Unveiling the Yin-Yang Balance of M1 and M2 Macrophages in Hepatocellular Carcinoma: Role of Exosomes in Tumor Microenvironment and Immune Modulation
Abstract
1. Introduction
2. Understanding the Biology and Biogenesis of Exosomes
2.1. Biogenesis of Exosomes
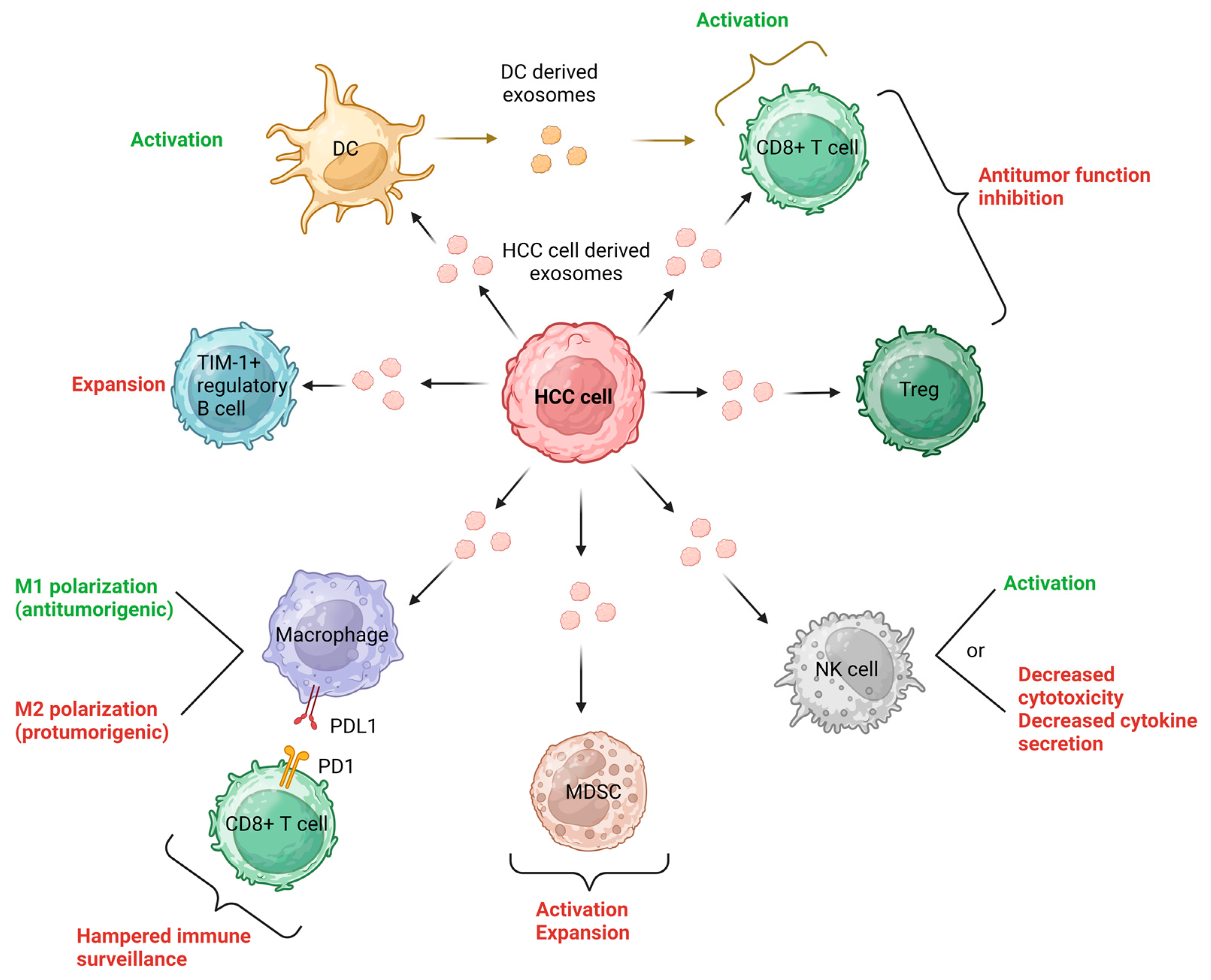
2.2. Exosomes and Cell-to-Cell Communication in the HCC Tumor Microenvironment (TME)
2.3. Exploring Exosomes as Efficient Drug Carriers: Advantages and Strategies for Loading Small-Molecule Drugs
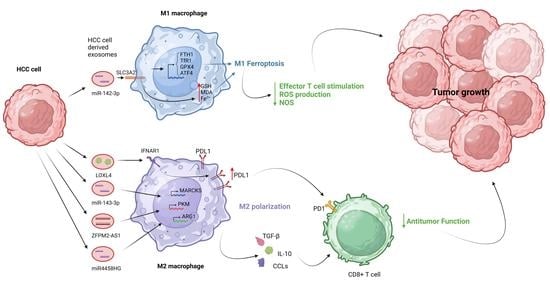
3. Macrophages as Targets of Exosomes in HCC
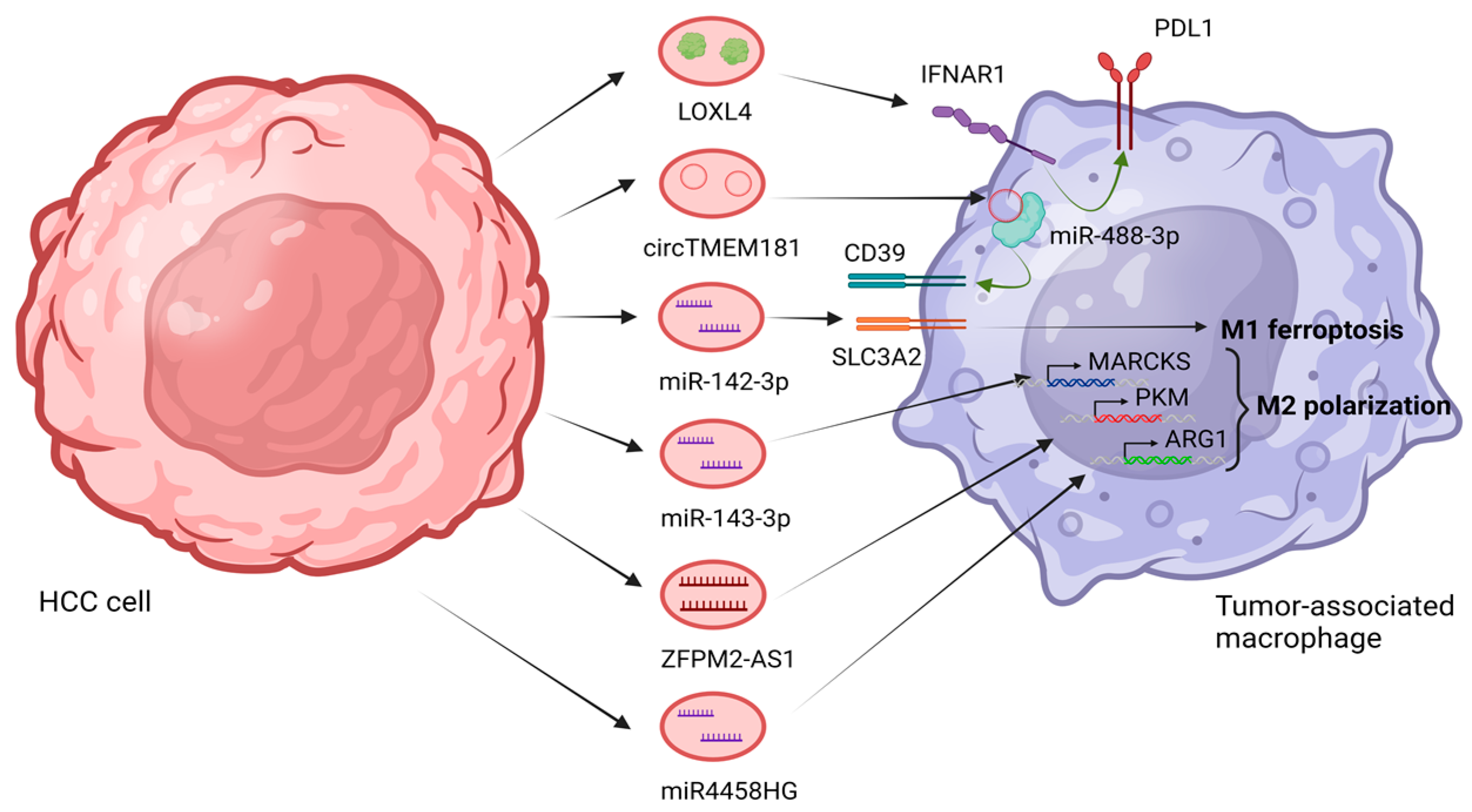
3.1. The Role of Exosomes in M1 Macrophage Polarization
3.2. The Role of Exosomes in M2 Macrophage Polarization
3.2.1. Metabolism Regulation: Harnessing Exosomes for M2 Polarization in Immune Response
3.2.2. Epigenetic Modifiers: Key Players in M2 Polarization in HCC
4. Exploring the Potential of Macrophage Exosomes in HCC Cell Targeting
4.1. Unveiling the Metabolic Landscape: Macrophage-Derived Exosomes Fueling Metabolic Alterations in HCC Cells
4.2. The Influence of Macrophage-Derived Exosomes in Various HCC Cellular Processes
4.3. Impact of Macrophage-Derived Exosomes on Signaling Pathways in HCC
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, E.; Viatour, P. Hepatocellular carcinoma: Old friends and new tricks. Exp. Mol. Med. 2020, 52, 1898–1907. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies from 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular carcinoma. Lancet 2022, 400, 1345–1362. [Google Scholar] [CrossRef]
- Kotsifa, E.; Vergadis, C.; Vailas, M.; Machairas, N.; Kykalos, S.; Damaskos, C.; Garmpis, N.; Lianos, G.D.; Schizas, D. Transarterial Chemoembolization for Hepatocellular Carcinoma: Why, When, How? J. Pers. Med. 2022, 12, 436. [Google Scholar] [CrossRef]
- Machairas, N.; Tsilimigras, D.I.; Pawlik, T.M. State-of-the-art surgery for hepatocellular carcinoma. Langenbeck’s Arch. Surg. 2021, 406, 2151–2162. [Google Scholar] [CrossRef] [PubMed]
- Sotiropoulos, G.C.; Prodromidou, A.; Kostakis, I.D.; Machairas, N. Meta-analysis of laparoscopic vs open liver resection for hepatocellular carcinoma. Updates Surg. 2017, 69, 291–311. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-C.; Lin, Y.-M.; Yen, A.M.-F.; Chen, S.L.-S.; Wu, W.Y.-Y.; Chiu, S.Y.-H.; Fann, J.C.-Y.; Lin, Y.-S.; Chen, H.-H.; Liao, C.-S. Predictors of long-term survival in hepatocellular carcinomas: A longitudinal follow-up of 108 patients with small tumors. Anticancer Res. 2013, 33, 5171–5178. [Google Scholar] [PubMed]
- Arvanitakis, K.; Koletsa, T.; Mitroulis, I.; Germanidis, G. Tumor-Associated Macrophages in Hepatocellular Carcinoma Pathogenesis, Prognosis and Therapy. Cancers 2022, 14, 226. [Google Scholar] [CrossRef]
- Feng, Y.; Ye, Z.; Song, F.; He, Y.; Liu, J. The Role of TAMs in Tumor Microenvironment and New Research Progress. Stem Cells Int. 2022, 2022, 5775696. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, C.; Lu, S.; Xu, Y.; Li, Z.; Jiang, H.; Ma, Y. Tumor-associated macrophages in cholangiocarcinoma: Complex interplay and potential therapeutic target. EbioMedicine 2021, 67, 103375. [Google Scholar] [CrossRef]
- Bai, R.; Li, Y.; Jian, L.; Yang, Y.; Zhao, L.; Wei, M. The hypoxia-driven crosstalk between tumor and tumor-associated macrophages: Mechanisms and clinical treatment strategies. Mol. Cancer 2022, 21, 177. [Google Scholar] [CrossRef] [PubMed]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [PubMed]
- Khanam, A.; Kottilil, S. New Therapeutics for HCC: Does Tumor Immune Microenvironment Matter? Int. J. Mol. Sci. 2022, 24, 437. [Google Scholar] [CrossRef]
- Shen, M.; Shen, Y.; Fan, X.; Men, R.; Ye, T.; Yang, L. Roles of Macrophages and Exosomes in Liver Diseases. Front. Med. 2020, 7, 583691. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Dedes, N.; Pergaris, A.; Gazouli, M.; Theocharis, S. Exosomes in the Treatment of Pancreatic Cancer: A Moonshot to PDAC Treatment? Int. J. Mol. Sci. 2022, 23, 3620. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, X.; Shi, Y.; Lu, Y.; Qiu, P.; Deng, Z.; Yao, W.; Wang, J. Exosomes in the tumor microenvironment of cholangiocarcinoma: Current status and future perspectives. J. Transl. Med. 2022, 20, 117. [Google Scholar] [CrossRef]
- Xiong, J.; Chi, H.; Yang, G.; Zhao, S.; Zhang, J.; Tran, L.J.; Xia, Z.; Yang, F.; Tian, G. Revolutionizing anti-tumor therapy: Unleashing the potential of B cell-derived exosomes. Front. Immunol. 2023, 14, 1188760. [Google Scholar] [CrossRef]
- Zeng, H.; Guo, S.; Ren, X.; Wu, Z.; Liu, S.; Yao, X. Current Strategies for Exosome Cargo Loading and Targeting Delivery. Cells 2023, 12, 1416. [Google Scholar] [CrossRef]
