MPP8 Governs the Activity of the LIF/STAT3 Pathway and Plays a Crucial Role in the Differentiation of Mouse Embryonic Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Differentiation
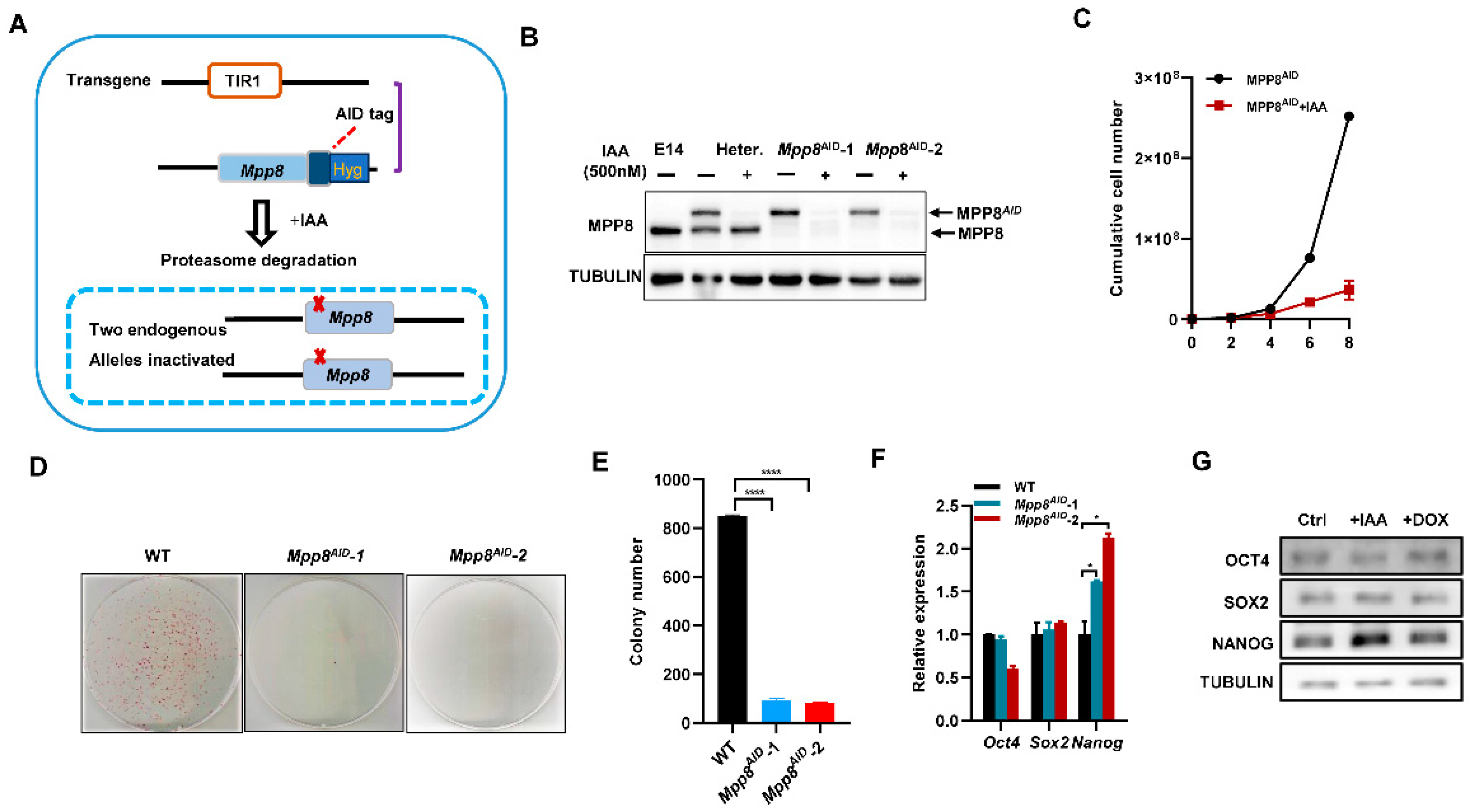
2.2. Generation of AID-Tagged ESCs and Mpp8-Inducible Overexpression ESCs
2.3. Western Blot Analysis
2.4. RT-PCR Analysis
2.5. Alkaline Phosphatase (AP) Staining
2.6. Cell Growth Curve Analysis
2.7. RNA-Seq Experiments
2.8. ChIP-Seq Data Analysis
2.9. Statistical Analysis
3. Results
3.1. Depletion of MPP8 Impairs the Self-Renewal of mESCs
3.2. Degradation of MPP8 Enhances the Activity of the LIF/STAT3 Pathway and Hinders the Transition from Naïve to Primed ESCs
3.3. MPP8 Deletion Impairs the Differentiation of ESCs
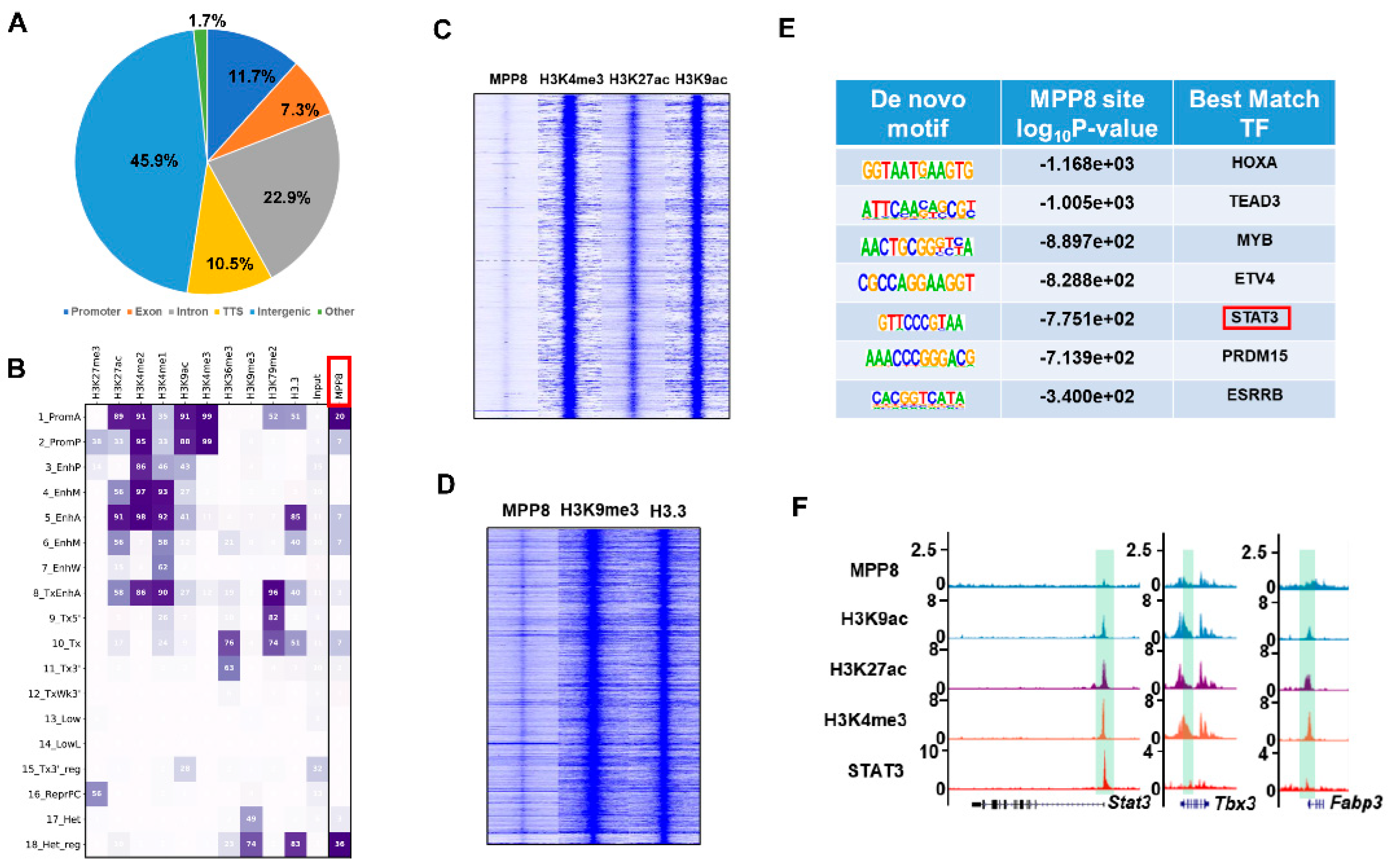
3.4. MPP8 Modulates Gene Expression by Influencing Epigenetic Modifications within the Promoter Region
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef]
- Tee, W.W.; Reinberg, D. Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development 2014, 141, 2376–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Belmonte, J.C. Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet. 2017, 18, 180–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divisato, G.; Passaro, F.; Russo, T.; Parisi, S. The Key Role of MicroRNAs in Self-Renewal and Differentiation of Embryonic Stem Cells. Int. J. Mol. Sci. 2020, 21, 6285. [Google Scholar] [CrossRef]
- Papatsenko, D.; Waghray, A.; Lemischka, I.R. Feedback control of pluripotency in embryonic stem cells: Signaling, transcription and epigenetics. Stem Cell Res. 2018, 29, 180–188. [Google Scholar] [CrossRef]
- Schlesinger, S.; Meshorer, E. Open Chromatin, Epigenetic Plasticity, and Nuclear Organization in Pluripotency. Dev. Cell 2019, 48, 135–150. [Google Scholar] [CrossRef] [Green Version]
- Geng, T.; Zhang, D.; Jiang, W. Epigenetic Regulation of Transition Among Different Pluripotent States: Concise Review. Stem Cells 2019, 37, 1372–1380. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, O.; Li, R.; Hung, J.H.; Chen, P.B.; Dong, X.; Ee, L.S.; Weng, Z.; Rando, O.J.; Fazzio, T.G. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 2011, 147, 1498–1510. [Google Scholar] [CrossRef] [Green Version]
- van Mierlo, G.; Dirks, R.A.M.; De Clerck, L.; Brinkman, A.B.; Huth, M.; Kloet, S.L.; Saksouk, N.; Kroeze, L.I.; Willems, S.; Farlik, M.; et al. Integrative Proteomic Profiling Reveals PRC2-Dependent Epigenetic Crosstalk Maintains Ground-State Pluripotency. Cell Stem Cell 2019, 24, 123–137.e128. [Google Scholar] [CrossRef] [Green Version]
