Altered Visual Function in Short-Wave-Sensitive 1 (sws1) Gene Knockout Japanese Medaka (Oryzias latipes) Larvae
Abstract
:1. Introduction
2. Results
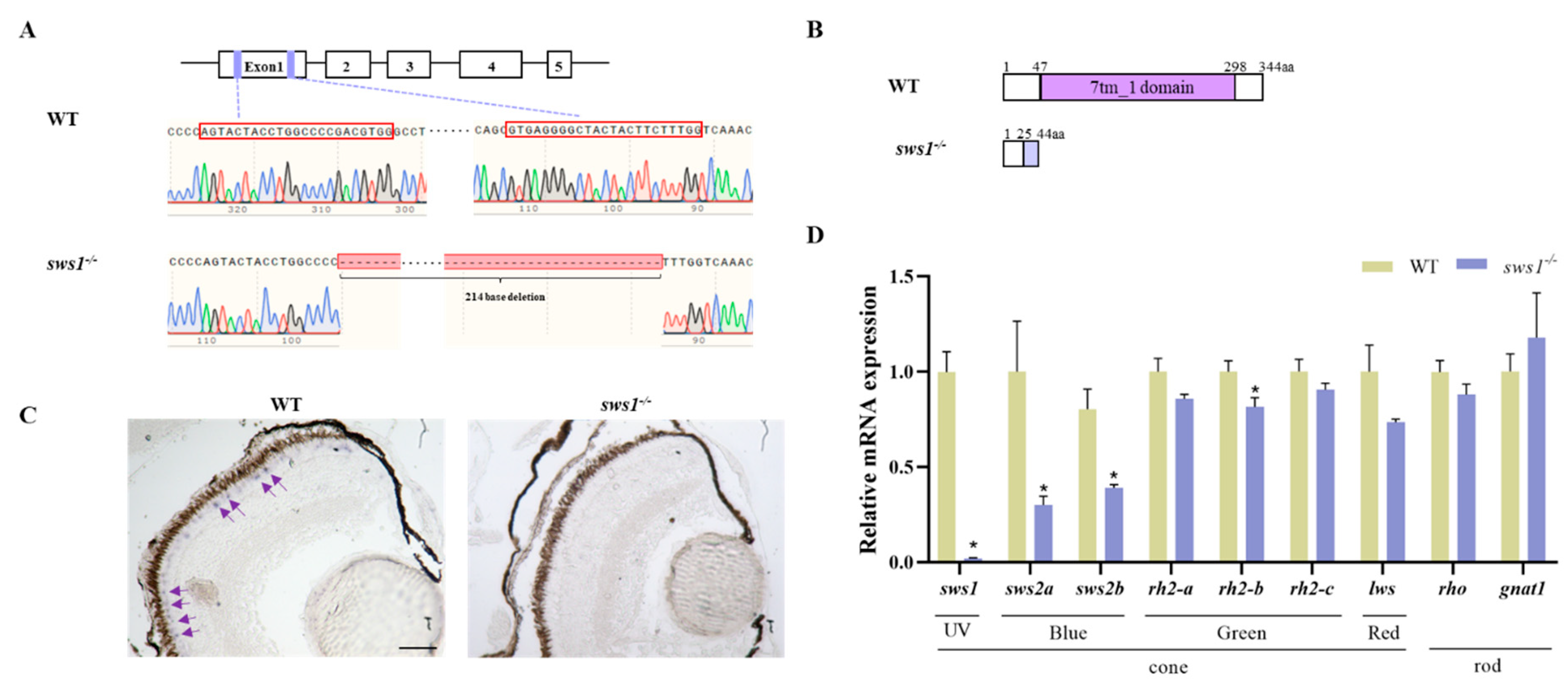
2.1. Generation of sws1 Knockout Medaka
2.2. Decreased Food Intake in the Larvae of sws1 Knockout Medaka
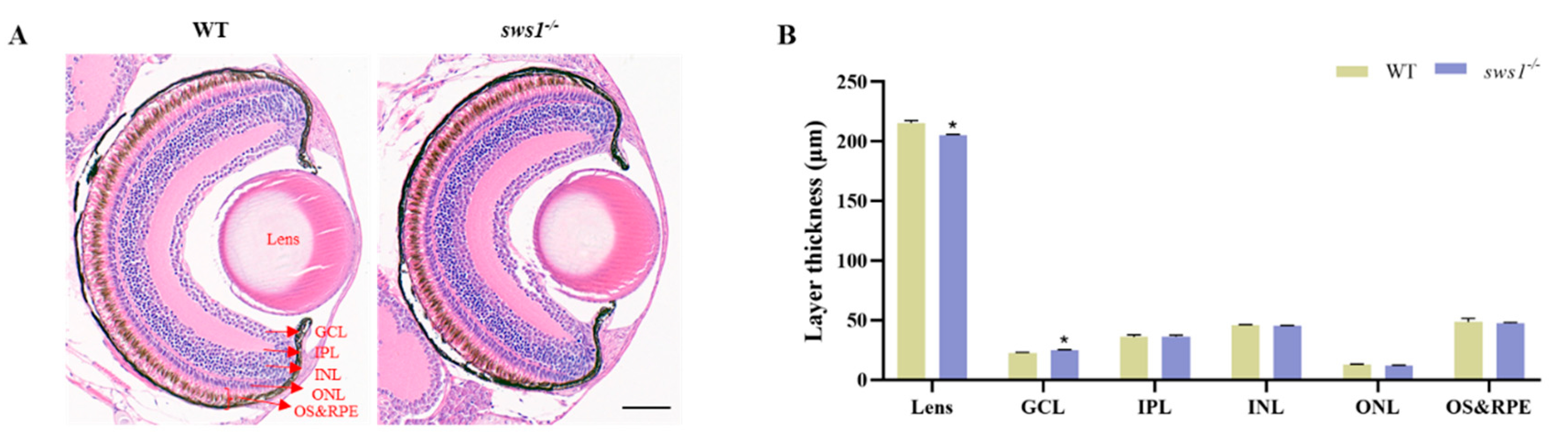
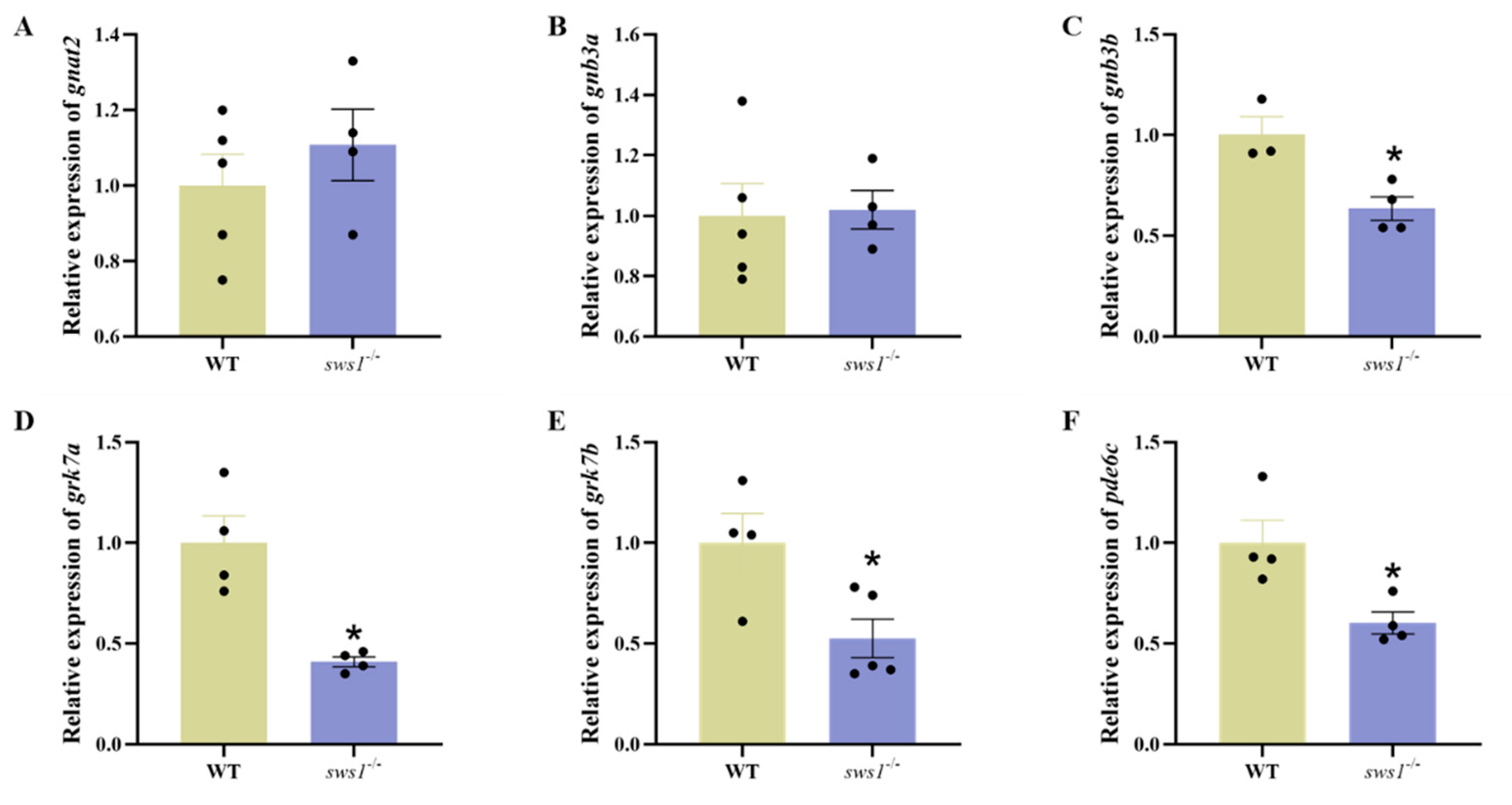
2.3. sws1 Deficiency in Larvae Affected Retinal Lamination and Reduced Expression of Phototransduction Genes
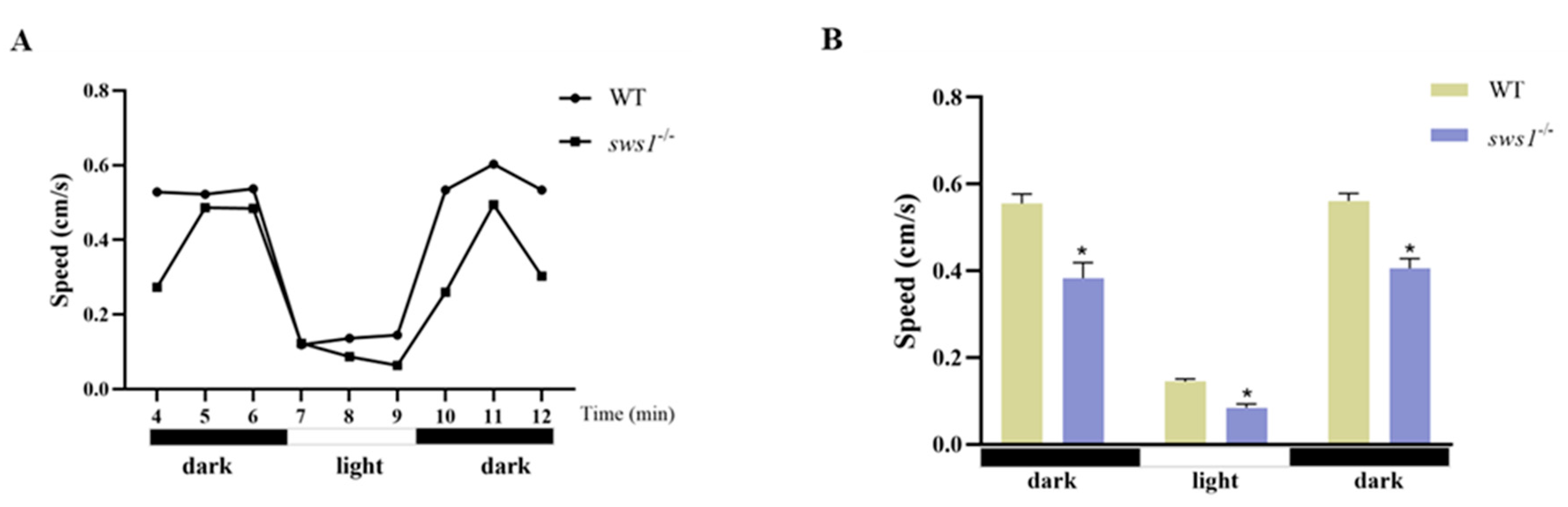
2.4. Disruption of sws1-Impaired Medaka Larvae Swimming Behavior
2.5. Induced Oxidative Stress in sws1-Deficient Medaka Larvae
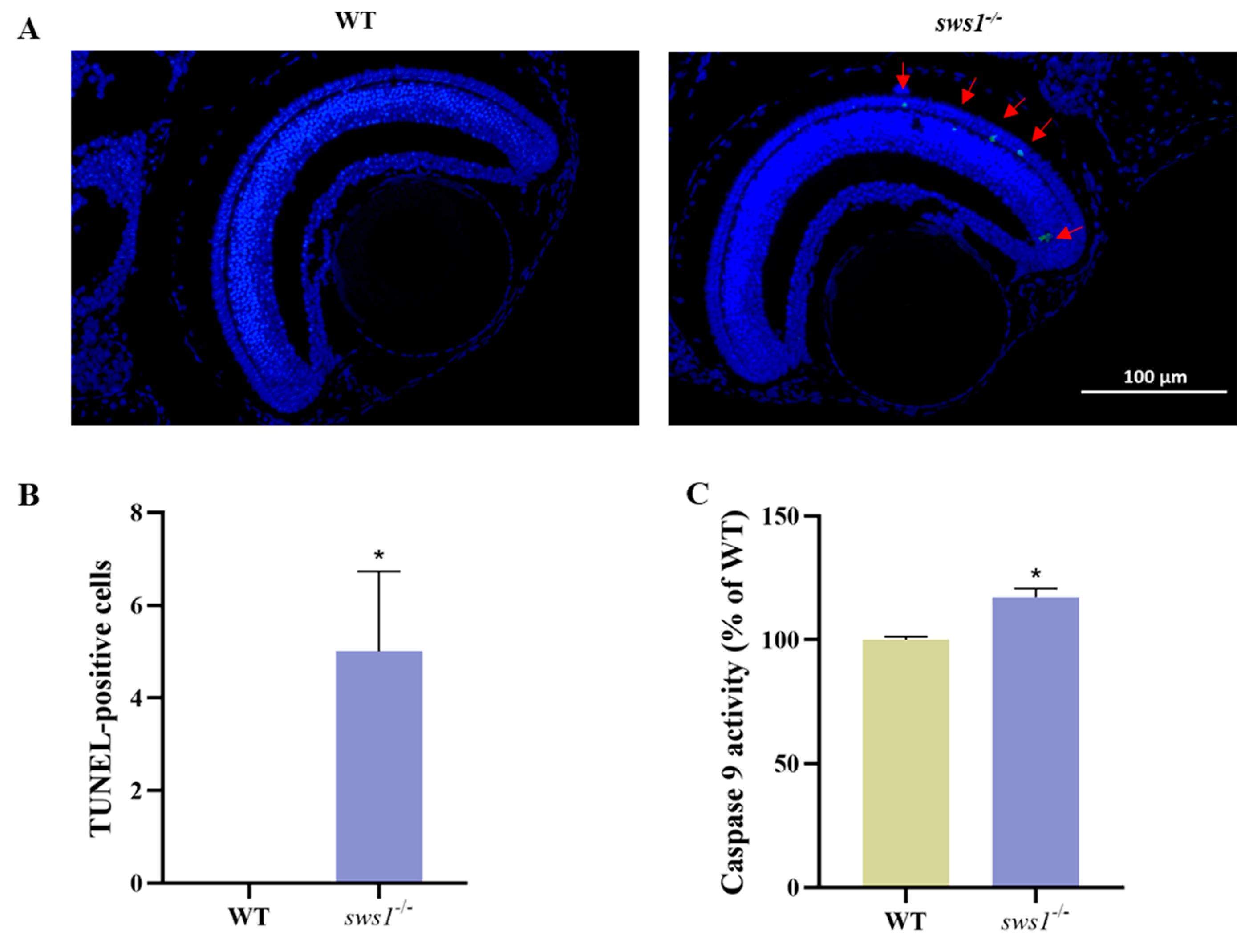
2.6. Depletion of sws1 Led to Retinal Cell Apoptosis in Medaka Larvae
3. Discussion
4. Materials and Methods
4.1. Medaka Lines and Maintenance
4.2. Generating sws1−/− Mutants Using CRISPR/Cas9 Technology
4.3. Histological Assessment, In Situ Hybridization (ISH), and TUNEL Staining
4.4. Light/Dark Behavior Analysis
4.5. Larvae Feeding Assays
4.6. Growth Performance and Survival Rate
4.7. Biochemical Analyses and Caspase Activity Assay
4.8. RNA Isolation and Quantitative RT-PCR
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baden, T.; Euler, T.; Berens, P. Understanding the retinal basis of vision across species. Nat. Rev. Neurosci. 2020, 21, 5–20. [Google Scholar] [CrossRef]
- Lamb, T.D. Evolution of the genes mediating phototransduction in rod and cone photoreceptors. Prog. Retin. Eye. Res. 2020, 76, 100823. [Google Scholar] [CrossRef]
- Ali, M.A.; Klyne, M.A. Vision in Vertebrates; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Carleton, K.L.; Yourick, M.R. Axes of Visual Adaptation in the Ecologically Diverse Family Cichlidae, Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 43–52. [Google Scholar]