- Sasaki, R.; Kanda, T.; Yokosuka, O.; Kato, N.; Matsuoka, S.; Moriyama, M. Exosomes and Hepatocellular Carcinoma: From Bench to Bedside. Int. J. Mol. Sci. 2019, 20, 1406. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Rashed, M.H.; Bayraktar, E.; Helal, G.K.; Abd-Ellah, M.F.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; McAndrews, K.M. The role of extracellular vesicles in cancer. Cell 2023, 186, 1610–1626. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; DeGeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Friand, V.; David, G.; Zimmermann, P. Syntenin and syndecan in the biogenesis of exosomes. Biol. Cell 2015, 107, 331–341. [Google Scholar] [CrossRef]
- Thery, C.; Clayton, A.; Amigorena, S.; Raposo, G. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 3, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Sykaras, A.G.; Christofidis, K.; Politi, E.; Theocharis, S. Exosomes on Endometrial Cancer: A Biomarkers Treasure Trove? Cancers 2022, 14, 1733. [Google Scholar] [CrossRef]
- Goutas, D.; Pergaris, A.; Goutas, N.; Theocharis, S. Utilizing Exosomal-EPHs/Ephrins as Biomarkers and as a Potential Platform for Targeted Delivery of Therapeutic Exosomes. Int. J. Mol. Sci. 2022, 23, 3551. [Google Scholar] [CrossRef] [PubMed]
- Hadjimichael, A.C.; Pergaris, A.; Kaspiris, A.; Foukas, A.F.; Theocharis, S.E. Liquid Biopsy: A New Translational Diagnostic and Monitoring Tool for Musculoskeletal Tumors. Int. J. Mol. Sci. 2021, 22, 11526. [Google Scholar] [CrossRef]
- Georgantzoglou, N.; Pergaris, A.; Masaoutis, C.; Theocharis, S. Extracellular Vesicles as Biomarkers Carriers in Bladder Cancer: Diagnosis, Surveillance, and Treatment. Int. J. Mol. Sci. 2021, 22, 2744. [Google Scholar] [CrossRef] [PubMed]
- Hanson, P.I.; Cashikar, A. Multivesicular Body Morphogenesis. Annu. Rev. Cell Dev. Biol. 2012, 28, 337–362. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2011, 40, D1241–D1244. [Google Scholar] [CrossRef]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Mignot, G.; Roux, S.; Thery, C.; Ségura, E.; Zitvogel, L. Prospects for exosomes in immunotherapy of cancer. J. Cell. Mol. Med. 2006, 10, 376–388. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.M.; Charrier, M.; Viaud, S.; André, F.; Besse, B.; Chaput, N.; Zitvogel, L. Dendritic Cell–Derived Exosomes as Immunotherapies in the Fight against Cancer. J. Immunol. 2014, 193, 1006–1011. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Gezer, U.; Özgür, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef]
- Liu, D.S.; Frampton, A.E. Plasma extracellular vesicles contain unannotated small RNA clusters suitable as biomarkers for detecting early hepatocellular carcinoma. Gut 2021, 71, 1935–1936. [Google Scholar] [CrossRef]
- Ostenfeld, M.S.; Jeppesen, D.K.; Laurberg, J.R.; Boysen, A.T.; Bramsen, J.B.; Primdal-Bengtson, B.; Hendrix, A.; Lamy, P.; Dagnaes-Hansen, F.; Rasmussen, M.H.; et al. Cellular Disposal of miR23b by RAB27-Dependent Exosome Release Is Linked to Acquisition of Metastatic Properties. Cancer Res. 2014, 74, 5758–5771. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Croce, C.M. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013, 10, 169–174. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Arvanitakis, K.; Stergiou, I.E.; Lekakis, V.; Davakis, S.; Christodoulou, M.-I.; Germanidis, G.; Theocharis, S. The Role of TLR4 in the Immunotherapy of Hepatocellular Carcinoma: Can We Teach an Old Dog New Tricks? Cancers 2023, 15, 2795. [Google Scholar] [CrossRef]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458, 445–452. [Google Scholar] [CrossRef]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef]
- Booth, A.M.; Fang, Y.; Fallon, J.K.; Yang, J.-M.; Hildreth, J.E.; Gould, S.J. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 2006, 172, 923–935. [Google Scholar] [CrossRef]
- Stuffers, S.; Wegner, C.S.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- Matsuo, H.; Chevallier, J.; Mayran, N.; Le Blanc, I.; Ferguson, C.; Fauré, J.; Blanc, N.S.; Matile, S.; Dubochet, J.; Sadoul, R.; et al. Role of LBPA and Alix in Multivesicular Liposome Formation and Endosome Organization. Science 2004, 303, 531–534. [Google Scholar] [CrossRef]
- Babst, M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 2011, 23, 452–457. [Google Scholar] [CrossRef]
- van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A Common Pathway for a Specialized Function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef]
