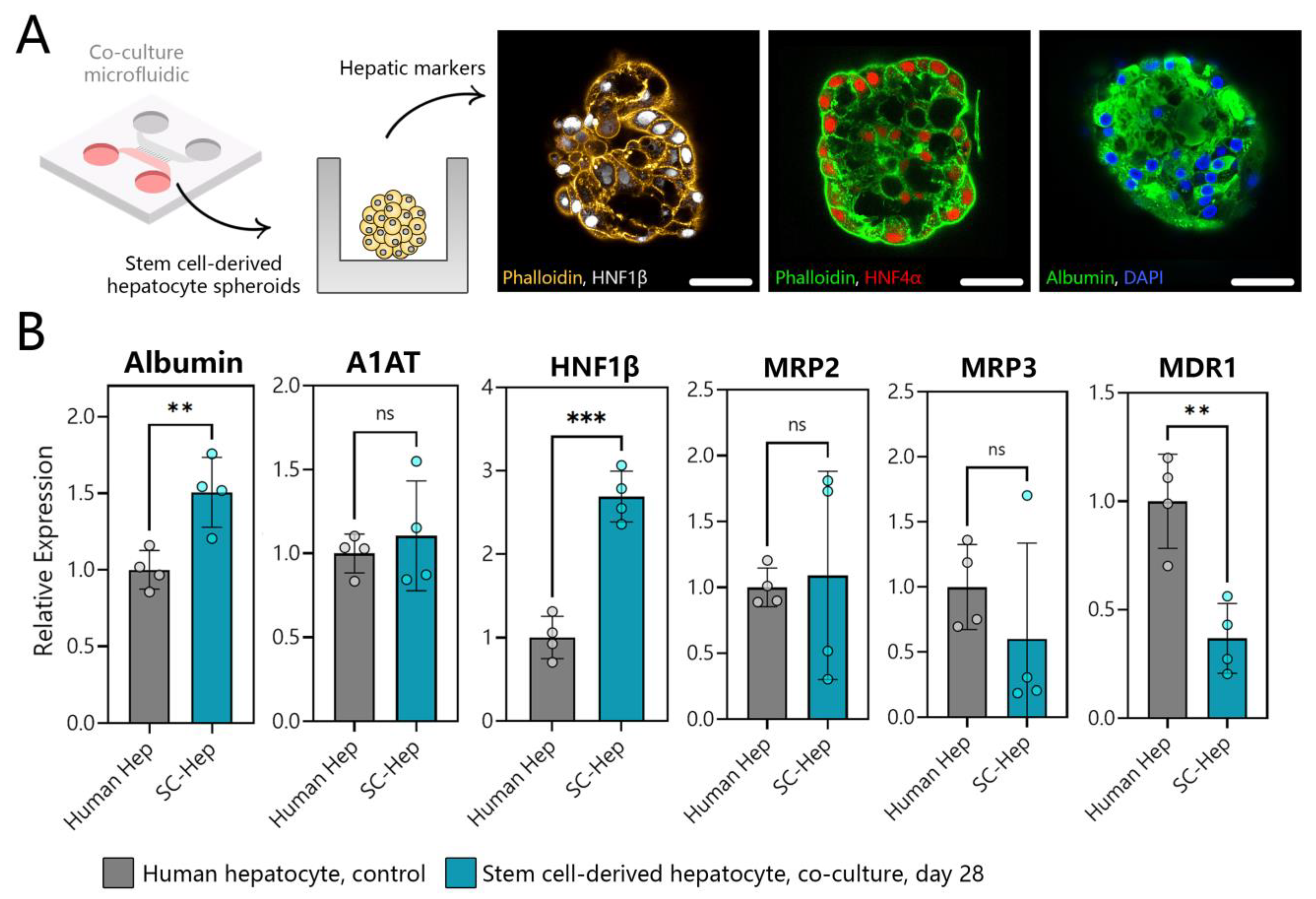
3.7. Benchmarking SC-Hepatocytes against Human Hepatocytes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, R.E.; Fleming, H.E.; Khetani, S.R.; Bhatia, S.N. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol. Adv. 2014, 32, 504–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szkolnicka, D.; Zhou, W.; Lucendo-Villarin, B.; Hay, D.C. Pluripotent Stem Cell–Derived Hepatocytes: Potential and Challenges in Pharmacology. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 147–159. [Google Scholar] [CrossRef]
- Kim, W.; Gwon, Y.; Park, S.; Kim, H.; Kim, J. Therapeutic strategies of three-dimensional stem cell spheroids and organoids for tissue repair and regeneration. Bioact. Mater. 2023, 19, 50–74. [Google Scholar] [CrossRef]
- Basma, H.; Soto–Gutiérrez, A.; Yannam, G.R.; Liu, L.; Ito, R.; Yamamoto, T.; Ellis, E.; Carson, S.D.; Sato, S.; Chen, Y.; et al. Differentiation and Transplantation of Human Embryonic Stem Cell–Derived Hepatocytes. Gastroenterology 2009, 136, 990–999.e994. [Google Scholar] [CrossRef] [Green Version]
- Si-Tayeb, K.; Noto, F.K.; Nagaoka, M.; Li, J.; Battle, M.A.; Duris, C.; North, P.E.; Dalton, S.; Duncan, S.A. Highly efficient generation of human hepatocyte–like cells from induced pluripotent stem cells. Hepatology 2010, 51, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Chen, S.; Duo, S.; Xiang, C.; Jia, J.; Guo, M.; Lai, W.; Lu, S.; Deng, H. Promotion of the efficient metabolic maturation of human pluripotent stem cell-derived hepatocytes by correcting specification defects. Cell Res. 2013, 23, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Zhao, Y.; Liu, Y.; Ye, F.; Song, Z.; Qin, H.; Meng, S.; Chen, Y.; Zhou, R.; Song, X.; et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 2007, 45, 1229–1239. [Google Scholar] [CrossRef]
- Carpentier, A.; Nimgaonkar, I.; Chu, V.; Xia, Y.; Hu, Z.; Liang, T.J. Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Res. 2016, 16, 640–650. [Google Scholar] [CrossRef] [Green Version]
- Rashid, S.T.; Corbineau, S.; Hannan, N.; Marciniak, S.J.; Miranda, E.; Alexander, G.; Huang-Doran, I.; Griffin, J.; Ahrlund-Richter, L.; Skepper, J.; et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Investig. 2010, 120, 3127–3136. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Cai, J.; Liu, Y.; Zhao, D.; Yong, J.; Duo, S.; Song, X.; Guo, Y.; Zhao, Y.; Qin, H.; et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009, 19, 1233–1242. [Google Scholar] [CrossRef]
- Touboul, T.; Hannan, N.R.F.; Corbineau, S.; Martinez, A.; Martinet, C.; Branchereau, S.; Mainot, S.; Strick-Marchand, H.; Pedersen, R.; Di Santo, J.; et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010, 51, 1754–1765. [Google Scholar] [CrossRef]
