Human Lung Organoids—A Novel Experimental and Precision Medicine Approach
Abstract
:1. Introduction
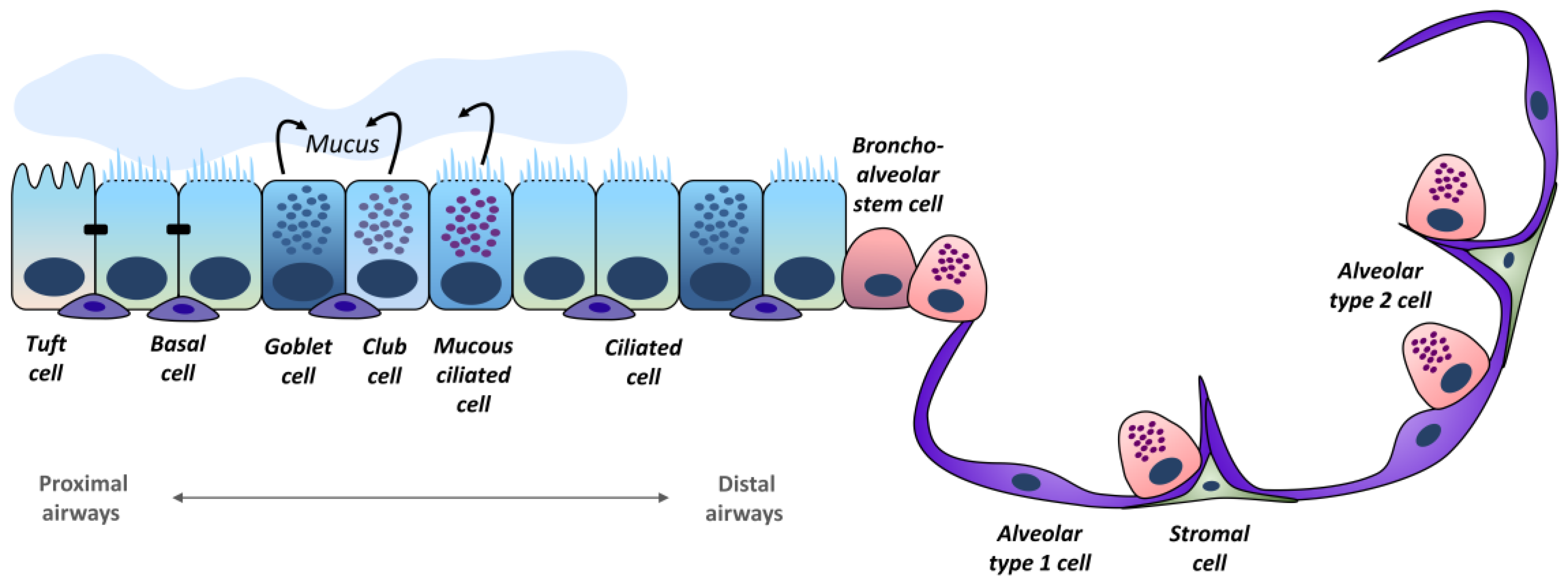
2. Lung Epithelium
3. Human Cell-/Tissue-Based Model Systems
3.1. 2D Cell Cultures
3.2. Air–Liquid Interface Cultures
3.3. Precision Cut Lung Slices
3.4. Organoids
3.5. Decellularized Lung Scaffolds
3.6. Microphysiological Systems
3.7. Animal Models
4. Human Lung Organoids
5. Lung Organoids in Translational Research and Personalized Respiratory Medicine
5.1. Cystic Fibrosis
5.2. Idiopathic Pulmonary Fibrosis
5.3. Chronic Obstructive Lung Diseases—COPD and Asthma
5.4. Infectious Diseases
5.5. Oncology
6. Future Perspectives for Lung Transplantation and Gene Therapy
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ferkol, T.; Schraufnagel, D. The global burden of respiratory disease. Ann. Am. Thorac. Soc. 2014, 11, 404–406. [Google Scholar] [CrossRef]
- Li, X.; Cao, X.; Guo, M.; Xie, M.; Liu, X. Trends and risk factors of mortality and disability adjusted life years for chronic respiratory diseases from 1990 to 2017: Systematic analysis for the Global Burden of Disease Study 2017. BMJ 2020, 368, m234. [Google Scholar] [CrossRef] [Green Version]
- GBD Chronic Respiratory Disease Collaborators. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2020, 8, 585–596. [Google Scholar] [CrossRef]
- WHO. Estimated Age-Standardized Mortality Rates (World) in 2020, World, Both Sexes, All Ages (excl. NMSC). Available online: https://gco.iarc.fr/today/online-analysis-multi-bars (accessed on 28 July 2023).