- Savina, A.; Furlán, M.; Vidal, M.; Colombo, M.I. Exosome Release Is Regulated by a Calcium-dependent Mechanism in K562 Cells. J. Biol. Chem. 2003, 278, 20083–20090. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH Is a Key Factor for Exosome Traffic in Tumor Cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.C.; Chaudhary, V.; Bartscherer, K.; Boutros, M. Active Wnt proteins are secreted on exosomes. Nature 2012, 14, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Südhof, T.C.; Rothman, J.E. Membrane Fusion: Grappling with SNARE and SM Proteins. Science 2009, 323, 474–477. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Zhao, W.-L.; Ye, Y.-Y.; Bai, X.-C.; Liu, R.-Q.; Chang, L.-F.; Zhou, Q.; Sui, S.-F. Cellular Internalization of Exosomes Occurs through Phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhu, Y.-L.; Zhou, Y.-Y.; Liang, G.-F.; Wang, Y.-Y.; Hu, F.-H.; Xiao, Z.-D. Exosome Uptake through Clathrin-mediated Endocytosis and Macropinocytosis and Mediating miR-21 Delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Mörgelin, M.; Belting, M. Exosome Uptake Depends on ERK1/2-Heat Shock Protein 27 Signaling and Lipid Raft-mediated Endocytosis Negatively Regulated by Caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.-W.; Han, C.-L.; Dong, Z.-R.; Tan, S.-Y.; Wang, D.-X.; Li, T. Role of Exosomes in Immunotherapy of Hepatocellular Carcinoma. Cancers 2022, 14, 4036. [Google Scholar] [CrossRef] [PubMed]
- Sinha, D.; Roy, S.; Saha, P.; Chatterjee, N.; Bishayee, A. Trends in Research on Exosomes in Cancer Progression and Anticancer Therapy. Cancers 2021, 13, 326. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, H.; Zhangyuan, G.; Huang, R.; Zhang, W.; He, Q.; Jin, K.; Zhuo, H.; Zhang, Z.; Wang, J.; et al. 14-3-3ζ delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death Dis. 2018, 9, 159. [Google Scholar] [CrossRef]
- Huang, M.; Huang, X.; Huang, N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022, 113, 1968–1983. [Google Scholar] [CrossRef]
- Zhang, H.-Y.; Liang, H.-X.; Wu, S.-H.; Jiang, H.-Q.; Wang, Q.; Yu, Z.-J. Overexpressed Tumor Suppressor Exosomal miR-15a-5p in Cancer Cells Inhibits PD1 Expression in CD8+T Cells and Suppresses the Hepatocellular Carcinoma Progression. Front. Oncol. 2021, 11, 622263. [Google Scholar] [CrossRef]
- Zhang, P.-F.; Gao, C.; Huang, X.-Y.; Lu, J.-C.; Guo, X.-J.; Shi, G.-M.; Cai, J.-B.; Ke, A.-W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef]
- Kouwaki, T.; Fukushima, Y.; Daito, T.; Sanada, T.; Yamamoto, N.; Mifsud, E.J.; Leong, C.R.; Tsukiyama-Kohara, K.; Kohara, M.; Matsumoto, M.; et al. Extracellular Vesicles Including Exosomes Regulate Innate Immune Responses to Hepatitis B Virus Infection. Front. Immunol. 2016, 7, 335. [Google Scholar] [CrossRef]
- Lu, Z.; Zuo, B.; Jing, R.; Gao, X.; Rao, Q.; Liu, Z.; Qi, H.; Guo, H.; Yin, H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 2017, 67, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Chen, K.; Lu, X.; Li, A.; Liu, C.; Wu, B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol. Int. 2021, 15, 444–458. [Google Scholar] [CrossRef]
- Liu, J.; Fan, L.; Yu, H.; Zhang, J.; He, Y.; Feng, D.; Wang, F.; Li, X.; Liu, Q.; Li, Y.; et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology 2019, 70, 241–258. [Google Scholar] [CrossRef]
- Wang, L.; Hu, Y.-Y.; Zhao, J.-L.; Huang, F.; Liang, S.-Q.; Dong, L.; Chen, Y.; Yu, H.-C.; Bai, J.; Yang, J.-M.; et al. Targeted delivery of miR-99b reprograms tumor-associated macrophage phenotype leading to tumor regression. J. Immunother. Cancer 2020, 8, e000517. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Han, Q.; Xu, D.; Zheng, B.; Zhao, X.; Zhang, J. SALL4-mediated upregulation of exosomal miR-146a-5p drives T-cell exhaustion by M2 tumor-associated macrophages in HCC. Oncoimmunology 2019, 8, e1601479. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, Q.; Cheng, Y.; Chen, X.; Wang, G.; Shi, M.; Zhang, T.; Cao, Y.; Pan, H.; Zhang, L.; et al. Tumor-derived exosomal HMGB1 fosters hepatocellular carcinoma immune evasion by promoting TIM-1+ regulatory B cell expansion. J. Immunother. Cancer 2018, 6, 145. [Google Scholar] [CrossRef]
- Tian, X.; Shen, H.; Li, Z.; Wang, T.; Wang, S. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J. Hematol. Oncol. 2019, 12, 84. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, L.; Lv, D.; Zhu, X.; Tang, H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J. Hematol. Oncol. 2019, 12, 53. [Google Scholar] [CrossRef]
- Li, M.; Lu, Y.; Xu, Y.; Wang, J.; Zhang, C.; Du, Y.; Wang, L.; Li, L.; Wang, B.; Shen, J.; et al. Horizontal transfer of exosomal CXCR4 promotes murine hepatocarcinoma cell migration, invasion and lymphangiogenesis. Gene 2018, 676, 101–109. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, W.; Zhuo, H.; Zhu, D.; Rong, D.; Sun, J.; Song, J. Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/ nuclear factor -kappa B (NF-κB) pathway. Bioengineered 2022, 13, 4786–4797. [Google Scholar] [CrossRef]
- Liu, Y.; Tan, J.; Ou, S.; Chen, J.; Chen, L. Adipose-derived exosomes deliver miR-23a/b to regulate tumor growth in hepatocellular cancer by targeting the VHL/HIF axis. J. Physiol. Biochem. 2019, 75, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, G.; Lin, X.; Xing, X.; Cai, Z.; Liu, X.; Liu, J. Role of exosomes in hepatocellular carcinoma cell mobility alteration. Oncol. Lett. 2017, 14, 8122–8131. [Google Scholar] [CrossRef]