- Ang, L.T.; Tan, A.K.Y.; Autio, M.I.; Goh, S.H.; Choo, S.H.; Lee, K.L.; Tan, J.; Pan, B.; Lee, J.J.H.; Lum, J.J.; et al. A Roadmap for Human Liver Differentiation from Pluripotent Stem Cells. Cell Rep. 2018, 22, 2190–2205. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, S.; Surapisitchat, J.; Virtanen, C.; Ogawa, M.; Niapour, M.; Sugamori, K.S.; Wang, S.; Tamblyn, L.; Guillemette, C.; Hoffmann, E.; et al. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development 2013, 140, 3285–3296. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Xu, D.; Garfin, P.M.; Ehmer, U.; Hurwitz, M.; Enns, G.; Michie, S.; Wu, M.; Zheng, M.; Nishimura, T.; et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2017, 2, e94954. [Google Scholar] [CrossRef] [Green Version]
- Akbari, S.; Sevinç, G.G.; Ersoy, N.; Basak, O.; Kaplan, K.; Sevinç, K.; Ozel, E.; Sengun, B.; Enustun, E.; Ozcimen, B.; et al. Robust, Long-Term Culture of Endoderm-Derived Hepatic Organoids for Disease Modeling. Stem Cell Rep. 2019, 13, 627–641. [Google Scholar] [CrossRef] [Green Version]
- Mun, S.J.; Ryu, J.-S.; Lee, M.-O.; Son, Y.S.; Oh, S.J.; Cho, H.-S.; Son, M.-Y.; Kim, D.-S.; Kim, S.J.; Yoo, H.J.; et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019, 71, 970–985. [Google Scholar] [CrossRef]
- Messina, A.; Luce, E.; Benzoubir, N.; Pasqua, M.; Pereira, U.; Humbert, L.; Eguether, T.; Rainteau, D.; Duclos-Vallée, J.-C.; Legallais, C.; et al. Evidence of Adult Features and Functions of Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells and Self-Organized as Organoids. Cells 2022, 11, 537. [Google Scholar] [CrossRef]
- Yu, Y.-D.; Kim, K.-H.; Lee, S.-G.; Choi, S.-Y.; Kim, Y.-C.; Byun, K.-S.; Cha, I.-H.; Park, K.-Y.; Cho, C.-H.; Choi, D.-H. Hepatic Differentiation from Human Embryonic Stem Cells Using Stromal Cells. J. Surg. Res. 2011, 170, e253–e261. [Google Scholar] [CrossRef]
- Elham, H.; Fardin, F.; Mahmod, H. The roles of the co-culture of mEScs with pancreatic islets and liver stromal cells in the differentiation of definitive endoderm cells. Biologicals 2017, 45, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.S.; Yaqoob, N.; Iwamuro, M.; Kobayashi, N.; Fujiwara, T. Generation of hepatocyte-like cells from human induced pluripotent stem (iPS) cells by co-culturing embryoid body cells with liver non-parenchymal cell line TWNT-1. J. Coll. Physicians Surg. Pak. 2014, 24, 91–96. [Google Scholar] [PubMed]
- Pettinato, G.; Lehoux, S.; Ramanathan, R.; Salem, M.M.; He, L.-X.; Muse, O.; Flaumenhaft, R.; Thompson, M.T.; Rouse, E.A.; Cummings, R.D.; et al. Generation of fully functional hepatocyte-like organoids from human induced pluripotent stem cells mixed with Endothelial Cells. Sci. Rep. 2019, 9, 8920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuleuova, N.; Lee, J.Y.; Lee, J.; Ramanculov, E.; Zern, M.A.; Revzin, A. Using growth factor arrays and micropatterned co-cultures to induce hepatic differentiation of embryonic stem cells. Biomaterials 2010, 31, 9221–9231. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Tuleuova, N.; Jones, C.N.; Ramanculov, E.; Zern, M.A.; Revzin, A. Directing hepatic differentiation of embryonic stem cells with protein microarray-based co-cultures. Integr. Biol. 2009, 1, 460–468. [Google Scholar] [CrossRef]