- Musilova, Z.; Cortesi, F.; Matschiner, M.; Davies, W.I.; Patel, J.S.; Stieb, S.M.; Busserolles, F.D.; Malmstrøm, M.; Tørresen, O.K.; Brown, C.J.; et al. Vision using multiple distinct rod opsins in deep-sea fishes. Science 2019, 364, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Hunt, D.M.; Hankins, M.W.; Collin, S.P.; Marshall, N.J. Evolution of Visual and Non-Visual Pigments; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Marshall, N.J.; Cortesi, F.; de Busserolles, F.; Siebeck, U.E.; Cheney, K.L. Colours and colour vision in reef fishes: Past, present and future research directions. J. Fish. Biol. 2019, 95, 5–38. [Google Scholar] [CrossRef] [PubMed]
- Musilova, Z.; Salzburger, W.; Cortesi, F. The visual opsin gene repertoires of teleost fishes: Evolution, ecology, and function. Annu. Rev. Cell Dev. Biol. 2021, 37, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Flamarique, I.N. Opsin switch reveals function of the ultraviolet cone in fish foraging. Proc. Biol. Sci. 2013, 280, 20122490. [Google Scholar]
- O’Quin, K.E.; Hofmann, C.M.; Hofmann, H.A.; Carleton, K.L. Parallel evolution of opsin gene expression in African cichlid fishes. Mol. Biol. Evol. 2010, 27, 2839–2854. [Google Scholar] [CrossRef]
- Cheng, C.L.; Flamarique, I.N. Chromatic organization of cone photoreceptors in the retina of rainbow trout: Single cones irreversibly switch from UV (SWS1) to blue (SWS2) light sensitive opsin during natural development. J. Exp. Biol. 2007, 210 Pt 23, 4123–4135. [Google Scholar] [CrossRef]
- Hunt, D.M.; Cowing, J.A.; Wilkie, S.E.; Parry, J.W.; Poopalasundaram, S.; Bowmaker, J.K. Divergent mechanisms for the tuning of shortwave sensitive visual pigments in vertebrates. Photoch. Photobio. Sci. 2004, 3, 713–720. [Google Scholar] [CrossRef]
- Hofmann, C.M.; Carleton, K.L. Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. Integr. Comp. Biol. 2009, 49, 630–643. [Google Scholar] [CrossRef]
- Daniele, L.L.; Insinna, C.; Chance, R.; Wang, J.; Nikonov, S.S.; Pugh, E.N., Jr. A mouse M-opsin monochromat: Retinal cone photoreceptors have increased M-opsin expression when S-opsin is knocked out. Vision. Res. 2011, 51, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Flamarique, N.I.; Fujihara, R.; Yazawa, R.; Bolstad, K.; Gowen, B.; Yoshizaki, G. Disrupted eye and head development in rainbow trout with reduced ultraviolet (sws1) opsin expression. J. Comp. Neurol. 