- Zhu, C.; Su, Y.; Liu, L.; Wang, S.; Liu, Y.; Wu, J. Circular RNA hsa_circ_0004277 Stimulates Malignant Phenotype of Hepatocellular Carcinoma and Epithelial-Mesenchymal Transition of Peripheral Cells. Front. Cell Dev. Biol. 2021, 8, 585565. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-S.; Liu, J.-B.; Lin, L.; Zhang, H.; Wu, J.-J.; Shi, Y.; Jia, C.-Y.; Zhang, D.-D.; Yu, F.; Wang, H.-M.; et al. Exosomal microRNA-15a from mesenchymal stem cells impedes hepatocellular carcinoma progression via downregulation of SALL4. Cell Death Discov. 2021, 7, 224. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Johnson, A.; Jacobs, E.; Zhang, K.; Chen, J.; Wei, Z.; Lian, Q.; Chen, H.J. Energy Sources for Exosome Communication in a Cancer Microenvironment. Cancers 2022, 14, 1698. [Google Scholar] [CrossRef]
- Bader, J.E.; Voss, K.; Rathmell, J.C. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol. Cell 2020, 78, 1019–1033. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Dedes, N.; Kouroumalis, E.; Theocharis, S. The Role of the NLRP3 Inflammasome in HCC Carcinogenesis and Treatment: Harnessing Innate Immunity. Cancers 2022, 14, 3150. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Stergiou, I.E.; Gkolemi, N.; Arvanitakis, K.; Theocharis, S. Unraveling the Significance of EPH/Ephrin Signaling in Liver Cancer: Insights into Tumor Progression and Therapeutic Implications. Cancers 2023, 15, 3434. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Ferraro, D.; Carbone, G.; Frampton, A.E.; Vennarecci, G.; Kykalos, S.; Schizas, D.; Theocharis, S.; Machairas, N. The Emerging Role of Metformin in the Treatment of Hepatocellular Carcinoma: Is There Any Value in Repurposing Metformin for HCC Immunotherapy? Cancers 2023, 15, 3161. [Google Scholar] [CrossRef]
- Luo, W. Nasopharyngeal carcinoma ecology theory: Cancer as multidimensional spatiotemporal “unity of ecology and evolution” pathological ecosystem. Theranostics 2023, 13, 1607–1631. [Google Scholar] [CrossRef]
- Tenchov, R.; Sasso, J.M.; Wang, X.; Liaw, W.-S.; Chen, C.-A.; Zhou, Q.A. Exosomes─Nature’s Lipid Nanoparticles, a Rising Star in Drug Delivery and Diagnostics. ACS Nano 2022, 16, 17802–17846. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, J.; Peng, Y.; Du, Y.; Yang, Z.; Qi, X. Dendritic cell derived exosomes loaded neoantigens for personalized cancer immunotherapies. J. Control. Release 2023, 353, 423–433. [Google Scholar] [CrossRef]
- Zhu, S.; Huang, H.; Liu, D.; Wen, S.; Shen, L.; Lin, Q. Augmented cellular uptake and homologous targeting of exosome-based drug loaded IOL for posterior capsular opacification prevention and biosafety improvement. Bioact. Mater. 2022, 15, 469–481. [Google Scholar] [CrossRef]
- Wu, G.; Zhang, J.; Zhao, Q.; Zhuang, W.; Ding, J.; Zhang, C.; Gao, H.; Pang, D.-W.; Pu, K.; Xie, H.-Y. Molecularly Engineered Macrophage-Derived Exosomes with Inflammation Tropism and Intrinsic Heme Biosynthesis for Atherosclerosis Treatment. Angew. Chem. Int. Ed. 2020, 59, 4068–4074. [Google Scholar] [CrossRef] [PubMed]
- Lennaárd, A.J.; Mamand, D.R.; Wiklander, R.J.; EL Andaloussi, S.; Wiklander, O.P.B. Optimised Electroporation for Loading of Extracellular Vesicles with Doxorubicin. Pharmaceutics 2021, 14, 38. [Google Scholar] [CrossRef]
- Le Saux, S.; Aarrass, H.; Lai-Kee-Him, J.; Bron, P.; Armengaud, J.; Miotello, G.; Bertrand-Michel, J.; Dubois, E.; George, S.; Faklaris, O.; et al. Post-production modifications of murine mesenchymal stem cell (mMSC) derived extracellular vesicles (EVs) and impact on their cellular interaction. Biomaterials 2019, 231, 119675. [Google Scholar] [CrossRef]
- Sun, L.; Fan, M.; Huang, D.; Li, B.; Xu, R.; Gao, F.; Chen, Y. Clodronate-loaded liposomal and fibroblast-derived exosomal hybrid system for enhanced drug delivery to pulmonary fibrosis. Biomaterials 2021, 271, 120761. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, D.; Ma, X.; Wang, J.; Hou, W.; Zhang, W. Exosomes as drug carriers for cancer therapy and challenges regarding exosome uptake. Biomed. Pharmacother. 2020, 128, 110237. [Google Scholar] [CrossRef]
- Machairas, N.; Tsilimigras, D.I.; Pawlik, T.M. Current Landscape of Immune Checkpoint Inhibitor Therapy for Hepatocellular Carcinoma. Cancers 2022, 14, 2018. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.-Y.; Wang, N.; Zhang, C.; Chan, Y.-T.; Yuen, M.-F.; Feng, Y. Lysyl Oxidase-Like 4 Fosters an Immunosuppressive Microenvironment During Hepatocarcinogenesis. Hepatology 2021, 73, 2326–2341. [Google Scholar] [CrossRef]
- Li, Y.-J.; Wu, J.-Y.; Liu, J.; Xu, W.; Qiu, X.; Huang, S.; Hu, X.-B.; Xiang, D.-X. Artificial exosomes for translational nanomedicine. J. Nanobiotechnol. 2021, 19, 242. [Google Scholar] [CrossRef]
- Arvanitakis, K.; Mitroulis, I.; Chatzigeorgiou, A.; Elefsiniotis, I.; Germanidis, G. The Liver Cancer Immune Microenvironment: Emerging Concepts for Myeloid Cell Profiling with Diagnostic and Therapeutic Implications. Cancers 2023, 15, 1522. [Google Scholar] [CrossRef]
