Glia Cells Control Olfactory Neurogenesis by Fine-Tuning CXCL12
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Breeding and Treatment
2.2. Immunofluorescence and Tissue Preparation
2.3. Heparanase Treatment
2.4. Proximity Ligation Assay (PLA)
2.5. PLA Signal Density Heatmap Generation
2.6. In Situ Hybridization
2.7. Immunosorbent Assay (ELISA)
2.8. RNA Sequencing and Transcriptomic Analysis
2.9. Statistical Analysis
3. Results
3.1. ACKR3 Is Expressed in Glia Cells of the OE
3.2. ACKR3 in Sustentacular Cells Regulates CXCR4 Activation
3.3. CXCL12 Scavenging by ACKR3 Regulates CXCR4 Signaling
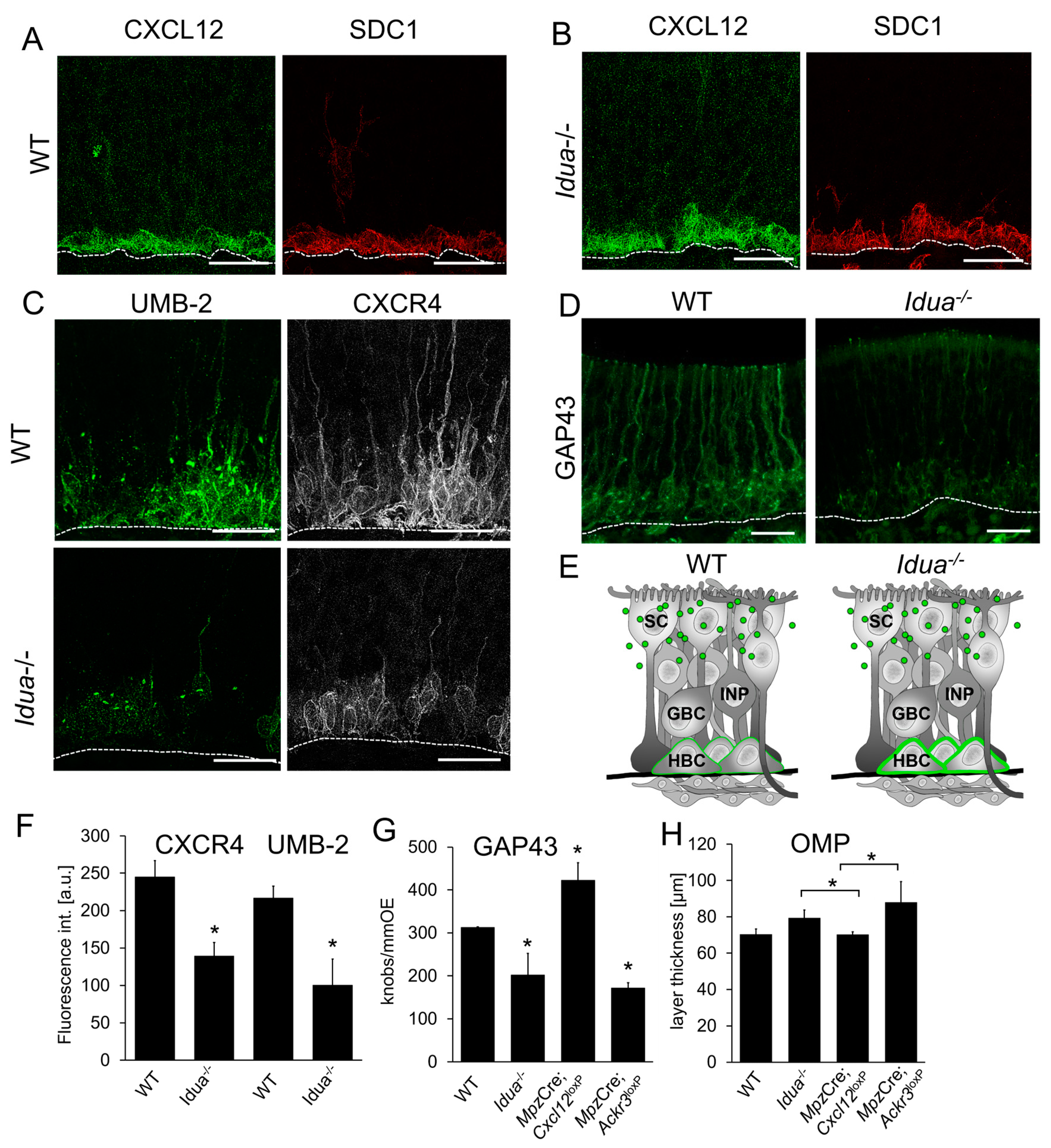
3.4. CXCL12 Scavenging by ACKR3 Regulates Stem Cell Proliferation and Neurogenesis
3.5. CXCL12 Availability Regulates CXCR4-Dependent Neurogenesis
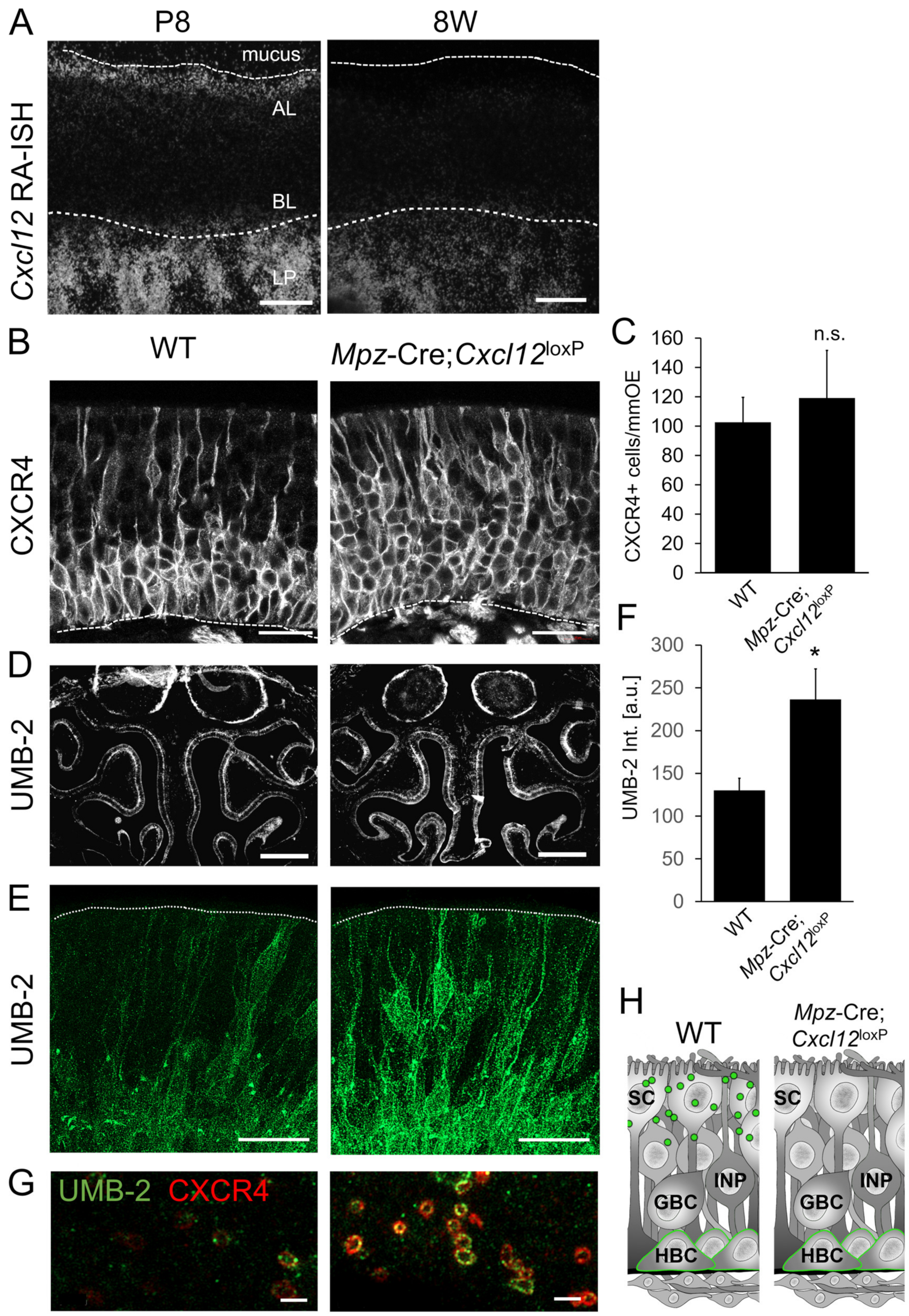
3.6. Cxcl12 Expression in Sustentacular Cells Is Necessary for Proper Regulation of Neurogenesis
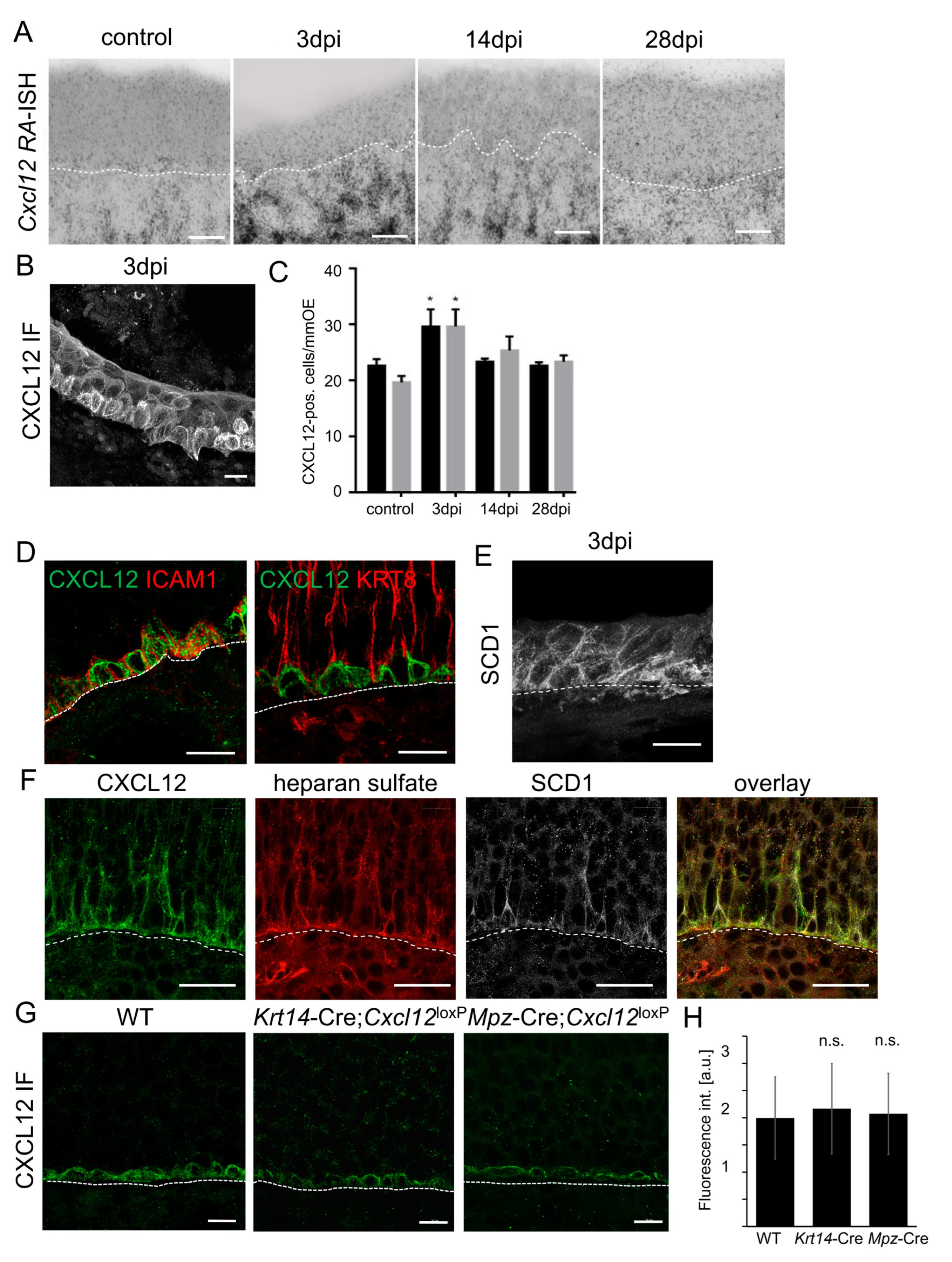
3.7. CXCL12 Detected in HBCs Is Derived from the Lamina Propria
3.8. CXCL12 Accumulates in the Extracellular Matrix Surrounding HBCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scadden, D.T. The stem-cell niche as an entity of action. Nature 2006, 441, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Cope, E.C.; Gould, E. Adult Neurogenesis, Glia, and the Extracellular Matrix. Cell Stem Cell 2019, 24, 690–705. [Google Scholar] [CrossRef] [PubMed]
- Mackay-Sim, A.; Kittel, P. Cell dynamics in the adult mouse olfactory epithelium: A quantitative autoradiographic study. J. Neurosci. Off. J. Soc. Neurosci. 1991, 11, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Graziadei, P.P.; Graziadei, G.A. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J. Neurocytol. 1979, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Schwob, J.E.; Youngentob, S.L.; Mezza, R.C. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J. Comp. Neurol. 1995, 359, 15–37. [Google Scholar] [CrossRef]
- Schwob, J.E.; Huard, J.M.; Luskin, M.B.; Youngentob, S.L. Retroviral lineage studies of the rat olfactory epithelium. Chem. Senses 1994, 19, 671–682. [Google Scholar] [CrossRef]
- Schwob, J.E.; Jang, W.; Holbrook, E.H.; Lin, B.; Herrick, D.B.; Peterson, J.N.; Hewitt Coleman, J. Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J. Comp. Neurol. 2017, 525, 1034–1054. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Oliver, J.; Gilmer, D.; Lovins, C.; Rodriguez-Gil, D.J.; Hagg, T. Inhibition of focal adhesion kinase increases adult olfactory stem cell self-renewal and neuroregeneration through ciliary neurotrophic factor. Stem Cell Res. 2020, 49, 102061. [Google Scholar] [CrossRef]