2021, 529, 3013–3031. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.S.; Noltie, D.B.; Tillitt, D.E. Ontogenetic improvement of visual function in the medaka Oryzias latipes based on an optomotor testing system for larval and adult fish. Anim. Behav. 2002, 64, 1–10. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Fukamachi, S.; Mitani, H.; Kawamura, S. Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes). Gene 2006, 371, 268–278. [Google Scholar] [CrossRef]
- Nakayasu, T.; Watanabe, E. Biological motion stimuli are attractive to medaka fish. Anim. Cogn. 2014, 17, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Tohya, S.; Mochizuki, A.; Iwasa, Y. Difference in the retinal cone mosaic pattern between zebrafish and medaka: Cell-rearrangement model. J. Theor. Biol. 2003, 221, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhou, L.; Zhao, T.; Liu, X.; Zhang, P.; Liu, Y.; Zheng, X.; Li, Q. Caspase-9: Structure, mechanisms and clinical application. Oncotarget 2017, 8, 23996. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, Y.; Ma, Z.; Qin, J.G. Feeding and development of warm water marine fish larvae in early life. In Emerging Issues in Fish Larvae Research; Springer: Cham, Switzerland, 2018; pp. 275–296. [Google Scholar]
- Mahavadiya, D.; Sapra, D.; Rathod, V.; Sarman, V. Effect of biotic and abiotic factors in feeding activity in teleost fish: A review. J. Entomol. Zool. Stud. 2018, 6, 387–390. [Google Scholar]
- Flamarique, I.N.; Cheng, C.L.; Bergstrom, C.; Reimchen, T.E. Pronounced heritable variation and limited phenotypic plasticity in visual pigments and opsin expression of threespine stickleback photoreceptors. J. Exp. Biol. 2013, 216, 656–667. [Google Scholar]
- O’Daniels, S.T.; Kesler, D.C.; Mihail, J.D.; Webb, E.B.; Werner, S.J. Functional visual sensitivity to ultraviolet wavelengths in the Pileated Woodpecker (Dryocopus pileatus), and its influence on foraging substrate selection. Physiol. Behav. 2017, 174, 144–154. [Google Scholar] [CrossRef]