- Loh, C.H.; van Genesen, S.; Perino, M.; Bark, M.R.; Veenstra, G.J.C. Loss of PRC2 subunits primes lineage choice during exit of pluripotency. Nat. Commun. 2021, 12, 6985. [Google Scholar] [CrossRef]
- Zhang, W.; Chronis, C.; Chen, X.; Zhang, H.; Spalinskas, R.; Pardo, M.; Chen, L.; Wu, G.; Zhu, Z.; Yu, Y.; et al. The BAF and PRC2 Complex Subunits Dpf2 and Eed Antagonistically Converge on Tbx3 to Control ESC Differentiation. Cell Stem Cell 2019, 24, 138–152.e138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brand, M.; Nakka, K.; Zhu, J.; Dilworth, F.J. Polycomb/Trithorax Antagonism: Cellular Memory in Stem Cell Fate and Function. Cell Stem Cell 2019, 24, 518–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Li, Y.; Li, B.; Zhang, B.; Wang, Y.; Li, L.; Gao, J.; Fu, Y.; Li, S.; Chen, C.; et al. DNMTs Play an Important Role in Maintaining the Pluripotency of Leukemia Inhibitory Factor-Dependent Embryonic Stem Cells. Stem Cell Rep. 2021, 16, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Tchasovnikarova, I.A.; Timms, R.T.; Matheson, N.J.; Wals, K.; Antrobus, R.; Gottgens, B.; Dougan, G.; Dawson, M.A.; Lehner, P.J. Gene Silencing. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 2015, 348, 1481–1485. [Google Scholar] [CrossRef] [Green Version]
- Tunbak, H.; Enriquez-Gasca, R.; Tie, C.H.C.; Gould, P.A.; Mlcochova, P.; Gupta, R.K.; Fernandes, L.; Holt, J.; van der Veen, A.G.; Giampazolias, E.; et al. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat Commun 2020, 11, 5387. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Guney, M.H.; Kim, K.; Goh, S.L.; McCauley, S.; Dauphin, A.; Diehl, W.E.; Luban, J. Primate immunodeficiency virus proteins Vpx and Vpr counteract transcriptional repression of proviruses by the HUSH complex. Nat. Microbiol. 2018, 3, 1354–1361. [Google Scholar] [CrossRef]
- Robbez-Masson, L.; Tie, C.H.C.; Conde, L.; Tunbak, H.; Husovsky, C.; Tchasovnikarova, I.A.; Timms, R.T.; Herrero, J.; Lehner, P.J.; Rowe, H.M. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018, 28, 836–845. [Google Scholar] [CrossRef] [Green Version]
