Differential Gene Expression in Activated Microglia Treated with Adenosine A2A Receptor Antagonists Highlights Olfactory Receptor 56 and T-Cell Activation GTPase-Activating Protein 1 as Potential Biomarkers of the Polarization of Activated Microglia
Abstract
:1. Introduction
2. Methodology
2.1. Reagents
2.2. Isolation and Activation of Microglia
2.3. RNAseq Data Processing
2.4. Differential Expression Analysis
2.5. Data Curation
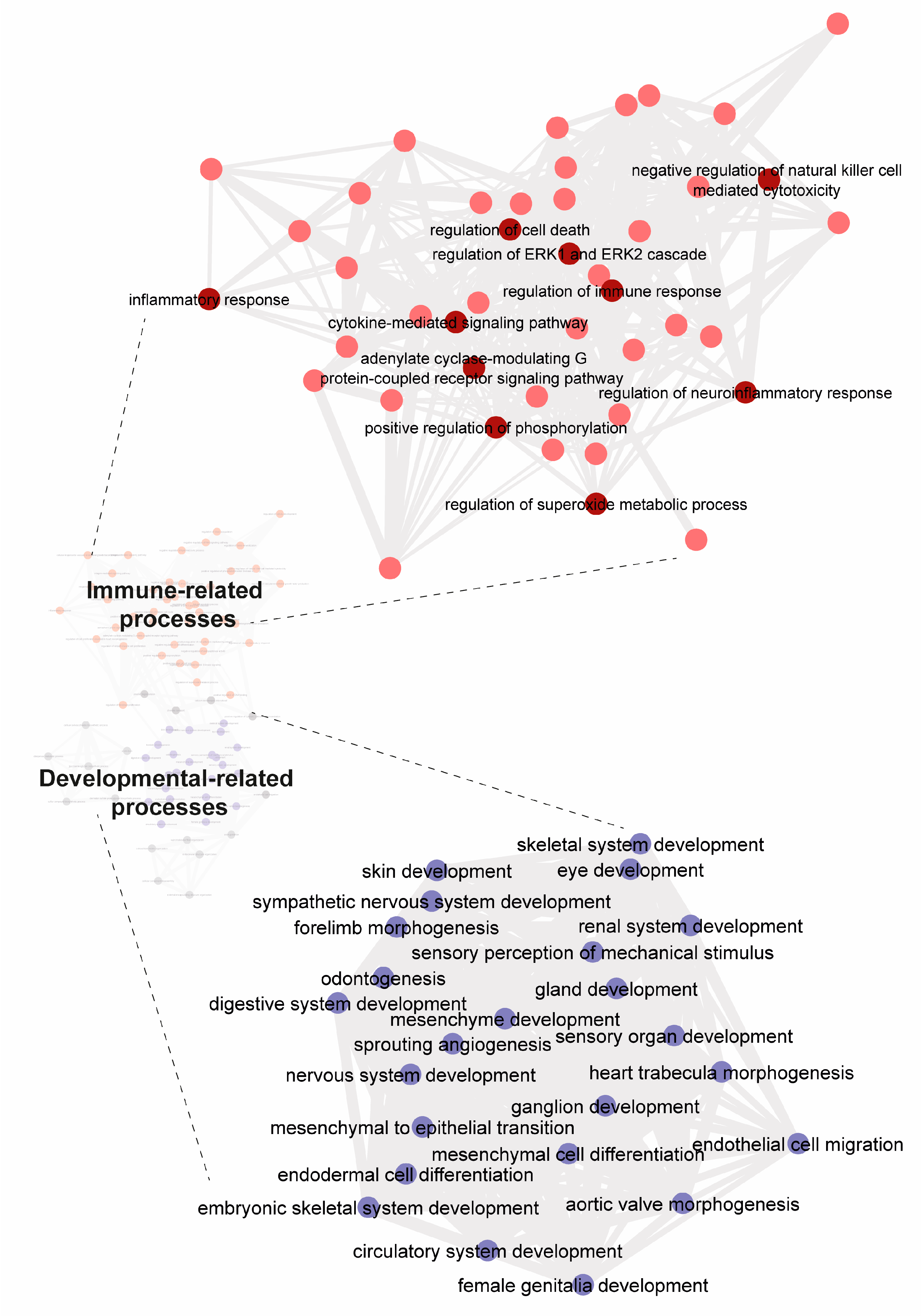
2.6. Gene Set Enrichment Analyses
3. Results
3.1. RNAseq in Activated Microglia in the Presence of an Antagonist of the A2AR
3.2. RNAseq in Activated Microglia Treated Simultaneously with an Antagonist of the A2AR and an Agonist of the A3R
3.3. Comparing RNAseq Data from Individual and Combined Treatments
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Angulo, E.; Casadó, V.; Mallol, J.; Canela, E.I.; Viñals, F.; Ferrer, I.; Lluis, C.; Franco, R. A1 adenosine receptors accumulate in neurodegenerative structures in Alzheimer disease and mediate both amyloid precursor protein processing and tau phosphorylation and translocation. Brain Pathol. 2003, 13, 440–451. [Google Scholar] [CrossRef]
- Franco, R.; Fernández-Suárez, D. Alternatively activated microglia and macrophages in the central nervous system. Prog. Neurobiol. 2015, 131, 65–86. [Google Scholar] [CrossRef] [PubMed]
- Girard, S.; Brough, D.; Lopez-Castejon, G.; Giles, J.; Rothwell, N.J.; Allan, S.M. Microglia and macrophages differentially modulate cell death after brain injury caused by oxygen-glucose deprivation in organotypic brain slices. Glia 2013, 61, 813–824. [Google Scholar] [CrossRef]
- Walker, D.G.; Lue, L.F. Immune phenotypes of microglia in human neurodegenerative disease: Challenges to detecting microglial polarization in human brains. Alzheimer’s Res. Ther. 2015, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Jaguin, M.; Houlbert, N.; Fardel, O.; Lecureur, V. Polarization profiles of human M-CSF-generated macrophages and comparison of M1-markers in classically activated macrophages from GM-CSF and M-CSF origin. Cell. Immunol. 2013, 281, 51–61. [Google Scholar] [CrossRef]