- Cheng, K.; Cai, N.; Zhu, J.; Yang, X.; Liang, H.; Zhang, W. Tumor-associated macrophages in liver cancer: From mechanisms to therapy. Cancer Commun. 2022, 42, 1112–1140. [Google Scholar] [CrossRef]
- Lu, J.-C.; Zhang, P.-F.; Huang, X.-Y.; Guo, X.-J.; Gao, C.; Zeng, H.-Y.; Zheng, Y.-M.; Wang, S.-W.; Cai, J.-B.; Sun, Q.-M.; et al. Amplification of spatially isolated adenosine pathway by tumor–macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J. Hematol. Oncol. 2021, 14, 200. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lin, Z.; Liu, L.; Zhang, R.; Geng, Y.; Fan, M.; Zhu, W.; Lu, M.; Jia, H.; Zhang, J.; et al. GOLM1 exacerbates CD8+ T cell suppression in hepatocellular carcinoma by promoting exosomal PD-L1 transport into tumor-associated macrophages. Signal Transduct. Target. Ther. 2021, 6, 397. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Hu, Z.; Yin, Y.; Jiang, J.; Yan, C.; Wang, Y.; Wang, D.; Li, L. Exosomal miR-142-3p secreted by hepatitis B virus (HBV)-hepatocellular carcinoma (HCC) cells promotes ferroptosis of M1-type macrophages through SLC3A2 and the mechanism of HCC progression. J. Gastrointest. Oncol. 2022, 13, 754–767. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.; Li, C.; Zhang, Z.; Gao, X.; Jia, Z.; Chen, H.; Jia, Q.; Zhao, X.; Liu, J.; et al. SLC3A2 is a novel endoplasmic reticulum stress-related signaling protein that regulates the unfolded protein response and apoptosis. PLoS ONE 2018, 13, e0208993. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, H.; Liu, W.; Yin, Y.; Jiang, J.; Yan, C.; Wang, Y.; Li, L. Mechanism of HBV-positive liver cancer cell exosomal miR-142-3p by inducing ferroptosis of M1 macrophages to promote liver cancer progression. Transl. Cancer Res. 2022, 11, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- Zanganeh, S.; Hutter, G.; Spitler, R.; Lenkov, O.; Mahmoudi, M.; Shaw, A.; Pajarinen, J.S.; Nejadnik, H.; Goodman, S.; Moseley, M.; et al. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat. Nanotechnol. 2016, 11, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Kuo, N.; Kryczek, I.; Zou, W.; Welling, T.H. Myeloid cells in hepatocellular carcinoma. Hepatology 2015, 62, 1304–1312. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Ju, Y.; Wang, C.; Wei, R.; Sun, H.; Zhang, Q. MARCKS on Tumor-Associated Macrophages is Correlated with Immune Infiltrates and Poor Prognosis in Hepatocellular Carcinoma. Cancer Investig. 2021, 39, 756–768. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef]
- Sun, Q.; Shen, M.; Zhu, S.; Liao, Y.; Zhang, D.; Sun, J.; Guo, Z.; Wu, L.; Xiao, L.; Liu, L. Targeting NAD+ metabolism of hepatocellular carcinoma cells by lenvatinib promotes M2 macrophages reverse polarization, suppressing the HCC progression. Hepatol. Int. 2023. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-X.; Xu, X.-H.; Jin, L. Effects of Metabolism on Macrophage Polarization Under Different Disease Backgrounds. Front. Immunol. 2022, 13, 880286. [Google Scholar] [CrossRef]
- Ji, W.; Bai, J.; Ke, Y. Exosomal ZFPM2-AS1 contributes to tumorigenesis, metastasis, stemness, macrophage polarization, and infiltration in hepatocellular carcinoma through PKM mediated glycolysis. Environ. Toxicol. 2023, 38, 1332–1346. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, M.; Wang, G.; Mai, Z.; Zhou, B.; Han, Y.; Zhuang, J.; Xia, W. lncRNA miR4458HG modulates hepatocellular carcinoma progression by activating m6A-dependent glycolysis and promoting the polarization of tumor-associated macrophages. Cell. Mol. Life Sci. 2023, 80, 99. [Google Scholar] [CrossRef]
- Han, Q.; Wang, M.; Dong, X.; Wei, F.; Luo, Y.; Sun, X. Non-coding RNAs in hepatocellular carcinoma: Insights into regulatory mechanisms, clinical significance, and therapeutic potential. Front. Immunol. 2022, 13, 985815. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, R.; Li, J.; Tang, S.; Li, S.; Tong, Q.; Li, S. Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int. J. Nanomed. 2021, 16, 2803–2818. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, Y.; Dong, K.; Zhang, H.; Gong, J.; Wang, S. ExosomallncRNA HMMR-AS1mediates macrophage polarization throughmiR-147a/ARID3Aaxis under hypoxia and affects the progression of hepatocellular carcinoma. Environ. Toxicol. 2022, 37, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lei, Y.; Wu, M.; Li, N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018, 19, 2958. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Li, D.; Mu, S.; Tian, G.; Yan, G. LncRNA MAPKAPK5_AS1 facilitates cell proliferation in hepatitis B virus -related hepatocellular carcinoma. Lab. Investig. 2022, 102, 494–504. [Google Scholar] [CrossRef]
- Lv, S.; Wang, J.; Li, L. Extracellular vesicular lncRNA FAL1 promotes hepatocellular carcinoma cell proliferation and invasion by inducing macrophage M2 polarization. J. Physiol. Biochem. 2023, 79, 669–682. [Google Scholar] [CrossRef]