- Foreman, K.J.; Marquez, N.; Dolgert, A.; Fukutaki, K.; Fullman, N.; McGaughey, M.; Pletcher, M.A.; Smith, A.E.; Tang, K.; Yuan, C.-W.; et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: Reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet 2018, 392, 2052–2090. [Google Scholar] [CrossRef] [Green Version]
- Hild, M.; Jaffe, A.B. Production of 3-D Airway Organoids From Primary Human Airway Basal Cells and Their Use in High-Throughput Screening. Curr. Protoc. Stem Cell Biol. 2016, 37, IE.9.1–IE.9.15. [Google Scholar] [CrossRef]
- Vieira Braga, F.A.; Kar, G.; Berg, M.; Carpaij, O.A.; Polanski, K.; Simon, L.M.; Brouwer, S.; Gomes, T.; Hesse, L.; Jiang, J.; et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019, 25, 1153–1163. [Google Scholar] [CrossRef] [Green Version]
- Barkauskas, C.E.; Chung, M.-I.; Fioret, B.; Gao, X.; Katsura, H.; Hogan, B.L.M. Lung organoids: Current uses and future promise. Development 2017, 144, 986–997. [Google Scholar] [CrossRef] [Green Version]
- Deprez, M.; Zaragosi, L.-E.; Truchi, M.; Becavin, C.; Ruiz García, S.; Arguel, M.-J.; Plaisant, M.; Magnone, V.; Lebrigand, K.; Abelanet, S.; et al. A Single-Cell Atlas of the Human Healthy Airways. Am. J. Respir. Crit. Care Med. 2020, 202, 1636–1645. [Google Scholar] [CrossRef]
- Vock, C.; Yildirim, A.Ö.; Wagner, C.; Schlick, S.; Lunding, L.P.; Lee, C.G.; Elias, J.A.; Fehrenbach, H.; Wegmann, M. Distal airways are protected from goblet cell metaplasia by diminished expression of IL-13 signalling components. Clin. Exp. Allergy 2015, 45, 1447–1458. [Google Scholar] [CrossRef]
- Whitsett, J.A. Airway Epithelial Differentiation and Mucociliary Clearance. Ann. Am. Thorac. Soc. 2018, 15, S143–S148. [Google Scholar] [CrossRef]
- Kim, H.-T.; Yin, W.; Nakamichi, Y.; Panza, P.; Grohmann, B.; Buettner, C.; Guenther, S.; Ruppert, C.; Kobayashi, Y.; Guenther, A.; et al. WNT/RYK signaling restricts goblet cell differentiation during lung development and repair. Proc. Natl. Acad. Sci. USA 2019, 116, 25697–25706. [Google Scholar] [CrossRef]
- Bustamante-Marin, X.M.; Ostrowski, L.E. Cilia and Mucociliary Clearance. Cold Spring Harb. Perspect. Biol. 2017, 9, a028241. [Google Scholar] [CrossRef]
- Ualiyeva, S.; Lemire, E.; Aviles, E.C.; Wong, C.; Boyd, A.A.; Lai, J.; Liu, T.; Matsumoto, I.; Barrett, N.A.; Boyce, J.A.; et al. Tuft cell-produced cysteinyl leukotrienes and IL-25 synergistically initiate lung type 2 inflammation. Sci. Immunol. 2021, 6, eabj0474. [Google Scholar] [CrossRef]
- Bankova, L.G.; Dwyer, D.F.; Yoshimoto, E.; Ualiyeva, S.; McGinty, J.W.; Raff, H.; von Moltke, J.; Kanaoka, Y.; Frank Austen, K.; Barrett, N.A. The cysteinyl leukotriene 3 receptor regulates expansion of IL-25-producing airway brush cells leading to type 2 inflammation. Sci. Immunol. 2018, 3, eaat9453. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Liu, K.; Cui, G.; Huang, X.; Yao, S.; Guo, W.; Qin, Z.; Li, Y.; Yang, R.; Pu, W.; et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat. Genet. 2019, 51, 728–738. [Google Scholar] [CrossRef]
- Montoro, D.T.; Haber, A.L.; Biton, M.; Vinarsky, V.; Lin, B.; Birket, S.E.; Yuan, F.; Chen, S.; Leung, H.M.; Villoria, J.; et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018, 560, 319–324. [Google Scholar] [CrossRef]
- Sui, P.; Wiesner, D.L.; Xu, J.; Zhang, Y.; Lee, J.; van Dyken, S.; Lashua, A.; Yu, C.; Klein, B.S.; Locksley, R.M.; et al. Pulmonary neuroendocrine cells amplify allergic asthma responses. Science 2018, 360, eaan8546. [Google Scholar] [CrossRef] [Green Version]
- Barkauskas, C.E.; Cronce, M.J.; Rackley, C.R.; Bowie, E.J.; Keene, D.R.; Stripp, B.R.; Randell, S.H.; Noble, P.W.; Hogan, B.L.M. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Investig. 2013, 123, 3025–3036. [Google Scholar] [CrossRef]
- Weibel, E.R. Lung morphometry: The link between structure and function. Cell Tissue Res. 2017, 367, 413–426. [Google Scholar] [CrossRef]
- Travaglini, K.J.; Nabhan, A.N.; Penland, L.; Sinha, R.; Gillich, A.; Sit, R.V.; Chang, S.; Conley, S.D.; Mori, Y.; Seita, J.; et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020, 587, 619–625. [Google Scholar] [CrossRef]
- Wang, K.; Man, K.; Liu, J.; Liu, Y.; Chen, Q.; Zhou, Y.; Yang, Y. Microphysiological Systems: Design, Fabrication, and Applications. ACS Biomater. Sci. Eng. 2020, 6, 3231–3257. [Google Scholar] [CrossRef] [PubMed]
- Reddel, R.R.; Ke, Y.; Gerwin, B.I.; McMenamin, M.G.; Lechner, J.F.; Su, R.T.; Brash, D.E.; Park, J.B.; Rhim, J.S.; Harris, C.C. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988, 48, 1904–1909. [Google Scholar] [PubMed]