- Kokura, K.; Sun, L.; Bedford, M.T.; Fang, J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010, 29, 3673–3687. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.; Horton, J.R.; Bedford, M.T.; Zhang, X.; Cheng, X. Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: Potential effect of phosphorylation on methyl-lysine binding. J. Mol. Biol. 2011, 408, 807–814. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.; Sun, L.; Kokura, K.; Horton, J.R.; Fukuda, M.; Espejo, A.; Izumi, V.; Koomen, J.M.; Bedford, M.T.; Zhang, X.; et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat. Commun. 2011, 2, 533. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Liu, T.; Zhang, W.; Zhang, K.; Guo, S.; Liang, J. Lentivirus-mediated knockdown of M-phase phosphoprotein 8 inhibits proliferation of colon cancer cells. Biotechnol. Appl. Biochem. 2017, 64, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Y.; Qiao, Y.L.; Zhang, Y.; Wang, J.; Shen, X.; Xu, C.W. Knockdown of MPP8 suppresses cell proliferation via regulation of HOXA5 in non-small cell lung cancer cells. Cell. Mol. Biol. 2018, 64, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, H.; Wang, C.; Wu, H.; He, H.; Yao, C.; Cui, J.; Li, W. M-phase phosphoprotein 8 promotes gastric cancer growth and metastasis via p53/Bcl-2 and EMT-related signaling pathways. J. Cell Biochem. 2020, 121, 2330–2342. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Kokura, K.; Izumi, V.; Koomen, J.M.; Seto, E.; Chen, J.; Fang, J. MPP8 and SIRT1 crosstalk in E-cadherin gene silencing and epithelial-mesenchymal transition. EMBO Rep. 2015, 16, 689–699. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wang, G.Z.; Cingoz, O.; Goff, S.P. NP220 mediates silencing of unintegrated retroviral DNA. Nature 2018, 564, 278–282. [Google Scholar] [CrossRef]
- Chougui, G.; Munir-Matloob, S.; Matkovic, R.; Martin, M.M.; Morel, M.; Lahouassa, H.; Leduc, M.; Ramirez, B.C.; Etienne, L.; Margottin-Goguet, F. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat. Microbiol. 2018, 3, 891–897. [Google Scholar] [CrossRef]
- Fukuda, K.; Okuda, A.; Yusa, K.; Shinkai, Y. A CRISPR knockout screen identifies SETDB1-target retroelement silencing factors in embryonic stem cells. Genome Res. 2018, 28, 846–858. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Lee, C.H.; Swigut, T.; Grow, E.; Gu, B.; Bassik, M.C.; Wysocka, J. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 2018, 553, 228–232. [Google Scholar] [CrossRef]
- Garland, W.; Muller, I.; Wu, M.; Schmid, M.; Imamura, K.; Rib, L.; Sandelin, A.; Helin, K.; Jensen, T.H. Chromatin modifier HUSH co-operates with RNA decay factor NEXT to restrict transposable element expression. Mol. Cell 2022, 82, 1691–1707.e1698. [Google Scholar] [CrossRef]