- Demirler, M.C.; Sakizli, U.; Bali, B.; Kocagöz, Y.; Eski, S.E.; Ergönen, A.; Alkiraz, A.S.; Bayramli, X.; Hassenklöver, T.; Manzini, I.; et al. Purinergic signalling selectively modulates maintenance but not repair neurogenesis in the zebrafish olfactory epithelium. FEBS J. 2020, 287, 2699–2722. [Google Scholar] [CrossRef]
- Leung, C.T.; Coulombe, P.A.; Reed, R.R. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat. Neurosci. 2007, 10, 720–726. [Google Scholar] [CrossRef]
- Gadye, L.; Das, D.; Sanchez, M.A.; Street, K.; Baudhuin, A.; Wagner, A.; Cole, M.B.; Choi, Y.G.; Yosef, N.; Purdom, E.; et al. Injury Activates Transient Olfactory Stem Cell States with Diverse Lineage Capacities. Cell Stem Cell 2017, 21, 775–790.e779. [Google Scholar] [CrossRef]
- Chen, M.; Reed, R.R.; Lane, A.P. Chronic Inflammation Directs an Olfactory Stem Cell Functional Switch from Neuroregeneration to Immune Defense. Cell Stem Cell 2019, 25, 501–513.e505. [Google Scholar] [CrossRef]
- Hansel, D.E.; Eipper, B.A.; Ronnett, G.V. Neuropeptide Y functions as a neuroproliferative factor. Nature 2001, 410, 940–944. [Google Scholar] [CrossRef] [PubMed]
- Breunig, E.; Manzini, I.; Piscitelli, F.; Gutermann, B.; Di Marzo, V.; Schild, D.; Czesnik, D. The endocannabinoid 2-arachidonoyl-glycerol controls odor sensitivity in larvae of Xenopus laevis. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 8965–8973. [Google Scholar] [CrossRef]
- Lacroix, M.C.; Badonnel, K.; Meunier, N.; Tan, F.; Schlegel-Le Poupon, C.; Durieux, D.; Monnerie, R.; Baly, C.; Congar, P.; Salesse, R.; et al. Expression of insulin system in the olfactory epithelium: First approaches to its role and regulation. J. Neuroendocr. 2008, 20, 1176–1190. [Google Scholar] [CrossRef]
- Senf, K.; Karius, J.; Stumm, R.; Neuhaus, E.M. Chemokine signaling is required for homeostatic and injury-induced neurogenesis in the olfactory epithelium. Stem Cells 2021, 39, 617–635. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.; Feitzinger, A.; Venkiteswaran, G.; Wang, J.; Lewellis, S.W.; Koplinski, C.A.; Peterson, F.C.; Volkman, B.F.; Meier-Schellersheim, M.; Knaut, H. A negative-feedback loop maintains optimal chemokine concentrations for directional cell migration. Nat. Cell Biol. 2020, 22, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Li, H.; Chang, T.; He, W.; Kong, Y.; Qi, C.; Li, R.; Huang, H.; Zhu, Z.; Zheng, P.; et al. Bone marrow hematopoiesis drives multiple sclerosis progression. Cell 2022, 185, 2234–2247. [Google Scholar] [CrossRef]