- Cabrini, G.; Rimessi, A.; Borgatti, M.; Lampronti, I.; Finotti, A.; Pinton, P.; Gambari, R. Role of Cystic Fibrosis Bronchial Epithelium in Neutrophil Chemotaxis. Front. Immunol. 2020, 11, 1438. [Google Scholar] [CrossRef]
- Veit, G.; Bossard, F.; Goepp, J.; Verkman, A.S.; Galietta, L.J.V.; Hanrahan, J.W.; Lukacs, G.L. Proinflammatory cytokine secretion is suppressed by TMEM16A or CFTR channel activity in human cystic fibrosis bronchial epithelia. Mol. Biol. Cell 2012, 23, 4188–4202. [Google Scholar] [CrossRef]
- Han, X.; Na, T.; Wu, T.; Yuan, B.-Z. Human lung epithelial BEAS-2B cells exhibit characteristics of mesenchymal stem cells. PLoS ONE 2020, 15, e0227174. [Google Scholar] [CrossRef]
- Sauler, M.; McDonough, J.E.; Adams, T.S.; Kothapalli, N.; Barnthaler, T.; Werder, R.B.; Schupp, J.C.; Nouws, J.; Robertson, M.J.; Coarfa, C.; et al. Characterization of the COPD alveolar niche using single-cell RNA sequencing. Nat. Commun. 2022, 13, 494. [Google Scholar] [CrossRef]
- Chen, K.G.; Park, K.; Spence, J.R. Studying SARS-CoV-2 infectivity and therapeutic responses with complex organoids. Nat. Cell Biol. 2021, 23, 822–833. [Google Scholar] [CrossRef]
- Lagowala, D.A.; Kwon, S.; Sidhaye, V.K.; Kim, D.-H. Human microphysiological models of airway and alveolar epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 321, L1072–L1088. [Google Scholar] [CrossRef]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Konar, D.; Devarasetty, M.; Yildiz, D.V.; Atala, A.; Murphy, S.V. Lung-On-A-Chip Technologies for Disease Modeling and Drug Development. Biomed. Eng. Comput. Biol. 2016, 7, 17–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cervena, T.; Vojtisek-Lom, M.; Vrbova, K.; Ambroz, A.; Novakova, Z.; Elzeinova, F.; Sima, M.; Beranek, V.; Pechout, M.; Macoun, D.; et al. Ordinary Gasoline Emissions Induce a Toxic Response in Bronchial Cells Grown at Air-Liquid Interface. Int. J. Mol. Sci. 2020, 22, 79. [Google Scholar] [CrossRef]
- Klein, S.G.; Serchi, T.; Hoffmann, L.; Blömeke, B.; Gutleb, A.C. An improved 3D tetraculture system mimicking the cellular organisation at the alveolar barrier to study the potential toxic effects of particles on the lung. Part. Fibre Toxicol. 2013, 10, 31. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Schoen, J. Air-liquid interface cell culture: From airway epithelium to the female reproductive tract. Reprod. Domest. Anim. 2019, 54 (Suppl. S3), 38–45. [Google Scholar] [CrossRef] [Green Version]
- Pezzulo, A.A.; Starner, T.D.; Scheetz, T.E.; Traver, G.L.; Tilley, A.E.; Harvey, B.-G.; Crystal, R.G.; McCray, P.B.; Zabner, J. The air-liquid interface and use of primary cell cultures are important to recapitulate the transcriptional profile of in vivo airway epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011, 300, L25–L31. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Adamcakova-Dodd, A.; Steines, B.R.; Jing, X.; Salem, A.K.; Thorne, P.S. Comparison of in vitro toxicity of aerosolized engineered nanomaterials using air-liquid interface mono-culture and co-culture models. NanoImpact 2020, 18, 100215. [Google Scholar] [CrossRef]
- Blom, R.A.M.; Erni, S.T.; Krempaská, K.; Schaerer, O.; van Dijk, R.M.; Amacker, M.; Moser, C.; Hall, S.R.R.; von Garnier, C.; Blank, F. A Triple Co-Culture Model of the Human Respiratory Tract to Study Immune-Modulatory Effects of Liposomes and Virosomes. PLoS ONE 2016, 11, e0163539. [Google Scholar] [CrossRef] [Green Version]
- Viana, F.; O’Kane, C.M.; Schroeder, G.N. Precision-cut lung slices: A powerful ex vivo model to investigate respiratory infectious diseases. Mol. Microbiol. 2022, 117, 578–588. [Google Scholar] [CrossRef]
- Alsafadi, H.N.; Uhl, F.E.; Pineda, R.H.; Bailey, K.E.; Rojas, M.; Wagner, D.E.; Königshoff, M. Applications and Approaches for Three-Dimensional Precision-Cut Lung Slices. Disease Modeling and Drug Discovery. Am. J. Respir. Cell Mol. Biol. 2020, 62, 681–691. [Google Scholar] [CrossRef]
- Liu, G.; Särén, L.; Douglasson, H.; Zhou, X.-H.; Åberg, P.M.; Ollerstam, A.; Betts, C.J.; Balogh Sivars, K. Precision cut lung slices: An ex vivo model for assessing the impact of immunomodulatory therapeutics on lung immune responses. Arch. Toxicol. 2021, 95, 2871–2877. [Google Scholar] [CrossRef]
- Wronski, S.; Beinke, S.; Obernolte, H.; Belyaev, N.N.; Saunders, K.A.; Lennon, M.G.; Schaudien, D.; Braubach, P.; Jonigk, D.; Warnecke, G.; et al. Rhinovirus-induced Human Lung Tissue Responses Mimic Chronic Obstructive Pulmonary Disease and Asthma Gene Signatures. Am. J. Respir. Cell Mol. Biol. 2021, 65, 544–554. [Google Scholar] [CrossRef]
- Lehmann, M.; Buhl, L.; Alsafadi, H.N.; Klee, S.; Hermann, S.; Mutze, K.; Ota, C.; Lindner, M.; Behr, J.; Hilgendorff, A.; et al. Differential effects of Nintedanib and Pirfenidone on lung alveolar epithelial cell function in ex vivo murine and human lung tissue cultures of pulmonary fibrosis. Respir. Res. 2018, 19, 175. [Google Scholar] [CrossRef]