- Carleton, K.L.; Escobar-Camacho, D.; Stieb, S.M.; Cortesi, F.; Marshall, N.J. Seeing the rainbow: Mechanisms underlying spectral sensitivity in teleost fishes. J. Exp. Biol. 2020, 223, jeb193334. [Google Scholar] [CrossRef] [PubMed]
- Flamarique, N.I. Diminished foraging performance of a mutant zebrafish with reduced population of ultraviolet cones. Proc. Biol. Sci. 2016, 283, 20160058. [Google Scholar]
- Yoshimatsu, T.; Schroder, C.; Nevala, N.E.; Berens, P.; Baden, T. Fovea-like Photoreceptor Specializations Underlie Single UV Cone Driven Prey-Capture Behavior in Zebrafish. Neuron 2020, 107, 320–337.e6. [Google Scholar] [CrossRef]
- Essner, R.A.; Smith, A.G.; Jamnik, A.A.; Ryba, A.R.; Trutner, Z.D.; Carter, M.E. AgRP neurons can increase food intake during conditions of appetite suppression and inhibit anorexigenic parabrachial neurons. J. Neurosci. 2017, 37, 8678–8687. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.-S.; Wen, H.-S. Neuropeptide Y and melanocortin receptors in fish: Regulators of energy homeostasis. Mar. Life. Sci. Tech. 2021, 4, 42–51. [Google Scholar] [CrossRef]
- Siemian, J.N.; Arenivar, M.A.; Sarsfield, S.; Aponte, Y. Hypothalamic control of interoceptive hunger. Curr. Biol. 2021, 31, 3797–3809.e5. [Google Scholar] [CrossRef]
- Cabral, A.; Fernandez, G.; Tolosa, M.J.; Moggia, Á.R.; Calfa, G.; De Francesco, P.N.; Perello, M. Fasting induces remodeling of the orexigenic projections from the arcuate nucleus to the hypothalamic paraventricular nucleus, in a growth hormone secretagogue receptor-dependent manner. Mol. Metab. 2020, 32, 69–84. [Google Scholar] [CrossRef]
- Santos-Carvalho, A.; Ambrósio, A.F.; Cavadas, C. Neuropeptide Y system in the retina: From localization to function. Prog. Retin. Eye. Res. 2015, 47, 19–37. [Google Scholar] [CrossRef]
- Sand, A.; Schmidt, T.M.; Kofuji, P. Diverse types of ganglion cell photoreceptors in the mammalian retina. Prog. Retin. Eye. Res. 2012, 31, 287–302. [Google Scholar] [CrossRef]