- López-Gil, J.C.; Martin-Hijano, L.; Hermann, P.C.; Sainz, B., Jr. The CXCL12 Crossroads in Cancer Stem Cells and Their Niche. Cancers 2021, 13, 469. [Google Scholar] [CrossRef]
- Maeda, Y.; Yonemochi, Y.; Nakajyo, Y.; Hidaka, H.; Ikeda, T.; Ando, Y. CXCL12 and osteopontin from bone marrow-derived mesenchymal stromal cells improve muscle regeneration. Sci. Rep. 2017, 7, 3305. [Google Scholar] [CrossRef]
- Balabanian, K.; Lagane, B.; Infantino, S.; Chow, K.Y.; Harriague, J.; Moepps, B.; Arenzana-Seisdedos, F.; Thelen, M.; Bachelerie, F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 2005, 280, 35760–35766. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.M.; Summers, B.C.; Wang, Y.; Melikian, A.; Berahovich, R.; Miao, Z.; Penfold, M.E.; Sunshine, M.J.; Littman, D.R.; Kuo, C.J.; et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J. Exp. Med. 2006, 203, 2201–2213. [Google Scholar] [CrossRef]
- Koenen, J.; Bachelerie, F.; Balabanian, K.; Schlecht-Louf, G.; Gallego, C. Atypical Chemokine Receptor 3 (ACKR3): A Comprehensive Overview of its Expression and Potential Roles in the Immune System. Mol. Pharmacol. 2019, 96, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Naumann, U.; Cameroni, E.; Pruenster, M.; Mahabaleshwar, H.; Raz, E.; Zerwes, H.G.; Rot, A.; Thelen, M. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS ONE 2010, 5, e9175. [Google Scholar] [CrossRef]
- Saaber, F.; Schutz, D.; Miess, E.; Abe, P.; Desikan, S.; Ashok Kumar, P.; Balk, S.; Huang, K.; Beaulieu, J.M.; Schulz, S.; et al. ACKR3 Regulation of Neuronal Migration Requires ACKR3 Phosphorylation, but Not beta-Arrestin. Cell Rep. 2019, 26, 1473–1488.e1479. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Su, J.; Zhang, Z. The CXCL12/CXCR4/ACKR3 Response Axis in Chronic Neurodegenerative Disorders of the Central Nervous System: Therapeutic Target and Biomarker. Cell. Mol. Neurobiol. 2021, 42, 2147–2156. [Google Scholar] [CrossRef]
- Handel, T.M.; Johnson, Z.; Crown, S.E.; Lau, E.K.; Sweeney, M.; Proudfoot, A.E. Regulation of protein function by glycosaminoglycans—As exemplified by chemokines. Annu. Rev. Biochem. 2005, 74, 385–410. [Google Scholar] [CrossRef]
- Netelenbos, T.; van den Born, J.; Kessler, F.L.; Zweegman, S.; Merle, P.A.; van Oostveen, J.W.; Zwaginga, J.J.; Huijgens, P.C.; Dräger, A.M. Proteoglycans on bone marrow endothelial cells bind and present SDF-1 towards hematopoietic progenitor cells. Leukemia 2003, 17, 175–184. [Google Scholar] [CrossRef]