- Rubio, K.; Singh, I.; Dobersch, S.; Sarvari, P.; Günther, S.; Cordero, J.; Mehta, A.; Wujak, L.; Cabrera-Fuentes, H.; Chao, C.-M.; et al. Inactivation of nuclear histone deacetylases by EP300 disrupts the MiCEE complex in idiopathic pulmonary fibrosis. Nat. Commun. 2019, 10, 2229. [Google Scholar] [CrossRef] [Green Version]
- Mertens, T.C.J.; Karmouty-Quintana, H.; Taube, C.; Hiemstra, P.S. Use of airway epithelial cell culture to unravel the pathogenesis and study treatment in obstructive airway diseases. Pulm. Pharmacol. Ther. 2017, 45, 101–113. [Google Scholar] [CrossRef]
- Alsafadi, H.N.; Staab-Weijnitz, C.A.; Lehmann, M.; Lindner, M.; Peschel, B.; Königshoff, M.; Wagner, D.E. An ex vivo model to induce early fibrosis-like changes in human precision-cut lung slices. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L896–L902. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, E.M.; Culha, S.; Menzen, M.H.; Bidan, C.M.; Gosens, R. Elastase-Induced Parenchymal Disruption and Airway Hyper Responsiveness in Mouse Precision Cut Lung Slices: Toward an Ex Vivo COPD Model. Front. Physiol. 2017, 7, 657. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Ohata, K.; Ott, H.C. Human-scale lung regeneration based on decellularized matrix scaffolds as a biologic platform. Surg. Today 2020, 50, 633–643. [Google Scholar] [CrossRef]
- Crabbé, A.; Liu, Y.; Sarker, S.F.; Bonenfant, N.R.; Barrila, J.; Borg, Z.D.; Lee, J.J.; Weiss, D.J.; Nickerson, C.A. Recellularization of decellularized lung scaffolds is enhanced by dynamic suspension culture. PLoS ONE 2015, 10, e0126846. [Google Scholar] [CrossRef]
- Gilpin, S.E.; Wagner, D.E. Acellular human lung scaffolds to model lung disease and tissue regeneration. Eur. Respir. Rev. 2018, 27, 180021. [Google Scholar] [CrossRef]
- Balijepalli, A.; Sivaramakrishan, V. Organs-on-chips: Research and commercial perspectives. Drug Discov. Today 2017, 22, 397–403. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 2018, 22, 310–324. [Google Scholar] [CrossRef] [Green Version]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef]
- Barreiro Carpio, M.; Dabaghi, M.; Ungureanu, J.; Kolb, M.R.; Hirota, J.A.; Moran-Mirabal, J.M. 3D Bioprinting Strategies, Challenges, and Opportunities to Model the Lung Tissue Microenvironment and Its Function. Front. Bioeng. Biotechnol. 2021, 9, 773511. [Google Scholar] [CrossRef] [PubMed]
- Akter, F.; Araf, Y.; Promon, S.K.; Zhai, J.; Zheng, C. 3D Bioprinting for Regenerating COVID-19-Mediated Irreversibly Damaged Lung Tissue. Int. J. Bioprint. 2022, 8, 616. [Google Scholar] [CrossRef]
- Matai, I.; Kaur, G.; Seyedsalehi, A.; McClinton, A.; Laurencin, C.T. Progress in 3D bioprinting technology for tissue/organ regenerative engineering. Biomaterials 2020, 226, 119536. [Google Scholar] [CrossRef]
- Coraux, C.; Hajj, R.; Lesimple, P.; Puchelle, E. In vivo models of human airway epithelium repair and regeneration. Eur. Respir. Rev. 2005, 14, 131–136. [Google Scholar] [CrossRef]
- Rawlins, E.L.; Okubo, T.; Xue, Y.; Brass, D.M.; Auten, R.L.; Hasegawa, H.; Wang, F.; Hogan, B.L. The Role of Scgb1a1+ Clara Cells in the Long-Term Maintenance and Repair of Lung Airway, but Not Alveolar, Epithelium. Cell Stem Cell 2009, 4, 525–534. [Google Scholar] [CrossRef] [Green Version]
- Vaughan, A.E.; Brumwell, A.N.; Xi, Y.; Gotts, J.E.; Brownfield, D.G.; Treutlein, B.; Tan, K.; Tan, V.; Liu, F.C.; Looney, M.R.; et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 2015, 517, 621–625. [Google Scholar] [CrossRef] [Green Version]
- Basil, M.C.; Morrisey, E.E. Lung regeneration: A tale of mice and men. Semin. Cell Dev. Biol. 2020, 100, 88–100. [Google Scholar] [CrossRef]
- Pan, H.; Deutsch, G.H.; Wert, S.E. Comprehensive anatomic ontologies for lung development: A comparison of alveolar formation and maturation within mouse and human lung. J. Biomed. Semant. 2019, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Potaczek, D.P.; Kabesch, M. Current concepts of IgE regulation and impact of genetic determinants. Clin. Exp. Allergy 2012, 42, 852–871. [Google Scholar] [CrossRef]
- Bukowy-Bieryłło, Z. Long-term differentiating primary human airway epithelial cell cultures: How far are we? Cell Commun. Signal. 2021, 19, 63. [Google Scholar] [CrossRef] [PubMed]
- Gunti, S.; Hoke, A.T.K.; Vu, K.P.; London, N.R. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers 2021, 13, 874. [Google Scholar] [CrossRef]
- Lu, T.; Cao, Y.; Zhao, P.; Shen, S.; Xi, Y. Organoid: A powerful tool to study lung regeneration and disease. Cell Regen. 2021, 10, 21. [Google Scholar] [CrossRef]