- Chen, L.; Huang, Y.; Huang, C.; Hu, B.; Hu, C.; Zhou, B. Acute exposure to DE-71 causes alterations in visual behavior in zebrafish larvae. Environ. Toxicol. Chem. 2013, 32, 1370–1375. [Google Scholar] [CrossRef]
- Bilotta, J.; Saszik, S. The zebrafish as a model visual system. Int. J. Dev. Neurosci. 2001, 19, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Chen, Z.; Fu, L.; Han, J.; Hu, J.; Zhou, B.; Zhou, B. Optical toxicity of triphenyl phosphate in zebrafish larvae. Aquat. Toxicol. 2019, 210, 139–147. [Google Scholar] [CrossRef]
- Chen, X.-F.; Chen, Z.F.; Lin, Z.C.; Liao, X.L.; Zou, T.; Qi, Z.; Cai, Z. Toxic effects of triclocarban on larval zebrafish: A focus on visual dysfunction. Aquat. Toxicol. 2021, 241, 106013. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Yimer, T.A.; Xie, S.; Wong, F.; Yu, S.; Liu, X.; Han, S.; Ma, J.; Lu, Z.; Hu, X.; et al. Knocking out lca5 in zebrafish causes cone-rod dystrophy due to impaired outer segment protein trafficking. BBA-Mol. Basis. Dis. 2019, 1865, 2694–2705. [Google Scholar] [CrossRef]
- Yu, S.; Li, C.; Biswas, L.; Hu, X.; Liu, F.; Reilly, J.; Liu, X.; Liu, Y.; Huang, Y.; Lu, Z.; et al. CERKL gene knockout disturbs photoreceptor outer segment phagocytosis and causes rod-cone dystrophy in zebrafish. Hum. Mol. Genet. 2017, 26, 2335–2345. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, A.; Montoya, A.; Vera, L.M.; Sánchez-Vázquez, F.J. Daily rhythms of clock gene expression, glycaemia and digestive physiology in diurnal/nocturnal European seabass. Physiol. Behav. 2012, 106, 446–450. [Google Scholar] [CrossRef]
- Lopez-Olmeda, J.F.; Tartaglione, E.V.; De La Iglesia, H.O.; Sanchez-Vazquez, F.J. Feeding entrainment of food-anticipatory activity and per1 expression in the brain and liver of zebrafish under different lighting and feeding conditions. Chronobiol. Int. 2010, 27, 1380–1400. [Google Scholar] [CrossRef]