- Zongqiang, H.; Jiapeng, C.; Yingpeng, Z.; Chuntao, Y.; Yiting, W.; Jiashun, Z.; Li, L. Exosomal miR-452-5p Induce M2 Macrophage Polarization to Accelerate Hepatocellular Carcinoma Progression by Targeting TIMP3. J. Immunol. Res. 2022, 2022, 1032106. [Google Scholar] [CrossRef]
- Yu, H.; Pan, J.; Zheng, S.; Cai, D.; Luo, A.; Xia, Z.; Huang, J. Hepatocellular Carcinoma Cell-Derived Exosomal miR-21-5p Induces Macrophage M2 Polarization by Targeting RhoB. Int. J. Mol. Sci. 2023, 24, 4593. [Google Scholar] [CrossRef]
- Xu, Y.; Luan, G.; Liu, F.; Zhang, Y.; Li, Z.; Liu, Z.; Yang, T. Exosomal miR-200b-3p induce macrophage polarization by regulating transcriptional repressor ZEB1 in hepatocellular carcinoma. Hepatol. Int. 2023, 17, 889–903. [Google Scholar] [CrossRef]
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020, 13, 156. [Google Scholar] [CrossRef]
- Xu, M.; Zhou, C.; Weng, J.; Chen, Z.; Zhou, Q.; Gao, J.; Shi, G.; Ke, A.; Ren, N.; Sun, H.; et al. Tumor associated macrophages-derived exosomes facilitate hepatocellular carcinoma malignance by transferring lncMMPA to tumor cells and activating glycolysis pathway. J. Exp. Clin. Cancer Res. 2022, 41, 253. [Google Scholar] [CrossRef]
- Li, W.; Xin, X.; Li, X.; Geng, J.; Sun, Y. Exosomes secreted by M2 macrophages promote cancer stemness of hepatocellular carcinoma via the miR-27a-3p/TXNIP pathways. Int. Immunopharmacol. 2021, 101, 107585. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tang, C.; Tan, C.; Wu, F.; Li, Z.; Ji, W.; Lu, L.; Xu, C.; Shen, Z.; Huang, Y. IL-2 Modulates TAMs Derived Exosomal MiRNAs to Ameliorate Hepatocellular Carcinoma Development and Progression. J. Oncol. 2022, 2022, 3445350. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, B.; Xiao, S.; Li, Y.; Chen, Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J. Cell. Biochem. 2018, 120, 3046–3055. [Google Scholar] [CrossRef]
- Tian, B.; Zhou, L.; Wang, J.; Yang, P. miR-660-5p-loaded M2 macrophages-derived exosomes augment hepatocellular carcinoma development through regulating KLF3. Int. Immunopharmacol. 2021, 101, 108157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, J.; Li, P.; Li, T.; Zhou, Z.; Wu, H. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 2022, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Yao, L.; Liu, G.; Liu, S.; Gong, L.; Xiao, Y. Loss of androgen receptor promotes HCC invasion and metastasis via activating circ-LNPEP/miR-532–3p/RAB9A signal under hypoxia. Biochem. Biophys. Res. Commun. 2021, 557, 26–32. [Google Scholar] [CrossRef]
- Liu, G.; Ouyang, X.; Sun, Y.; Xiao, Y.; You, B.; Gao, Y.; Yeh, S.; Li, Y.; Chang, C. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death Differ. 2020, 27, 3258–3272. [Google Scholar] [CrossRef]
- Wang, L.; Yi, X.; Xiao, X.; Zheng, Q.; Ma, L.; Li, B. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell. Mol. Biol. Lett. 2022, 27, 106. [Google Scholar] [CrossRef]
- Arvanitakis, K.; Mitroulis, I.; Germanidis, G. Tumor-Associated Neutrophils in Hepatocellular Carcinoma Pathogenesis, Prognosis, and Therapy. Cancers 2021, 13, 2899. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Bonapace, L.; Coissieux, M.-M.; Wyckoff, J.; Mertz, K.D.; Varga, Z.; Junt, T.; Bentires-Alj, M. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature 2014, 515, 130–133. [Google Scholar] [CrossRef]
- Hume, D.A.; MacDonald, K.P.A. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood 2012, 119, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Zeisberger, S.M.; Odermatt, B.; Marty, C.; Zehnder-Fjällman, A.H.M.; Ballmer-Hofer, K.; A Schwendener, R. Clodronate-liposome-mediated depletion of tumour-associated macrophages: A new and highly effective antiangiogenic therapy approach. Br. J. Cancer 2006, 95, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, P.; Tan, H.; Chen, X.; Wang, Q.; Chen, T. Exosomes as Smart Nanoplatforms for Diagnosis and Therapy of Cancer. Front. Oncol. 2021, 11, 743189. [Google Scholar] [CrossRef]
- Shang, L.; Jiang, X.; Yang, T.; Xu, H.; Xie, Q.; Hu, M.; Yang, C.; Kong, L.; Zhang, Z. Enhancing cancer chemo-immunotherapy by biomimetic nanogel with tumor targeting capacity and rapid drug-releasing in tumor microenvironment. Acta Pharm. Sin. B 2021, 12, 2550–2567. [Google Scholar] [CrossRef]
- Pippa, N.; Skouras, A.; Naziris, N.; Biondo, F.; Tiboni, M.; Katifelis, H.; Gazouli, M.; Demetzos, C.; Casettari, L. Incorporation of PEGylated δ-decalactone into lipid bilayers: Thermodynamic study and chimeric liposomes development. J. Liposome Res. 2019, 30, 209–217. [Google Scholar] [CrossRef]
- Papadopoulou, S.; Kolokithas-Ntoukas, A.; Salvanou, E.-A.; Gaitanis, A.; Xanthopoulos, S.; Avgoustakis, K.; Gazouli, M.; Paravatou-Petsotas, M.; Tsoukalas, C.; Bakandritsos, A.; et al. Chelator-Free/Chelator-Mediated Radiolabeling of Colloidally Stabilized Iron Oxide Nanoparticles for Biomedical Imaging. Nanomaterials 2021, 11, 1677. [Google Scholar] [CrossRef]