- Rock, J.R.; Onaitis, M.W.; Rawlins, E.L.; Lu, Y.; Clark, C.P.; Xue, Y.; Randell, S.H.; Hogan, B.L.M. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 12771–12775. [Google Scholar] [CrossRef]
- Tadokoro, T.; Wang, Y.; Barak, L.S.; Bai, Y.; Randell, S.H.; Hogan, B.L.M. IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, E3641–E3649. [Google Scholar] [CrossRef]
- Danahay, H.; Pessotti, A.D.; Coote, J.; Montgomery, B.E.; Xia, D.; Wilson, A.; Yang, H.; Wang, Z.; Bevan, L.; Thomas, C.; et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep. 2015, 10, 239–252. [Google Scholar] [CrossRef]
- Ekanger, C.T.; Zhou, F.; Bohan, D.; Lotsberg, M.L.; Ramnefjell, M.; Hoareau, L.; Røsland, G.V.; Lu, N.; Aanerud, M.; Gärtner, F.; et al. Human Organotypic Airway and Lung Organoid Cells of Bronchiolar and Alveolar Differentiation Are Permissive to Infection by Influenza and SARS-CoV-2 Respiratory Virus. Front. Cell. Infect. Microbiol. 2022, 12, 841447. [Google Scholar] [CrossRef]
- Kathiriya, J.J.; Brumwell, A.N.; Jackson, J.R.; Tang, X.; Chapman, H.A. Distinct Airway Epithelial Stem Cells Hide among Club Cells but Mobilize to Promote Alveolar Regeneration. Cell Stem Cell 2020, 26, 346–358.e4. [Google Scholar] [CrossRef]
- Lee, J.-H.; Tammela, T.; Hofree, M.; Choi, J.; Marjanovic, N.D.; Han, S.; Canner, D.; Wu, K.; Paschini, M.; Bhang, D.H.; et al. Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 2017, 170, 1149–1163.e12. [Google Scholar] [CrossRef] [Green Version]
- Rabata, A.; Fedr, R.; Soucek, K.; Hampl, A.; Koledova, Z. 3D Cell Culture Models Demonstrate a Role for FGF and WNT Signaling in Regulation of Lung Epithelial Cell Fate and Morphogenesis. Front. Cell Dev. Biol. 2020, 8, 574. [Google Scholar] [CrossRef]
- Alysandratos, K.-D.; Garcia-de-Alba, C.; Yao, C.; Pessina, P.; Huang, J.; Villacorta-Martin, C.; Hix, O.T.; Minakin, K.; Burgess, C.L.; Bawa, P.; et al. Culture impact on the transcriptomic programs of primary and iPSC-derived human alveolar type 2 cells. JCI Insight 2023, 8, e158937. [Google Scholar] [CrossRef]
- Choi, J.; Park, J.-E.; Tsagkogeorga, G.; Yanagita, M.; Koo, B.-K.; Han, N.; Lee, J.-H. Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration. Cell Stem Cell 2020, 27, 366–382.e7. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; van der Vaart, J.; Knoops, K.; Riesebosch, S.; Breugem, T.I.; Mykytyn, A.Z.; Beumer, J.; Schipper, D.; Bezstarosti, K.; Koopman, C.D.; et al. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. EMBO J. 2021, 40, e105912. [Google Scholar] [CrossRef]
- Youk, J.; Kim, T.; Evans, K.V.; Jeong, Y.-I.; Hur, Y.; Hong, S.P.; Kim, J.H.; Yi, K.; Kim, S.Y.; Na, K.J.; et al. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell 2020, 27, 905–919.e10. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Tata, A.; Konkimalla, A.; Katsura, H.; Lee, R.F.; Ou, J.; Banovich, N.E.; Kropski, J.A.; Tata, P.R. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 2020, 22, 934–946. [Google Scholar] [CrossRef]
- Sun, T.; Huang, Z.; Zhang, H.; Posner, C.; Jia, G.; Ramalingam, T.R.; Xu, M.; Brightbill, H.; Egen, J.G.; Dey, A.; et al. TAZ is required for lung alveolar epithelial cell differentiation after injury. JCI Insight 2019, 5, e128674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoareau, L.; Engelsen, A.S.T.; Aanerud, M.; Ramnefjell, M.P.; Salminen, P.-R.; Gärtner, F.; Halvorsen, T.; Raeder, H.; Bentsen, M.H.L. Induction of alveolar and bronchiolar phenotypes in human lung organoids. Physiol. Rep. 2021, 9, e14857. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-W.; Huang, S.X.; de Carvalho, A.L.R.T.; Ho, S.-H.; Islam, M.N.; Volpi, S.; Notarangelo, L.D.; Ciancanelli, M.; Casanova, J.-L.; Bhattacharya, J.; et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017, 19, 542–549. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.J.; Hill, D.R.; Nagy, M.S.; Aoki, Y.; Dye, B.R.; Chin, A.M.; Huang, S.; Zhu, F.; White, E.S.; Lama, V.; et al. In Vitro Induction and In Vivo Engraftment of Lung Bud Tip Progenitor Cells Derived from Human Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 101–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, A.J.; Dye, B.R.; Ferrer-Torres, D.; Hill, D.R.; Overeem, A.W.; Shea, L.D.; Spence, J.R. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019, 14, 518–540. [Google Scholar] [CrossRef]
- Strikoudis, A.; Cieślak, A.; Loffredo, L.; Chen, Y.-W.; Patel, N.; Saqi, A.; Lederer, D.J.; Snoeck, H.-W. Modeling of Fibrotic Lung Disease Using 3D Organoids Derived from Human Pluripotent Stem Cells. Cell Rep. 2019, 27, 3709–3723.e5. [Google Scholar] [CrossRef] [Green Version]