- Martín-Robles, Á.J.; Whitmore, D.; Sánchez-Vázquez, F.J.; Pendón, C.; Muñoz-Cueto, J.A. Cloning, tissue expression pattern and daily rhythms of Period1, Period2, and Clock transcripts in the flatfish Senegalese sole, Solea senegalensis. J. Comp. Physiol. B 2012, 82, 673–685. [Google Scholar] [CrossRef]
- Mata-Sotres, J.A.; Martínez-Rodríguez, G.; Pérez-Sánchez, J.; Sánchez-Vázquez, F.J.; Yúfera, M. Daily rhythms of clock gene expression and feeding behavior during the larval development in gilthead seabream, Sparus aurata. Chronobiol. Int. 2015, 32, 1061–1074. [Google Scholar] [CrossRef]
- Aranda, M.L.; Schmidt, T.M. Diversity of intrinsically photosensitive retinal ganglion cells: Circuits and functions. Cell. Mol. Life Sci. 2021, 78, 889–907. [Google Scholar] [CrossRef]
- Contreras, E.; Nobleman, A.P.; Robinson, P.R.; Schmidt, T.M. Melanopsin phototransduction: Beyond canonical cascades. J. Exp. Biol. 2021, 224, jeb226522. [Google Scholar] [CrossRef] [PubMed]
- Do, M.T.H.; Yau, K.W. Intrinsically photosensitive retinal ganglion cells. Physiol. Rev. 2010, 90, 1547–1581. [Google Scholar] [CrossRef] [PubMed]
- Shichida, Y.; Matsuyama, T. Evolution of opsins and phototransduction. Philos. T. R. Soc. B 2009, 364, 2881–2895. [Google Scholar] [CrossRef] [PubMed]
- Arshavsky, V.Y.; Burns, M.E. Photoreceptor signaling: Supporting vision across a wide range of light intensities. J. Biol. Chem. 2012, 287, 1620–1626. [Google Scholar] [CrossRef]
- Fu, Y.; Yau, K.-W. Phototransduction in mouse rods and cones. Pflug Arch. Eur. J. Phy. 2007, 454, 805–819. [Google Scholar]
- Winkler, C.W. Functional Characterization of gnb3b and guk1b for Photoreceptor Integrity in a Zebrafish Model for Retinitis Pigmentosa. Ph.D. Thesis, National University of Singapore, Singapore, 2016. [Google Scholar]
- Rinner, O.; Makhankov, Y.V.; Biehlmaier, O.; Neuhauss, S.C. Knockdown of cone-specific kinase GRK7 in larval zebrafish leads to impaired cone response recovery and delayed dark adaptation. Neuron 2005, 47, 231–242. [Google Scholar] [CrossRef]