- Niculescu, A.G.; Chircov, C.; Bîrcă, A.C.; Grumezescu, A.M. Fabrication and Applications of Microfluidic Devices: A Review. Int. J. Mol. Sci. 2021, 22, 2011. [Google Scholar] [CrossRef]
- Paguirigan, A.L.; Beebe, D.J. From the cellular perspective: Exploring differences in the cellular baseline in macroscale and microfluidic cultures. Integr. Biol. 2009, 1, 182–195. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Meyvantsson, I.; Shkel, I.A.; Beebe, D.J. Diffusion dependent cell behavior in microenvironments. Lab A Chip 2005, 5, 1089–1095. [Google Scholar] [CrossRef]
- Giobbe, G.G.; Michielin, F.; Luni, C.; Giulitti, S.; Martewicz, S.; Dupont, S.; Floreani, A.; Elvassore, N. Functional differentiation of human pluripotent stem cells on a chip. Nat. Methods 2015, 12, 637–640. [Google Scholar] [CrossRef]
- Luni, C.; Gagliano, O.; Elvassore, N. Derivation and Differentiation of Human Pluripotent Stem Cells in Microfluidic Devices. Annu. Rev. Biomed. Eng. 2022, 24, 231–248. [Google Scholar] [CrossRef]
- Blagovic, K.; Kim, L.Y.; Voldman, J. Microfluidic Perfusion for Regulating Diffusible Signaling in Stem Cells. PLoS ONE 2011, 6, e22892. [Google Scholar] [CrossRef] [Green Version]
- Ellison, D.; Munden, A.; Levchenko, A. Computational model and microfluidic platform for the investigation of paracrine and autocrine signaling in mouse embryonic stem cells. Mol. BioSystems 2009, 5, 1004–1012. [Google Scholar] [CrossRef] [Green Version]
- Przybyla, L.; Voldman, J. Probing Embryonic Stem Cell Autocrine and Paracrine Signaling Using Microfluidics. Annu. Rev. Anal. Chem. 2012, 5, 293–315. [Google Scholar] [CrossRef] [Green Version]
- Fattahi, P.; Haque, A.; Son, K.J.; Guild, J.; Revzin, A. Microfluidic devices, accumulation of endogenous signals and stem cell fate selection. Differentiation 2020, 112, 39–46. [Google Scholar] [CrossRef]
- Guild, J.; Haque, A.; Gheibi, P.; Gao, Y.; Son, K.J.; Foster, E.; Dumont, S.; Revzin, A. Embryonic Stem Cells Cultured in Microfluidic Chambers Take Control of Their Fate by Producing Endogenous Signals Including LIF. Stem Cells 2016, 34, 1501–1512. [Google Scholar] [CrossRef] [Green Version]
- Haque, A.; Gheibi, P.; Stybayeva, G.; Gao, Y.; Torok, N.; Revzin, A. Ductular reaction-on-a-chip: Microfluidic co-cultures to study stem cell fate selection during liver injury. Sci. Rep. 2016, 6, 36077. [Google Scholar] [CrossRef]