- Kontogiannis, O.; Selianitis, D.; Lagopati, N.; Pippa, N.; Pispas, S.; Gazouli, M. Surfactant and Block Copolymer Nanostructures: From Design and Development to Nanomedicine Preclinical Studies. Pharmaceutics 2023, 15, 501. [Google Scholar] [CrossRef]
- Nteli, P.; Bajwa, D.E.; Politakis, D.; Michalopoulos, C.; Kefala-Narin, A.; Efstathopoulos, E.P.; Gazouli, M. Nanomedicine approaches for treatment of hematologic and oncologic malignancies. World J. Clin. Oncol. 2022, 13, 553–566. [Google Scholar] [CrossRef]
- Wang, J.; Sun, X.; Mao, W.; Sun, W.; Tang, J.; Sui, M.; Shen, Y.; Gu, Z. Tumor Redox Heterogeneity-Responsive Prodrug Nanocapsules for Cancer Chemotherapy. Adv. Mater. 2013, 25, 3670–3676. [Google Scholar] [CrossRef] [PubMed]
- Papazoglou, S.; Tsagkaris, C.; Moysidis, D.V.; Papadakos, S.; Galkin, O.Y.; Orel, V.E.; Syvak, L.A. Nanotherapy based on magneto-mechanochemical modulation of tumor redox state. WIREs Nanomed. Nanobiotechnol. 2022, 15, e1868. [Google Scholar] [CrossRef]
- Lapitz, A.; Azkargorta, M.; Milkiewicz, P.; Olaizola, P.; Zhuravleva, E.; Grimsrud, M.M.; Schramm, C.; Arbelaiz, A.; O’Rourke, C.J.; La Casta, A.; et al. Liquid biopsy-based protein biomarkers for risk prediction, early diagnosis, and prognostication of cholangiocarcinoma. J. Hepatol. 2023, 79, 93–108. [Google Scholar] [CrossRef]
- Guo, X.; Peng, Y.; Song, Q.; Wei, J.; Wang, X.; Ru, Y.; Xu, S.; Cheng, X.; Li, X.; Wu, D.; et al. A Liquid Biopsy Signature for the Early Detection of Gastric Cancer in Patients. Gastroenterology 2023, 165, 402–413.e13. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]

| Author, Year | Molecule/Type of Study | Mechanisms | Outcomes | Ref. |
|---|---|---|---|---|
| Wang, 2021 | hsa_circ_0074854/preclinical | Interaction with human antigen R (HuR). | Downregulation of hsa_circ_0074854 hinders the migration and invasion of HCC cells. | [121] |
| Exosomes containing decreased levels of hsa_circ_0074854 exhibited the ability to inhibit M2 polarization. | ||||
| Wang, 2021 | LncRNA HMMR-AS1/preclinical | HMMR-AS1 is competitively bound to miR-147a, preventing ARID3A degradation. | Inhibiting HMMR-AS1 expression substantially suppresses tumor growth in vitro and in vivo. | [122] |
| Exosomes carrying HMMR-AS1 facilitated M2 polarization. | ||||
| HIF-1α enhanced HMMR-AS1 transcription, leading to an increased secretion of exosomes. | ||||
| Li, 2018 | lncRNA TUC339/in vitro | Tumor-derived exosomes containing elevated levels of the lncRNA TUC339 are taken up by THP-1 cells (macrophages). | TUC339 is involved in the regulation of macrophage activation and polarization, specifically M1/M2 polarization. | [123] |
| Suppression of TUC339 in macrophages leads to increased pro-inflammatory cytokine production, enhanced co-stimulatory molecule expression and improved phagocytosis. | TUC339 affects various cellular processes and pathways related to cytokine signaling, chemokine receptor binding, toll-like receptor signaling, phagocytosis, cytoskeleton regulation, and cell proliferation in macrophages. | |||
| Overexpression of TUC339 in macrophages has the opposite effect, reducing pro-inflammatory cytokine production, co-stimulatory molecule expression, and phagocytosis. | ||||
| TUC339 is involved in cytokine-cytokine receptor interaction, CXCR chemokine receptor binding, Toll-like receptor signaling, FcγR-mediated phagocytosis, regulation of the actin cytoskeleton, and cell proliferation in macrophages. | ||||
| Tao, 2022 | LncRNA MAPKAPK5_AS1 (MAAS)/preclinical | Upregulation of MAAS in HBV-related HCC cancerous tissues. Promotion of c-Myc-induced transcriptional activation. | Poor survival probability in patients with high MAAS expression. Facilitated proliferation of HBV+HCC cells in vitro and in vivo. | [124] |
| Stabilization of the c-Myc protein. Facilitation of the G1/S transition. Enhancing the N6-methyladenosine modification of MAAS mediated by methyltransferase-like 3. | Activation of cyclin-dependent kinase 4 (CDK4), CDK6, and S-phase kinase-associated protein 2. Promotion of cell proliferation in HBV and HCC cells. | |||
| Transfer of MAAS to HBV + HCC cells via exosomes derived from M2 macrophages. | Transfer of MAAS from M2 macrophages to HBV + HCC cells via exosomes. Establishment of a positive feedback loop between HBeAg, MAAS expression, and M2 macrophages. | |||
| Lv, 2022 | lncRNA FAL1/in vitro | Extracellular vesicular lncRNA FAL1 induces macrophage M2 polarization. | Macrophage M2 polarization is promoted by FAL1-enriched EVs. Co-culture of FAL1-overexpressing macrophages with HepG2 cells facilitates the malignant progression of HepG2 cells. | [125] |
| FAL1-enriched EVs stimulate the activation of the Wnt/β-catenin signaling pathway in HCC cells. | Activation of the Wnt/β-catenin signaling pathway in HCC cells is observed when co-cultured with EVs-incubated macrophages. Mouse xenograft tumor growth is increased by FAL1-enriched EVs-incubated macrophages. | |||