- Zhang, L.; Xiang, L.; Liu, Y.; Venkatraman, P.; Chong, L.; Cho, J.; Bonilla, S.; Jin, Z.B.; Pang, C.P.; Ko, K.M.; et al. A naturally-derived compound schisandrin B enhanced light sensation in the pde6c zebrafish model of retinal degeneration. PLoS ONE 2016, 11, e0149663. [Google Scholar]
- Huang, L.; Wang, C.; Zhang, Y.; Wu, M.; Zuo, Z. Phenanthrene causes ocular developmental toxicity in zebrafish embryos and the possible mechanisms involved. J. Haz. Mat. 2013, 261, 172–180. [Google Scholar] [CrossRef]
- Huang, L.; Zuo, Z.; Zhang, Y.; Wu, M.; Lin, J.J.; Wang, C. Use of toxicogenomics to predict the potential toxic effect of benzo (a) pyrene on zebrafish embryos: Ocular developmental toxicity. Chemosphere 2014, 108, 55–61. [Google Scholar] [CrossRef]
- Del Rio, D.; Stewart, A.J.; Pellegrini, N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 2005, 15, 316–328. [Google Scholar] [CrossRef]
- Kannan, K.; Jain, S.K. Oxidative stress and apoptosis. Pathophysiology 2000, 7, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. BBA-Mol. Cell. Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, J. Initiator caspases in apoptosis signaling pathways. Apoptosis 2002, 7, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kuida, K. Caspase-9. Int. J. Biochem. Cell. Biol. 2000, 32, 121–124. [Google Scholar] [CrossRef]
- Würstle, M.L.; Laussmann, M.A.; Rehm, M. The central role of initiator caspase-9 in apoptosis signal transduction and the regulation of its activation and activity on the apoptosome. Exp. Cell. Res. 2012, 318, 1213–1220. [Google Scholar] [CrossRef]
- Blanch, R.J.; Ahmed, Z.; Thompson, A.R.; Akpan, N.; Snead, D.R.; Berry, M.; Troy, C.M.; Scott, R.A.H.; Logan, A. Caspase-9 mediates photoreceptor death after blunt ocular trauma. Investig. Ophth. Vis. Sci. 2014, 55, 6350–6357. [Google Scholar] [CrossRef]
- Chen, J.; Xie, P.; Li, L.; Xu, J. First identification of the hepatotoxic microcystins in the serum of a chronically exposed human population together with indication of hepatocellular damage. Toxicol. Sci. 2009, 108, 81–89. [Google Scholar] [CrossRef]