- De Hoyos-Vega, J.M.; Hong, H.J.; Loutherback, K.; Stybayeva, G.; Revzin, A. A Microfluidic Device for Long-Term Maintenance of Organotypic Liver Cultures. Adv. Mater. Technol. 2023, 8, 2201121. [Google Scholar] [CrossRef]
- Zhou, Q.; Patel, D.; Kwa, T.; Haque, A.; Matharu, Z.; Stybayeva, G.; Gao, Y.; Diehl, A.M.; Revzin, A. Liver injury-on-a-chip: Microfluidic co-cultures with integrated biosensors for monitoring liver cell signaling during injury. Lab A Chip 2015, 15, 4467–4478. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.; Gao, Y.; Son, K.; Siltanen, C.; Neve, R.M.; Ferrara, K.; Revzin, A. Microfluidic co-cultures with hydrogel-based ligand trap to study paracrine signals giving rise to cancer drug resistance. Lab A Chip 2015, 15, 4614–4624. [Google Scholar] [CrossRef]
- Kakuni, M.; Yamasaki, C.; Tachibana, A.; Yoshizane, Y.; Ishida, Y.; Tateno, C. Chimeric Mice with Humanized Livers: A Unique Tool for in Vivo and in Vitro Enzyme Induction Studies. Int. J. Mol. Sci. 2014, 15, 58–74. [Google Scholar] [CrossRef]
- Tateno, C.; Yoshizane, Y.; Saito, N.; Kataoka, M.; Utoh, R.; Yamasaki, C.; Tachibana, A.; Soeno, Y.; Asahina, K.; Hino, H.; et al. Near Completely Humanized Liver in Mice Shows Human-Type Metabolic Responses to Drugs. Am. J. Pathol. 2004, 165, 901–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugahara, G.; Ishida, Y.; Sun, J.; Tateno, C.; Saito, T. Art of Making Artificial Liver: Depicting Human Liver Biology and Diseases in Mice. In Seminars in Liver Disease; Thieme Medical Publishers: New York, NY, USA, 2020; Volume 40, pp. 189–212. [Google Scholar] [CrossRef]
- Sugahara, G.; Ishida, Y.; Lee, J.J.; Li, M.; Tanaka, Y.; Eoh, H.; Higuchi, Y.; Saito, T. Long-term cell fate and functional maintenance of human hepatocyte through stepwise culture configuration. FASEB J. 2023, 37, e22750. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, P.; Rahimian, A.; Slama, M.Q.; Gwon, K.; Gonzalez-Suarez, A.M.; Wolf, J.; Baskaran, H.; Duffy, C.D.; Stybayeva, G.; Peterson, Q.P.; et al. Core–shell hydrogel microcapsules enable formation of human pluripotent stem cell spheroids and their cultivation in a stirred bioreactor. Sci. Rep. 2021, 11, 7177. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Gheibi, P.; Gao, Y.; Foster, E.; Son, K.J.; You, J.; Stybayeva, G.; Patel, D.; Revzin, A. Cell biology is different in small volumes: Endogenous signals shape phenotype of primary hepatocytes cultured in microfluidic channels. Sci. Rep. 2016, 6, 33980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.H.; Loarca, L.; Hoyos-Vega, J.M.D.; Dadgar, N.; Loutherback, K.; Shah, V.H.; Stybayeva, G.; Revzin, A. Microfluidic confinement enhances phenotype and function of hepatocyte spheroids. Am. J. Physiol.-Cell Physiol. 2020, 319, C552–C560. [Google Scholar] [CrossRef]