- Rueda, P.; Richart, A.; Récalde, A.; Gasse, P.; Vilar, J.; Guérin, C.; Lortat-Jacob, H.; Vieira, P.; Baleux, F.; Chretien, F.; et al. Homeostatic and tissue reparation defaults in mice carrying selective genetic invalidation of CXCL12/proteoglycan interactions. Circulation 2012, 126, 1882–1895. [Google Scholar] [CrossRef]
- Abe, P.; Mueller, W.; Schutz, D.; MacKay, F.; Thelen, M.; Zhang, P.; Stumm, R. CXCR7 prevents excessive CXCL12-mediated downregulation of CXCR4 in migrating cortical interneurons. Development 2014, 141, 1857–1863. [Google Scholar] [CrossRef]
- Sánchez-Alcañiz, J.A.; Haege, S.; Mueller, W.; Pla, R.; Mackay, F.; Schulz, S.; López-Bendito, G.; Stumm, R.; Marín, O. Cxcr7 Controls Neuronal Migration by Regulating Chemokine Responsiveness. Neuron 2011, 69, 77–90. [Google Scholar] [CrossRef]
- Memi, F.; Abe, P.; Cariboni, A.; MacKay, F.; Parnavelas, J.G.; Stumm, R. CXC chemokine receptor 7 (CXCR7) affects the migration of GnRH neurons by regulating CXCL12 availability. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 17527–17537. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hirata, T.; Mackay, F.; Murakami, F. Chemokine receptor CXCR7 non-cell-autonomously controls pontine neuronal migration and nucleus formation. Sci. Rep. 2020, 10, 11830. [Google Scholar] [CrossRef]
- Lewellis, S.W.; Nagelberg, D.; Subedi, A.; Staton, A.; LeBlanc, M.; Giraldez, A.; Knaut, H. Precise SDF1-mediated cell guidance is achieved through ligand clearance and microRNA-mediated decay. J. Cell Biol. 2013, 200, 337–355. [Google Scholar] [CrossRef]
- Donà, E.; Barry, J.D.; Valentin, G.; Quirin, C.; Khmelinskii, A.; Kunze, A.; Durdu, S.; Newton, L.R.; Fernandez-Minan, A.; Huber, W.; et al. Directional tissue migration through a self-generated chemokine gradient. Nature 2013, 503, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Dassule, H.R.; Lewis, P.; Bei, M.; Maas, R.; McMahon, A.P. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 2000, 127, 4775–4785. [Google Scholar] [CrossRef]
- Feltri, M.L.; D’Antonio, M.; Previtali, S.; Fasolini, M.; Messing, A.; Wrabetz, L. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann. N. Y. Acad. Sci. 1999, 883, 116–123. [Google Scholar] [CrossRef]
- Sierro, F.; Biben, C.; Martinez-Munoz, L.; Mellado, M.; Ransohoff, R.M.; Li, M.; Woehl, B.; Leung, H.; Groom, J.; Batten, M.; et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc. Natl. Acad. Sci. USA 2007, 104, 14759–14764. [Google Scholar] [CrossRef]