- Yi, M.; Hong, N.; Hong, Y. Derivation and characterization of haploid embryonic stem cell cultures in medaka fish. Nat. Protoc. 2010, 5, 1418–1430. [Google Scholar] [CrossRef]
- Tang, S.-L.; Liang, X.-F.; Li, L.; Wu, J.; Lu, K. Genome-wide identification and expression patterns of opsin genes during larval development in Chinese perch (Siniperca chuatsi). Gene 2022, 825, 146434. [Google Scholar] [CrossRef]
- Huang, H.; Huang, C.; Wang, L.; Ye, X.; Bai, C.; Simonich, M.T.; Tanguay, R.L.; Dong, Q. Toxicity, uptake kinetics and behavior assessment in zebrafish embryos following exposure to perfluorooctanesulphonicacid (PFOS). Aquat. Toxicol. 2010, 98, 139–147. [Google Scholar] [CrossRef]
- Shimada, Y.; Hirano, M.; Nishimura, Y.; Tanaka, T. A high-throughput fluorescence-based assay system for appetite-regulating gene and drug screening. PLoS ONE 2012, 7, e52549. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chisada, S.-I.; Kurokawa, T.; Murashita, K.; Rønnestad, I.; Taniguchi, Y.; Toyoda, A.; Sakaki, Y.; Takeda, S.; Yoshiura, Y. Leptin receptor-deficient (knockout) medaka, Oryzias latipes, show chronical up-regulated levels of orexigenic neuropeptides, elevated food intake and stage specific effects on growth and fat allocation. Gen. Comp. Endocr. 2014, 195, 9–20. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, K.; Wu, J.; Tang, S.; Wang, Y.; Zhang, L.; Chai, F.; Liang, X.-F. Altered Visual Function in Short-Wave-Sensitive 1 (sws1) Gene Knockout Japanese Medaka (Oryzias latipes) Larvae. Cells 2023, 12, 2157. https://doi.org/10.3390/cells12172157
Lu K, Wu J, Tang S, Wang Y, Zhang L, Chai F, Liang X-F. Altered Visual Function in Short-Wave-Sensitive 1 (sws1) Gene Knockout Japanese Medaka (Oryzias latipes) Larvae. Cells. 2023; 12(17):2157. https://doi.org/10.3390/cells12172157
Chicago/Turabian StyleLu, Ke, Jiaqi Wu, Shulin Tang, Yuye Wang, Lixin Zhang, Farui Chai, and Xu-Fang Liang. 2023. "Altered Visual Function in Short-Wave-Sensitive 1 (sws1) Gene Knockout Japanese Medaka (Oryzias latipes) Larvae" Cells 12, no. 17: 2157. https://doi.org/10.3390/cells12172157