- Mercer, D.F.; Schiller, D.E.; Elliott, J.F.; Douglas, D.N.; Hao, C.; Rinfret, A.; Addison, W.R.; Fischer, K.P.; Churchill, T.A.; Lakey, J.R.T.; et al. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 2001, 7, 927–933. [Google Scholar] [CrossRef]
- Slim, C.L.; van Ijzendoorn, S.C.D.; Lázaro-Diéguez, F.; Müsch, A. The special case of hepatocytes. BioArchitecture 2014, 4, 47–52. [Google Scholar] [CrossRef] [Green Version]
- Keppler, D. The Roles of MRP2, MRP3, OATP1B1, and OATP1B3 in Conjugated Hyperbilirubinemia. Drug Metab. Dispos. 2014, 42, 561–565. [Google Scholar] [CrossRef] [Green Version]
- Kubitz, R.; Dröge, C.; Stindt, J.; Weissenberger, K.; Häussinger, D. The bile salt export pump (BSEP) in health and disease. Clin. Res. Hepatol. Gastroenterol. 2012, 36, 536–553. [Google Scholar] [CrossRef]
- Ros, J.E.; Libbrecht, L.; Geuken, M.; Jansen, P.L.; Roskams, T.A. High expression of MDR1, MRP1, and MRP3 in the hepatic progenitor cell compartment and hepatocytes in severe human liver disease. J. Pathol. 2003, 200, 553–560. [Google Scholar] [CrossRef]
- Ji, J.-Z.; Tai, T.; Huang, B.-B.; Gu, T.-T.; Mi, Q.-Y.; Xie, H.-G. Mrp3 Transports Clopidogrel Acyl Glucuronide from the Hepatocytes into Blood. Drug Metab. Dispos. 2018, 46, 151–154. [Google Scholar] [CrossRef]
- Jørgensen, L.; Van Beek, J.; Lund, S.; Schousboe, A.; Badolo, L. Evidence of Oatp and Mdr1 in cryopreserved rat hepatocytes. Eur. J. Pharm. Sci. 2007, 30, 181–189. [Google Scholar] [CrossRef]
- Maeda, K. Organic anion transporting polypeptide (OATP)1B1 and OATP1B3 as important regulators of the pharmacokinetics of substrate drugs. Biol. Pharm. Bull. 2015, 38, 155–168. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Zhou, S.F. Synthetic and natural compounds that interact with human cytochrome P450 1A2 and implications in drug development. Curr. Med. Chem. 2009, 16, 4066–4218. [Google Scholar] [CrossRef]
- Lynch, T.; Price, A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician 2007, 76, 391–396. [Google Scholar]
- Bauters, T. Clinically Relevant Drug Interactions in HSCT. In The EBMT Handbook: Hematopoietic Stem Cell Transplantation and Cellular Therapies; Carreras, E., Dufour, C., Mohty, M., Kröger, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 229–235. [Google Scholar] [CrossRef]
- VandenBrink, B.M.; Foti, R.S.; Rock, D.A.; Wienkers, L.C.; Wahlstrom, J.L. Prediction of CYP2D6 Drug Interactions from In Vitro Data: Evidence for Substrate-Dependent Inhibition. Drug Metab. Dispos. 2012, 40, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, C.; Bladt, F.; Goedecke, S.; Brinkmann, V.; Zschiesche, W.; Sharpe, M.; Gherardi, E.; Birchmeler, C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature 1995, 373, 699–702. [Google Scholar] [CrossRef]
- Hay, D.C.; Zhao, D.; Fletcher, J.; Hewitt, Z.A.; McLean, D.; Urruticoechea-Uriguen, A.; Black, J.R.; Elcombe, C.; Ross, J.A.; Wolf, R.; et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells 2008, 26, 894–902. [Google Scholar] [CrossRef] [Green Version]