- Nikolić, M.Z.; Caritg, O.; Jeng, Q.; Johnson, J.-A.; Sun, D.; Howell, K.J.; Brady, J.L.; Laresgoiti, U.; Allen, G.; Butler, R.; et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. Elife 2017, 6, e26575. [Google Scholar] [CrossRef]
- Hein, R.F.C.; Conchola, A.S.; Fine, A.S.; Xiao, Z.; Frum, T.; Brastrom, L.K.; Akinwale, M.A.; Childs, C.J.; Tsai, Y.-H.; Holloway, E.M.; et al. Stable iPSC-derived NKX2-1+ lung bud tip progenitor organoids give rise to airway and alveolar cell types. Development 2022, 149, dev200693. [Google Scholar] [CrossRef]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Translational applications of adult stem cell-derived organoids. Development 2017, 144, 968–975. [Google Scholar] [CrossRef] [Green Version]
- Salahudeen, A.A.; Choi, S.S.; Rustagi, A.; Zhu, J.; van Unen, V.; de la O, S.M.; Flynn, R.A.; Margalef-Català, M.; Santos, A.J.M.; Ju, J.; et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 2020, 588, 670–675. [Google Scholar] [CrossRef]
- Chiu, M.C.; Zhang, S.; Li, C.; Liu, X.; Yu, Y.; Huang, J.; Wan, Z.; Zhu, X.; Zhou, J. Apical-Out Human Airway Organoids Modeling SARS-CoV-2 Infection. Viruses 2023, 15, 1166. [Google Scholar] [CrossRef] [PubMed]
- Lechner, A.J.; Driver, I.H.; Lee, J.; Conroy, C.M.; Nagle, A.; Locksley, R.M.; Rock, J.R. Recruited Monocytes and Type 2 Immunity Promote Lung Regeneration following Pneumonectomy. Cell Stem Cell 2017, 21, 120–134.e7. [Google Scholar] [CrossRef] [Green Version]
- Konda, B.; Mulay, A.; Yao, C.; Beil, S.; Israely, E.; Stripp, B.R. Isolation and Enrichment of Human Lung Epithelial Progenitor Cells for Organoid Culture. J. Vis. Exp. 2020, 161, e61541. [Google Scholar] [CrossRef]
- Miller, A.J.; Yu, Q.; Czerwinski, M.; Tsai, Y.-H.; Conway, R.F.; Wu, A.; Holloway, E.M.; Walker, T.; Glass, I.A.; Treutlein, B.; et al. In Vitro and In Vivo Development of the Human Airway at Single-Cell Resolution. Dev. Cell 2020, 53, 117–128.e6. [Google Scholar] [CrossRef]
- Sun, D.; Evans, L.; Perrone, F.; Sokleva, V.; Lim, K.; Rezakhani, S.; Lutolf, M.; Zilbauer, M.; Rawlins, E.L. A functional genetic toolbox for human tissue-derived organoids. Elife 2021, 10, e67886. [Google Scholar] [CrossRef]
- Szabo, M.; Svensson Akusjärvi, S.; Saxena, A.; Liu, J.; Chandrasekar, G.; Kitambi, S.S. Cell and small animal models for phenotypic drug discovery. Drug Des. Dev. Ther. 2017, 11, 1957–1967. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Wei, X.; Pan, Y.; Zou, Y.; Hu, N.; Wang, P. Bionic 3D spheroids biosensor chips for high-throughput and dynamic drug screening. Biomed. Microdevices 2018, 20, 82. [Google Scholar] [CrossRef]
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- Plasschaert, L.W.; Žilionis, R.; Choo-Wing, R.; Savova, V.; Knehr, J.; Roma, G.; Klein, A.M.; Jaffe, A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018, 560, 377–381. [Google Scholar] [CrossRef]
- Schwank, G.; Koo, B.-K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef] [Green Version]
- McCauley, K.B.; Hawkins, F.; Serra, M.; Thomas, D.C.; Jacob, A.; Kotton, D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 2017, 20, 844–857.e6. [Google Scholar] [CrossRef] [Green Version]
- Dekkers, J.F.; Wiegerinck, C.L.; de Jonge, H.R.; Bronsveld, I.; Janssens, H.M.; de Winter-de Groot, K.M.; Brandsma, A.M.; de Jong, N.W.M.; Bijvelds, M.J.C.; Scholte, B.J.; et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013, 19, 939–945. [Google Scholar] [CrossRef]
- Hirai, H.; Liang, X.; Sun, Y.; Zhang, Y.; Zhang, J.; Chen, Y.E.; Mou, H.; Zhao, Y.; Xu, J. The sodium/glucose cotransporters as potential therapeutic targets for CF lung diseases revealed by human lung organoid swelling assay. Mol. Ther. Methods Clin. Dev. 2022, 24, 11–19. [Google Scholar] [CrossRef]
- Richeldi, L.; Collard, H.R.; Jones, M.G. Idiopathic pulmonary fibrosis. Lancet 2017, 389, 1941–1952. [Google Scholar] [CrossRef] [PubMed]
- El Agha, E.; Wygrecka, M. State of the Art in Idiopathic Pulmonary Fibrosis. Cells 2022, 11, 2487. [Google Scholar] [CrossRef]
- Jaeger, B.; Schupp, J.C.; Plappert, L.; Terwolbeck, O.; Artysh, N.; Kayser, G.; Engelhard, P.; Adams, T.S.; Zweigerdt, R.; Kempf, H.; et al. Airway basal cells show a dedifferentiated KRT17highPhenotype and promote fibrosis in idiopathic pulmonary fibrosis. Nat. Commun. 2022, 13, 5637. [Google Scholar] [CrossRef]
- Velázquez-Díaz, P.; Nakajima, E.; Sorkhdini, P.; Hernandez-Gutierrez, A.; Eberle, A.; Yang, D.; Zhou, Y. Hermansky-Pudlak Syndrome and Lung Disease: Pathogenesis and Therapeutics. Front. Pharmacol. 2021, 12, 644671. [Google Scholar] [CrossRef]