- Tzeng, Y.S.; Li, H.; Kang, Y.L.; Chen, W.C.; Cheng, W.C.; Lai, D.M. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood 2011, 117, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, B.J.; Banisadr, G.; Jung, H.; Ren, D.; Cronshaw, D.G.; Zou, Y.; Miller, R.J. The Chemokine Stromal Cell-Derived Factor-1 Regulates GABAergic Inputs to Neural Progenitors in the Postnatal Dentate Gyrus. J. Neurosci. 2008, 28, 6720–6730. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Stumm, R.K.; Zhou, C.; Ara, T.; Lazarini, F.; Dubois-Dalcq, M.; Nagasawa, T.; Hollt, V.; Schulz, S. CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 5123–5130. [Google Scholar] [CrossRef]
- Tashiro, K.; Tada, H.; Heilker, R.; Shirozu, M.; Nakano, T.; Honjo, T. Signal sequence trap: A cloning strategy for secreted proteins and type I membrane proteins. Science 1993, 261, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Oh, S.Y.; Zhu, Z.; Lee, J.; Lane, A.P. Spontaneous eosinophilic nasal inflammation in a genetically-mutant mouse: Comparative study with an allergic inflammation model. PLoS ONE 2012, 7, e35114. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.H.; Murray, E.; Andrews, G.; Jiang, H.C.; Park, S.J.; Donnard, E.; Durán-Laforet, V.; Bear, D.M.; Faust, T.E.; Garber, M.; et al. Spatial transcriptomic reconstruction of the mouse olfactory glomerular map suggests principles of odor processing. Nat. Neurosci. 2022, 25, 484–492. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R.; Shiny, G.O. a graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef]
- Boldajipour, B.; Mahabaleshwar, H.; Kardash, E.; Reichman-Fried, M.; Blaser, H.; Minina, S.; Wilson, D.; Xu, Q.; Raz, E. Control of chemokine-guided cell migration by ligand sequestration. Cell 2008, 132, 463–473. [Google Scholar] [CrossRef]
- Berlutti, F.; Pilloni, A.; Pietropaoli, M.; Polimeni, A.; Valenti, P. Lactoferrin and oral diseases: Current status and perspective in periodontitis. Ann. Stomatol. 2011, 2, 10–18. [Google Scholar]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gústafsdóttir, S.M.; Ostman, A.; Landegren, U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef]
- Kunz, L.; Schroeder, T. A 3D Tissue-wide Digital Imaging Pipeline for Quantitation of Secreted Molecules Shows Absence of CXCL12 Gradients in Bone Marrow. Cell Stem Cell 2019, 25, 846–854.e844. [Google Scholar] [CrossRef]
- Schaffrath, D.; Stuhlsatz, H.W.; Greiling, H. Interactions of glycosaminoglycans with DNA and RNA synthesizing enzymes in vitro. Hoppe Seylers Z. Physiol. Chem. 1976, 357, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Milho, R.; Frederico, B.; Efstathiou, S.; Stevenson, P.G. A Heparan-Dependent Herpesvirus Targets the Olfactory Neuroepithelium for Host Entry. PLoS Pathog. 2012, 8, e1002986. [Google Scholar] [CrossRef]