- Kawaida, K.; Matsumoto, K.; Shimazu, H.; Nakamura, T. Hepatocyte growth factor prevents acute renal failure and accelerates renal regeneration in mice. Proc. Natl. Acad. Sci. USA 1994, 91, 4357–4361. [Google Scholar] [CrossRef]
- Friedman, S.L. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008, 88, 125–172. [Google Scholar] [CrossRef]
- Bissell, D.M.; Wang, S.S.; Jarnagin, W.R.; Roll, F.J. Cell-specific expression of transforming growth factor-beta in rat liver. Evidence for autocrine regulation of hepatocyte proliferation. J. Clin. Investig. 1995, 96, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okano, J.; Shiota, G.; Kawasaki, H. Expression of hepatocyte growth factor (HGF) and HGF receptor (c-met) proteins in liver diseases: An immunohistochemical study. Liver 1999, 19, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Gordillo, M.; Evans, T.; Gouon-Evans, V. Orchestrating liver development. Development 2015, 142, 2094–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, D.; Kong, X.; Weng, H.; Park, O.; Wang, H.; Dooley, S.; Gershwin, M.E.; Gao, B. Interleukin-22 promotes proliferation of liver stem/progenitor cells in mice and patients with chronic hepatitis B virus infection. Gastroenterology 2012, 143, 188–198.e187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillot, A.; Gasmi, I.; Brouillet, A.; Ait-Ahmed, Y.; Calderaro, J.; Ruiz, I.; Gao, B.; Lotersztajn, S.; Pawlotsky, J.-M.; Lafdil, F. Interleukins-17 and 27 promote liver regeneration by sequentially inducing progenitor cell expansion and differentiation. Hepatol. Commun. 2018, 2, 329–343. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.L.; Pang, B.Y.; Zhu, Y.; Wang, L.; Leng, A.J.; Chen, H.L. Yi Guan Jian decoction may enhance hepatic differentiation of bone marrow-derived mesenchymal stem cells via SDF-1 in vitro. Mol. Med. Rep. 2017, 16, 2511–2521. [Google Scholar] [CrossRef] [Green Version]
- Tsuzuki, S.; Yamaguchi, T.; Okumura, T.; Kasai, T.; Ueno, Y.; Taniguchi, H. PDGF Receptors and Signaling Are Required for 3D-Structure Formation and Differentiation of Human iPSC-Derived Hepatic Spheroids. Int. J. Mol. Sci. 2023, 24, 7075. [Google Scholar] [CrossRef]
- Lysy, P.A.; Smets, F.; Najimi, M.; Sokal, E.M. Leukemia inhibitory factor contributes to hepatocyte-like differentiation of human bone marrow mesenchymal stem cells. Differentiation 2008, 76, 1057–1067. [Google Scholar] [CrossRef]
- Tao, L.; Fang, S.Y.; Zhao, L.; He, T.C.; He, Y.; Bi, Y. Indocyanine Green Uptake and Periodic Acid-Schiff Staining Method for Function Detection of Liver Cells are Affected by Different Cell Confluence. Cytotechnology 2021, 73, 159–167. [Google Scholar] [CrossRef]