| Zongqiang, 2022 | miR-452-5p/preclinical | HCC cells secrete exosomal miR-452-5p. | Exosomal miR-452-5p promotes the progression of HCC. HCC cell-derived exosomes, along with miR-452-5p overexpression, accelerate HCC migration and invasion. | [126] |
| Exosomal miR-452-5p induces polarization of M2 macrophages. MiR-452-5p targets TIMP3, leading to its downregulation. | In vivo experiments demonstrate that miR-452-5p accelerates HCC growth and metastasis. Overexpression of TIMP3 inhibits the pro-invasive and migratory effects of HCC cell-derived exosomes. | |||
| Yu, 2023 | miR-21-5p/clinical and preclinical | HCC-derived exosomes mediate intercellular communication and promote TAMs’ phenotypic differentiation into M2-like macrophages. | HCC cell-derived exosomes significantly induce the differentiation of THP-1 macrophages into M2-like macrophages, characterized by increased production of TGF-β and IL-10. | [127] |
| Exosomal miR-21-5p is closely related to TAM differentiation and directly targets the 3′-UTR of RhoB in THP-1 cells. | Overexpression of miR-21-5p in THP-1 cells leads to downregulation of IL-1β levels, enhanced production of IL-10, and promotes malignant growth of HCC cells in vitro. | |||
| Downregulation of RhoB weakens the MAPK signaling pathways in THP-1 cells. | Tumor-derived miR-21-5p facilitates the malignant progression of HCC by mediating intercellular crosstalk between tumor cells and macrophages. | |||
| Liu, 2019 | miR-23a-3p/in vitro and preclinical | Exosomes derived from ER-stressed HCC cells stimulate macrophages to upregulate the expression of PD-L1. | [74] | |
| The induction of ER stress leads to the upregulation of ER stress markers, including glucose-regulated protein 78 (GRP78), activating transcription factor 6 (ATF6), PKR-like endoplasmic reticulum kinase (PERK), and inositol-requiring enzyme 1α (IRE1α), in HCC cells. | Increased PD-L1 expression on macrophages inhibits T-cell function by interacting with the PD-1 receptor on T cells, leading to a decreased CD8+ T-cell ratio, reduced IL2 production, and increased T-cell apoptosis. | |||
| ER stress induces HCC cells to release exosomes containing high levels of miR-23a-3p. | The release of exosomal miR-23a-3p and subsequent upregulation of PD-L1 on macrophages contribute to the evasion of antitumor immunity by HCC cells. | |||
| Xu, 2022 | miR-200b-3p/in vitro and in vivo | HCC cell-derived miR-200b-3p exosomes downregulate ZEB1. | Induction of M2 polarization in macrophages. | [128] |
| MiR-200b-3p exosomes upregulate IL4. | Enhanced proliferation and metastasis of HCC cells. | |||
| Activation of the JAK/STAT signaling pathway in M2 macrophages. | Establishment of a feedback loop between HCC cells and M2 macrophages. | |||
| Increased expression of PIM1 and VEGFα. | Promotion of tumor growth and progression in the tumor microenvironment. | |||
| Zhao, 2020 | miR-934/in vivo and in vitro | Tumor cells release exosomal miR-934. | Induction of M2 macrophage polarization. | [129] |
| Exosomal miR-934 is internalized by macrophages. | Activation of the CXCL13/CXCR5/NFκB/p65/miR-934 positive feedback loop. | |||
| Exosomal miR-934 downregulates PTEN expression. | Promotion of premetastatic niche formation. | |||
| Downregulation of PTEN activates the PI3K/AKT signaling pathway. | Facilitation of colorectal cancer liver metastasis (CRLM). | |||
| Polarized M2 macrophages secrete CXCL13. | Correlation of miR-934 overexpression with poor prognosis in CRC patients. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadakos, S.P.; Machairas, N.; Stergiou, I.E.; Arvanitakis, K.; Germanidis, G.; Frampton, A.E.; Theocharis, S. Unveiling the Yin-Yang Balance of M1 and M2 Macrophages in Hepatocellular Carcinoma: Role of Exosomes in Tumor Microenvironment and Immune Modulation. Cells 2023, 12, 2036. https://doi.org/10.3390/cells12162036
Papadakos SP, Machairas N, Stergiou IE, Arvanitakis K, Germanidis G, Frampton AE, Theocharis S. Unveiling the Yin-Yang Balance of M1 and M2 Macrophages in Hepatocellular Carcinoma: Role of Exosomes in Tumor Microenvironment and Immune Modulation. Cells. 2023; 12(16):2036. https://doi.org/10.3390/cells12162036
Chicago/Turabian StylePapadakos, Stavros P., Nikolaos Machairas, Ioanna E. Stergiou, Konstantinos Arvanitakis, Georgios Germanidis, Adam Enver Frampton, and Stamatios Theocharis. 2023. "Unveiling the Yin-Yang Balance of M1 and M2 Macrophages in Hepatocellular Carcinoma: Role of Exosomes in Tumor Microenvironment and Immune Modulation" Cells 12, no. 16: 2036. https://doi.org/10.3390/cells12162036
APA StylePapadakos, S. P., Machairas, N., Stergiou, I. E., Arvanitakis, K., Germanidis, G., Frampton, A. E., & Theocharis, S. (2023). Unveiling the Yin-Yang Balance of M1 and M2 Macrophages in Hepatocellular Carcinoma: Role of Exosomes in Tumor Microenvironment and Immune Modulation. Cells, 12(16), 2036. https://doi.org/10.3390/cells12162036