- Valdoz, J.C.; Franks, N.A.; Cribbs, C.G.; Jacobs, D.J.; Dodson, E.L.; Knight, C.J.; Poulson, P.D.; Garfield, S.R.; Johnson, B.C.; Hemeyer, B.M.; et al. Soluble ECM promotes organotypic formation in lung alveolar model. Biomaterials 2022, 283, 121464. [Google Scholar] [CrossRef]
- Kathiriya, J.J.; Wang, C.; Zhou, M.; Brumwell, A.; Cassandras, M.; Le Saux, C.J.; Cohen, M.; Alysandratos, K.-D.; Wang, B.; Wolters, P.; et al. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5+ basal cells. Nat. Cell Biol. 2022, 24, 10–23. [Google Scholar] [CrossRef]
- Rabe, K.F.; Watz, H. Chronic obstructive pulmonary disease. Lancet 2017, 389, 1931–1940. [Google Scholar] [CrossRef]
- Basil, M.C.; Cardenas-Diaz, F.L.; Kathiriya, J.J.; Morley, M.P.; Carl, J.; Brumwell, A.N.; Katzen, J.; Slovik, K.J.; Babu, A.; Zhou, S.; et al. Human distal airways contain a multipotent secretory cell that can regenerate alveoli. Nature 2022, 604, 120–126. [Google Scholar] [CrossRef]
- Song, S.; Liu, B.; Habibie, H.; van den Bor, J.; Smit, M.J.; Gosens, R.; Wu, X.; Brandsma, C.-A.; Cool, R.H.; Haisma, H.J.; et al. D-dopachrome tautomerase contributes to lung epithelial repair via atypical chemokine receptor 3-dependent Akt signaling. EBioMedicine 2021, 68, 103412. [Google Scholar] [CrossRef]
- Fahy, J.V.; Dickey, B.F. Airway mucus function and dysfunction. N. Engl. J. Med. 2010, 363, 2233–2247. [Google Scholar] [CrossRef] [Green Version]
- Holgate, S.T.; Wenzel, S.; Postma, D.S.; Weiss, S.T.; Renz, H.; Sly, P.D. Asthma. Nat. Rev. Dis. Primers 2015, 1, 15025. [Google Scholar] [CrossRef]
- Lafkas, D.; Shelton, A.; Chiu, C.; de Leon Boenig, G.; Chen, Y.; Stawicki, S.S.; Siltanen, C.; Reichelt, M.; Zhou, M.; Wu, X.; et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 2015, 528, 127–131. [Google Scholar] [CrossRef]
- Porotto, M.; Ferren, M.; Chen, Y.-W.; Siu, Y.; Makhsous, N.; Rima, B.; Briese, T.; Greninger, A.L.; Snoeck, H.-W.; Moscona, A. Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. mBio 2019, 10, e00723-19. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Li, C.; Sachs, N.; Chiu, M.C.; Wong, B.H.-Y.; Chu, H.; Poon, V.K.-M.; Wang, D.; Zhao, X.; Wen, L.; et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. USA 2018, 115, 6822–6827. [Google Scholar] [CrossRef] [Green Version]
- Van der Sanden, S.M.G.; Sachs, N.; Koekkoek, S.M.; Koen, G.; Pajkrt, D.; Clevers, H.; Wolthers, K.C. Enterovirus 71 infection of human airway organoids reveals VP1-145 as a viral infectivity determinant. Emerg. Microbes Infect. 2018, 7, 84. [Google Scholar] [CrossRef]
- Shen, Y.; Chen, L.; Wang, M.; Lin, D.; Liang, Z.; Song, P.; Yuan, Q.; Tang, H.; Li, W.; Duan, K.; et al. Flagellar Hooks and Hook Protein FlgE Participate in Host Microbe Interactions at Immunological Level. Sci. Rep. 2017, 7, 1433. [Google Scholar] [CrossRef] [Green Version]
- Hong, K.-J.; Seo, S.-H. Organoid as a culture system for viral vaccine strains. Clin. Exp. Vaccine Res. 2018, 7, 145–148. [Google Scholar] [CrossRef]
- Katsura, H.; Sontake, V.; Tata, A.; Kobayashi, Y.; Edwards, C.E.; Heaton, B.E.; Konkimalla, A.; Asakura, T.; Mikami, Y.; Fritch, E.J.; et al. Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell 2020, 27, 890–904.e8. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Monkhorst, K.; Schipper, L.J.; Hartemink, K.J.; Smit, E.F.; Kaing, S.; de Groot, R.; Wolkers, M.C.; Clevers, H.; Cuppen, E.; et al. Challenges in Establishing Pure Lung Cancer Organoids Limit Their Utility for Personalized Medicine. Cell Rep. 2020, 31, 107588. [Google Scholar] [CrossRef]
- Shi, R.; Radulovich, N.; Ng, C.; Liu, N.; Notsuda, H.; Cabanero, M.; Martins-Filho, S.N.; Raghavan, V.; Li, Q.; Mer, A.S.; et al. Organoid Cultures as Preclinical Models of Non-Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 1162–1174. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, R.W.; Aref, A.R.; Lizotte, P.H.; Ivanova, E.; Stinson, S.; Zhou, C.W.; Bowden, M.; Deng, J.; Liu, H.; Miao, D.; et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018, 8, 196–215. [Google Scholar] [CrossRef] [Green Version]
- Veelken, C.; Bakker, G.-J.; Drell, D.; Friedl, P. Single cell-based automated quantification of therapy responses of invasive cancer spheroids in organotypic 3D culture. Methods 2017, 128, 139–149. [Google Scholar] [CrossRef]
- Leibel, S.L.; Winquist, A.; Tseu, I.; Wang, J.; Luo, D.; Shojaie, S.; Nathan, N.; Snyder, E.; Post, M. Reversal of Surfactant Protein B Deficiency in Patient Specific Human Induced Pluripotent Stem Cell Derived Lung Organoids by Gene Therapy. Sci. Rep. 2019, 9, 13450. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Gao, J.; Garcia, I.M.; Chen, H.J.; Castaldi, A.; Chen, Y.-W. Human pluripotent stem cell-derived lung organoids: Potential applications in development and disease modeling. Wiley Interdiscip. Rev. Dev. Biol. 2021, 10, e399. [Google Scholar] [CrossRef]