- Muller, I.; Moroni, A.S.; Shlyueva, D.; Sahadevan, S.; Schoof, E.M.; Radzisheuskaya, A.; Hojfeldt, J.W.; Tatar, T.; Koche, R.P.; Huang, C.; et al. MPP8 is essential for sustaining self-renewal of ground-state pluripotent stem cells. Nat. Commun. 2021, 12, 3034. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 2009, 6, 917–922. [Google Scholar] [CrossRef]
- Holland, A.J.; Fachinetti, D.; Han, J.S.; Cleveland, D.W. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 2012, 109, E3350–E3357. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dai, C.; Xie, W.; Zhang, H.; Huang, X.; Chronis, C.; Ye, Y.; Zhang, W. A One-step strategy to target essential factors with auxin-inducible degron system in mouse embryonic stem cells. Front. Cell. Dev. Biol. 2022, 10, 964119. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.L.; Hilton, D.J.; Pease, S.; Willson, T.A.; Stewart, C.L.; Gearing, D.P.; Wagner, E.F.; Metcalf, D.; Nicola, N.A.; Gough, N.M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 1988, 336, 684–687. [Google Scholar] [CrossRef]
- Matsuda, T.; Nakamura, T.; Nakao, K.; Arai, T.; Katsuki, M.; Heike, T.; Yokota, T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999, 18, 4261–4269. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, D.; Xu, J.; Wang, Y.; Ma, F.; Li, Z.; Liu, N. Stat3 activation is critical for pluripotency maintenance. J. Cell. Physiol. 2019, 234, 1044–1051. [Google Scholar] [CrossRef]
- Chen, H.; Aksoy, I.; Gonnot, F.; Osteil, P.; Aubry, M.; Hamela, C.; Rognard, C.; Hochard, A.; Voisin, S.; Fontaine, E.; et al. Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat. Commun. 2015, 6, 7095. [Google Scholar] [CrossRef] [Green Version]
- Kalkan, T.; Bornelov, S.; Mulas, C.; Diamanti, E.; Lohoff, T.; Ralser, M.; Middelkamp, S.; Lombard, P.; Nichols, J.; Smith, A. Complementary Activity of ETV5, RBPJ, and TCF3 Drives Formative Transition from Naive Pluripotency. Cell Stem Cell 2019, 24, 785–801.e787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lackner, A.; Sehlke, R.; Garmhausen, M.; Giuseppe Stirparo, G.; Huth, M.; Titz-Teixeira, F.; van der Lelij, P.; Ramesmayer, J.; Thomas, H.F.; Ralser, M.; et al. Cooperative genetic networks drive embryonic stem cell transition from naive to formative pluripotency. EMBO J. 2021, 40, e105776. [Google Scholar] [CrossRef] [PubMed]
- Osnato, A.; Brown, S.; Krueger, C.; Andrews, S.; Collier, A.J.; Nakanoh, S.; Quiroga Londono, M.; Wesley, B.T.; Muraro, D.; Brumm, A.S.; et al. TGFbeta signalling is required to maintain pluripotency of human naive pluripotent stem cells. Elife 2021, 10, e67259. [Google Scholar] [CrossRef] [PubMed]