- Mueller, W.; Schutz, D.; Nagel, F.; Schulz, S.; Stumm, R. Hierarchical organization of multi-site phosphorylation at the CXCR4 C terminus. PLoS ONE 2013, 8, e64975. [Google Scholar] [CrossRef]
- Lapidot, T.; Kollet, O. The essential roles of the chemokine SDF-1 and its receptor CXCR4 in human stem cell homing and repopulation of transplanted immune-deficient NOD/SCID and NOD/SCID/B2m(null) mice. Leukemia 2002, 16, 1992–2003. [Google Scholar] [CrossRef]
- Mimura-Yamamoto, Y.; Shinohara, H.; Kashiwagi, T.; Sato, T.; Shioda, S.; Seki, T. Dynamics and function of CXCR4 in formation of the granule cell layer during hippocampal development. Sci. Rep. 2017, 7, 5647. [Google Scholar] [CrossRef]
- Gerits, N.; Kostenko, S.; Shiryaev, A.; Johannessen, M.; Moens, U. Relations between the mitogen-activated protein kinase and the cAMP-dependent protein kinase pathways: Comradeship and hostility. Cell. Signal. 2008, 20, 1592–1607. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Park, I.W.; Groopman, J.E. Stromal cell-derived factor-1alpha stimulates tyrosine phosphorylation of multiple focal adhesion proteins and induces migration of hematopoietic progenitor cells: Roles of phosphoinositide-3 kinase and protein kinase C. Blood 2000, 95, 2505–2513. [Google Scholar] [CrossRef]
- Goo, B.S.; Mun, D.J.; Kim, S.; Nhung, T.T.M.; Lee, S.B.; Woo, Y.; Kim, S.J.; Suh, B.K.; Park, S.J.; Lee, H.E.; et al. Schizophrenia-associated Mitotic Arrest Deficient-1 (MAD1) regulates the polarity of migrating neurons in the developing neocortex. Mol. Psychiatry 2023, 28, 856–870. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Q.; Yang, P.; Wang, C.; Liu, J.; Ding, W.; Liu, W.; Bai, Y.; Yang, Y.; Wang, H.; et al. LSD1 co-repressor Rcor2 orchestrates neurogenesis in the developing mouse brain. Nat. Commun. 2016, 7, 10481. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, C.E.; Nechiporuk, T.; Jeng, S.; McWeeney, S.K.; Wang, J.; Rosenfeld, M.G.; Mandel, G. REST corepressors RCOR1 and RCOR2 and the repressor INSM1 regulate the proliferation-differentiation balance in the developing brain. Proc. Natl. Acad. Sci. USA 2017, 114, e406–e415. [Google Scholar] [CrossRef]
- Coleman, J.H.; Lin, B.; Schwob, J.E. Dissecting LSD1-Dependent Neuronal Maturation in the Olfactory Epithelium. J. Comp. Neurol. 2017, 525, 3391–3413. [Google Scholar] [CrossRef]
- Krolewski, R.C.; Packard, A.; Schwob, J.E. Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium. J. Comp. Neurol. 2013, 521, 833–859. [Google Scholar] [CrossRef]