- Finkbeiner, S.R.; Hill, D.R.; Altheim, C.H.; Dedhia, P.H.; Taylor, M.J.; Tsai, Y.-H.; Chin, A.M.; Mahe, M.M.; Watson, C.L.; Freeman, J.J.; et al. Transcriptome-wide Analysis Reveals Hallmarks of Human Intestine Development and Maturation In Vitro and In Vivo. Stem Cell Rep. 2015, 4, 1140–1155. [Google Scholar] [CrossRef] [Green Version]
- Múnera, J.O.; Sundaram, N.; Rankin, S.A.; Hill, D.; Watson, C.; Mahe, M.; Vallance, J.E.; Shroyer, N.F.; Sinagoga, K.L.; Zarzoso-Lacoste, A.; et al. Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling. Cell Stem Cell 2017, 21, 51–64.e6. [Google Scholar] [CrossRef] [Green Version]
- Trecartin, A.; Danopoulos, S.; Spurrier, R.; Knaneh-Monem, H.; Hiatt, M.; Driscoll, B.; Hochstim, C.; Al-Alam, D.; Grikscheit, T.C. Establishing Proximal and Distal Regional Identities in Murine and Human Tissue-Engineered Lung and Trachea. Tissue Eng. Part C Methods 2016, 22, 1049–1057. [Google Scholar] [CrossRef] [Green Version]
- Hayes, D.; Kopp, B.T.; Hill, C.L.; Lallier, S.W.; Schwartz, C.M.; Tadesse, M.; Alsudayri, A.; Reynolds, S.D. Cell Therapy for Cystic Fibrosis Lung Disease: Regenerative Basal Cell Amplification. Stem Cells Transl. Med. 2019, 8, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Petersen, T.H.; Calle, E.A.; Zhao, L.; Lee, E.J.; Gui, L.; Raredon, M.B.; Gavrilov, K.; Yi, T.; Zhuang, Z.W.; Breuer, C.; et al. Tissue-engineered lungs for in vivo implantation. Science 2010, 329, 538–541. [Google Scholar] [CrossRef] [Green Version]
- Song, J.J.; Kim, S.S.; Liu, Z.; Madsen, J.C.; Mathisen, D.J.; Vacanti, J.P.; Ott, H.C. Enhanced in vivo function of bioartificial lungs in rats. Ann. Thorac. Surg. 2011, 92, 998–1005; discussion 1005–1006. [Google Scholar] [CrossRef]
- Ren, X.; Moser, P.T.; Gilpin, S.E.; Okamoto, T.; Wu, T.; Tapias, L.F.; Mercier, F.E.; Xiong, L.; Ghawi, R.; Scadden, D.T.; et al. Engineering pulmonary vasculature in decellularized rat and human lungs. Nat. Biotechnol. 2015, 33, 1097–1102. [Google Scholar] [CrossRef]
- Wanczyk, H.; Jensen, T.; Weiss, D.J.; Finck, C. Advanced single-cell technologies to guide the development of bioengineered lungs. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 320, L1101–L1117. [Google Scholar] [CrossRef]
- Wagar, L.E.; Salahudeen, A.; Constantz, C.M.; Wendel, B.S.; Lyons, M.M.; Mallajosyula, V.; Jatt, L.P.; Adamska, J.Z.; Blum, L.K.; Gupta, N.; et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 2021, 27, 125–135. [Google Scholar] [CrossRef]
- Nerger, B.A.; Nelson, C.M. 3D culture models for studying branching morphogenesis in the mammary gland and mammalian lung. Biomaterials 2019, 198, 135–145. [Google Scholar] [CrossRef]



| Organoid Type | Description/Origin | Cell Types & Markers | Modeling | Sources |
|---|---|---|---|---|
| Tracheospheres |
| Basal cells: p63+, KRT5+, KRT14+, NGFR+, ITGA6+ Secretory cells: KRT8+ Goblet cells: MUC5AC+ Ciliated cells: α-tubulin+ |
| Human and murine:
|
| Bronchiospheres/ Bronchial organoids |
| Basal cells: p63+, KRT5+, NGFR+, ITGA6+, PDPN+, KRT14+ Club cells: SCGB1A1+, SCGB3A2+, SPLUNC1+ Goblet cells: MUC5AC+ Ciliated cells: acetylated tubulin+, α-tubulin+, FOXJ1+ Neuroendocrine cells: CGRP+ Pulmonary ionocytes: FOXI1+ General epithelial cell markers: KRT8+, E-cadherin+ |
| Human: Murine: Human and murine:
|
| Alveolar organoids |
| AT1: AQP5+, PDPN+, HTI-56+; flat form AT2: EPCAM+, HTII-280+, pro-SPB+ and pro-SPC+; cuboidal form, stem cell capacity |
| Human:
Human and murine:
|
| Bronchioalveolar organoids |
|
|
| Human:
|
| Lung bud organoids |
|
|
| Human: |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kühl, L.; Graichen, P.; von Daacke, N.; Mende, A.; Wygrecka, M.; Potaczek, D.P.; Miethe, S.; Garn, H. Human Lung Organoids—A Novel Experimental and Precision Medicine Approach. Cells 2023, 12, 2067. https://doi.org/10.3390/cells12162067
Kühl L, Graichen P, von Daacke N, Mende A, Wygrecka M, Potaczek DP, Miethe S, Garn H. Human Lung Organoids—A Novel Experimental and Precision Medicine Approach. Cells. 2023; 12(16):2067. https://doi.org/10.3390/cells12162067
Chicago/Turabian StyleKühl, Laura, Pauline Graichen, Nele von Daacke, Anne Mende, Malgorzata Wygrecka, Daniel P. Potaczek, Sarah Miethe, and Holger Garn. 2023. "Human Lung Organoids—A Novel Experimental and Precision Medicine Approach" Cells 12, no. 16: 2067. https://doi.org/10.3390/cells12162067
APA StyleKühl, L., Graichen, P., von Daacke, N., Mende, A., Wygrecka, M., Potaczek, D. P., Miethe, S., & Garn, H. (2023). Human Lung Organoids—A Novel Experimental and Precision Medicine Approach. Cells, 12(16), 2067. https://doi.org/10.3390/cells12162067