- Chronis, C.; Fiziev, P.; Papp, B.; Butz, S.; Bonora, G.; Sabri, S.; Ernst, J.; Plath, K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell 2017, 168, 442–459.e420. [Google Scholar] [CrossRef] [Green Version]
- Chambers, I.; Smith, A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene 2004, 23, 7150–7160. [Google Scholar] [CrossRef] [Green Version]
- Niwa, H.; Burdon, T.; Chambers, I.; Smith, A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998, 12, 2048–2060. [Google Scholar] [CrossRef] [Green Version]
- Rodda, D.J.; Chew, J.L.; Lim, L.H.; Loh, Y.H.; Wang, B.; Ng, H.H.; Robson, P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005, 280, 24731–24737. [Google Scholar] [CrossRef] [Green Version]
- Bhargava, S.; Cox, B.; Polydorou, C.; Gresakova, V.; Korinek, V.; Strnad, H.; Sedlacek, R.; Epp, T.A.; Chawengsaksophak, K. The epigenetic modifier Fam208a is required to maintain epiblast cell fitness. Sci. Rep. 2017, 7, 9322. [Google Scholar] [CrossRef] [Green Version]
- Murata, K.; Sato, S.; Haruta, M.; Goshima, T.; Chiba, Y.; Takahashi, S.; Sharif, J.; Koseki, H.; Nakanishi, M.; Shimada, M. Physical interaction between MPP8 and PRC1 complex and its implication for regulation of spermatogenesis. Biochem. Biophys. Res. Commun. 2015, 458, 470–475. [Google Scholar] [CrossRef]
- Ye, Y.; Chen, X.; Zhang, W. Mammalian SWI/SNF Chromatin Remodeling Complexes in Embryonic Stem Cells: Regulating the Balance Between Pluripotency and Differentiation. Front. Cell Dev. Biol. 2020, 8, 626383. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Yang, T.; Wu, H.; Yi, W.; Dai, C.; Chen, X.; Zhang, W.; Ye, Y. MPP8 Governs the Activity of the LIF/STAT3 Pathway and Plays a Crucial Role in the Differentiation of Mouse Embryonic Stem Cells. Cells 2023, 12, 2023. https://doi.org/10.3390/cells12162023
Zhang H, Yang T, Wu H, Yi W, Dai C, Chen X, Zhang W, Ye Y. MPP8 Governs the Activity of the LIF/STAT3 Pathway and Plays a Crucial Role in the Differentiation of Mouse Embryonic Stem Cells. Cells. 2023; 12(16):2023. https://doi.org/10.3390/cells12162023
Chicago/Turabian StyleZhang, Heyao, Tenghui Yang, Hao Wu, Wen Yi, Chunhong Dai, Xi Chen, Wensheng Zhang, and Ying Ye. 2023. "MPP8 Governs the Activity of the LIF/STAT3 Pathway and Plays a Crucial Role in the Differentiation of Mouse Embryonic Stem Cells" Cells 12, no. 16: 2023. https://doi.org/10.3390/cells12162023