- Rodgarkia-Dara, C.; Vejda, S.; Erlach, N.; Losert, A.; Bursch, W.; Berger, W.; Schulte-Hermann, R.; Grusch, M. The activin axis in liver biology and disease. Mutat. Res. 2006, 613, 123–137. [Google Scholar] [CrossRef]
- Sasaki, T.; Suzuki, Y.; Kakisaka, K.; Wang, T.; Ishida, K.; Suzuki, A.; Abe, H.; Sugai, T.; Takikawa, Y. IL-8 induces transdifferentiation of mature hepatocytes toward the cholangiocyte phenotype. FEBS Open Bio 2019, 9, 2105–2116. [Google Scholar] [CrossRef] [Green Version]
- Ng, E.S.; Azzola, L.; Bruveris, F.F.; Calvanese, V.; Phipson, B.; Vlahos, K.; Hirst, C.; Jokubaitis, V.J.; Yu, Q.C.; Maksimovic, J.; et al. Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 2016, 34, 1168–1179. [Google Scholar] [CrossRef]
- Duan, Y.; Ma, X.; Zou, W.; Wang, C.; Bahbahan, I.S.; Ahuja, T.P.; Tolstikov, V.; Zern, M.A. Differentiation and Characterization of Metabolically Functioning Hepatocytes from Human Embryonic Stem Cells. Stem Cells 2010, 28, 674–686. [Google Scholar] [CrossRef]
- Costa, M.; Sourris, K.; Hatzistavrou, T.; Elefanty, A.G.; Stanley, E.G. Expansion of Human Embryonic Stem Cells In Vitro. Curr. Protoc. Stem Cell Biol. 2008, 5, 1C.1.1–1C.1.7. [Google Scholar] [CrossRef]
- Chetty, S.; Pagliuca, F.W.; Honore, C.; Kweudjeu, A.; Rezania, A.; Melton, D.A. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods 2013, 10, 553–556. [Google Scholar] [CrossRef] [Green Version]
- Gage, B.K.; Webber, T.D.; Kieffer, T.J. Initial Cell Seeding Density Influences Pancreatic Endocrine Development During in vitro Differentiation of Human Embryonic Stem Cells. PLoS ONE 2013, 8, e82076. [Google Scholar] [CrossRef] [Green Version]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fattahi, P.; de Hoyos-Vega, J.M.; Choi, J.H.; Duffy, C.D.; Gonzalez-Suarez, A.M.; Ishida, Y.; Nguyen, K.M.; Gwon, K.; Peterson, Q.P.; Saito, T.; et al. Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes. Cells 2023, 12, 1982. https://doi.org/10.3390/cells12151982
Fattahi P, de Hoyos-Vega JM, Choi JH, Duffy CD, Gonzalez-Suarez AM, Ishida Y, Nguyen KM, Gwon K, Peterson QP, Saito T, et al. Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes. Cells. 2023; 12(15):1982. https://doi.org/10.3390/cells12151982
Chicago/Turabian StyleFattahi, Pouria, Jose M. de Hoyos-Vega, Jong Hoon Choi, Caden D. Duffy, Alan M. Gonzalez-Suarez, Yuji Ishida, Kianna M. Nguyen, Kihak Gwon, Quinn P. Peterson, Takeshi Saito, and et al. 2023. "Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes" Cells 12, no. 15: 1982. https://doi.org/10.3390/cells12151982
APA StyleFattahi, P., de Hoyos-Vega, J. M., Choi, J. H., Duffy, C. D., Gonzalez-Suarez, A. M., Ishida, Y., Nguyen, K. M., Gwon, K., Peterson, Q. P., Saito, T., Stybayeva, G., & Revzin, A. (2023). Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes. Cells, 12(15), 1982. https://doi.org/10.3390/cells12151982