- Gauthier-Kemper, A.; Igaev, M.; Sündermann, F.; Janning, D.; Brühmann, J.; Moschner, K.; Reyher, H.J.; Junge, W.; Glebov, K.; Walter, J.; et al. Interplay between phosphorylation and palmitoylation mediates plasma membrane targeting and sorting of GAP43. Mol. Biol. Cell 2014, 25, 3284–3299. [Google Scholar] [CrossRef] [PubMed]
- McNamara, R.K.; Lenox, R.H. Distribution of the protein kinase C substrates MARCKS and MRP in the postnatal developing rat brain. J. Comp. Neurol. 1998, 397, 337–356. [Google Scholar] [CrossRef]
- Hartman, B.K.; Margolis, F.L. Immunofluorescence localization of the olfactory marker protein. Brain Res. 1975, 96, 176–180. [Google Scholar] [CrossRef]
- Brittebo, E.B. Metabolism-dependent toxicity of methimazole in the olfactory nasal mucosa. Pharm. Toxicol. 1995, 76, 76–79. [Google Scholar] [CrossRef]
- Muenzer, J.; Wraith, J.E.; Clarke, L.A.; Mucopolysaccharidosis, I. management and treatment guidelines. Pediatrics 2009, 123, 19–29. [Google Scholar] [CrossRef]
- Watson, H.A.; Holley, R.J.; Langford-Smith, K.J.; Wilkinson, F.L.; van Kuppevelt, T.H.; Wynn, R.F.; Wraith, J.E.; Merry, C.L.; Bigger, B.W. Heparan sulfate inhibits hematopoietic stem and progenitor cell migration and engraftment in mucopolysaccharidosis I. J. Biol. Chem. 2014, 289, 36194–36203. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, X.; Jiang, Y.; Chen, Z.; Zhang, Y.; Hao, D.; Yang, H. The Anti-inflammation Property of Olfactory Ensheathing Cells in Neural Regeneration After Spinal Cord Injury. Mol. Neurobiol. 2022, 59, 6447–6459. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.R.; McCandless, E.E.; Dorsey, D.; Klein, R.S. CXCR4 promotes differentiation of oligodendrocyte progenitors and remyelination. Proc. Natl. Acad. Sci. USA 2010, 107, 11062–11067. [Google Scholar] [CrossRef] [PubMed]
- Calderon, T.M.; Eugenin, E.A.; Lopez, L.; Kumar, S.S.; Hesselgesser, J.; Raine, C.S.; Berman, J.W. A role for CXCL12 (SDF-1alpha) in the pathogenesis of multiple sclerosis: Regulation of CXCL12 expression in astrocytes by soluble myelin basic protein. J. Neuroimmunol. 2006, 177, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Packard, A.; Schnittke, N.; Romano, R.A.; Sinha, S.; Schwob, J.E. DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 8748–8759. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Takeda, M.; Farbman, A.I. Supporting cells as phagocytes in the olfactory epithelium after bulbectomy. J. Comp. Neurol. 1996, 376, 509–517. [Google Scholar] [CrossRef]
- Thiebaud, N.; Veloso Da Silva, S.; Jakob, I.; Sicard, G.; Chevalier, J.; Ménétrier, F.; Berdeaux, O.; Artur, Y.; Heydel, J.M.; Le Bon, A.M. Odorant metabolism catalyzed by olfactory mucosal enzymes influences peripheral olfactory responses in rats. PLoS ONE 2013, 8, e59547. [Google Scholar] [CrossRef]
- Neiers, F.; Jarriault, D.; Menetrier, F.; Briand, L.; Heydel, J.-M. The odorant metabolizing enzyme UGT2A1: Immunolocalization and impact of the modulation of its activity on the olfactory response. PLoS ONE 2021, 16, e0249029. [Google Scholar] [CrossRef] [PubMed]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; Van den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.J.; Fletcher, R.B.; et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020, 6, eabc5801. [Google Scholar] [CrossRef]
- Finlay, J.B.; Brann, D.H.; Abi Hachem, R.; Jang, D.W.; Oliva, A.D.; Ko, T.; Gupta, R.; Wellford, S.A.; Moseman, E.A.; Jang, S.S.; et al. Persistent post-COVID-19 smell loss is associated with immune cell infiltration and altered gene expression in olfactory epithelium. Sci. Transl. Med. 2022, 14, eadd0484. [Google Scholar] [CrossRef]
- Zazhytska, M.; Kodra, A.; Hoagland, D.A.; Frere, J.; Fullard, J.F.; Shayya, H.; McArthur, N.G.; Moeller, R.; Uhl, S.; Omer, A.D.; et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell 2022, 185, 1052–1064.e1012. [Google Scholar] [CrossRef]
- Janssens, R.; Struyf, S.; Proost, P. The unique structural and functional features of CXCL12. Cell Mol. Immunol. 2018, 15, 299–311. [Google Scholar] [CrossRef]
- Ballas, N.; Battaglioli, E.; Atouf, F.; Andres, M.E.; Chenoweth, J.; Anderson, M.E.; Burger, C.; Moniwa, M.; Davie, J.R.; Bowers, W.J.; et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron 2001, 31, 353–365. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dietz, A.; Senf, K.; Karius, J.; Stumm, R.; Neuhaus, E.M. Glia Cells Control Olfactory Neurogenesis by Fine-Tuning CXCL12. Cells 2023, 12, 2164. https://doi.org/10.3390/cells12172164
Dietz A, Senf K, Karius J, Stumm R, Neuhaus EM. Glia Cells Control Olfactory Neurogenesis by Fine-Tuning CXCL12. Cells. 2023; 12(17):2164. https://doi.org/10.3390/cells12172164
Chicago/Turabian StyleDietz, André, Katja Senf, Julia Karius, Ralf Stumm, and Eva Maria Neuhaus. 2023. "Glia Cells Control Olfactory Neurogenesis by Fine-Tuning CXCL12" Cells 12, no. 17: 2164. https://doi.org/10.3390/cells12172164
APA StyleDietz, A., Senf, K., Karius, J., Stumm, R., & Neuhaus, E. M. (2023). Glia Cells Control Olfactory Neurogenesis by Fine-Tuning CXCL12. Cells, 12(17), 2164. https://doi.org/10.3390/cells12172164