- Mesquida-Veny, F.; Del Río, J.A.; Hervera, A. Macrophagic and microglial complexity after neuronal injury. Prog. Neurobiol. 2021, 200, 101970. [Google Scholar] [CrossRef]
- Devanney, N.A.; Stewart, A.N.; Gensel, J.C. Microglia and macrophage metabolism in CNS injury and disease: The role of immunometabolism in neurodegeneration and neurotrauma. Exp. Neurol. 2020, 329, 113310. [Google Scholar] [CrossRef]
- Du, L.; Zhang, Y.; Chen, Y.; Zhu, J.; Yang, Y.; Zhang, H.L. Role of Microglia in Neurological Disorders and Their Potentials as a Therapeutic Target. Mol. Neurobiol. 2017, 54, 7567–7584. [Google Scholar] [CrossRef]
- Peña-Altamira, E.; Prati, F.; Massenzio, F.; Virgili, M.; Contestabile, A.; Bolognesi, M.L.; Monti, B. Changing paradigm to target microglia in neurodegenerative diseases: From anti-inflammatory strategy to active immunomodulation. Expert Opin. Ther. Targets 2016, 20, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Song, G.J.; Suk, K. Pharmacological Modulation of Functional Phenotypes of Microglia in Neurodegenerative Diseases. Front. Aging Neurosci. 2017, 9, 139. [Google Scholar] [CrossRef]
- L’Episcopo, F.; Tirolo, C.; Serapide, M.F.; Caniglia, S.; Testa, N.; Leggio, L.; Vivarelli, S.; Iraci, N.; Pluchino, S.; Marchetti, B. Microglia Polarization, Gene-Environment Interactions and Wnt/β-Catenin Signaling: Emerging Roles of Glia-Neuron and Glia-Stem/Neuroprogenitor Crosstalk for Dopaminergic Neurorestoration in Aged Parkinsonian Brain. Front. Aging Neurosci. 2018, 10, 12. [Google Scholar] [CrossRef]
- Rebola, N.; Simões, A.P.; Canas, P.M.; Tomé, A.R.; Andrade, G.M.; Barry, C.E.; Agostinho, P.M.; Lynch, M.A.; Cunha, R.A. Adenosine A2A receptors control neuroinflammation and consequent hippocampal neuronal dysfunction. J. Neurochem. 2011, 117, 100–111. [Google Scholar] [CrossRef]
- Frau, L.; Borsini, F.; Wardas, J.; Khairnar, A.S.; Schintu, N.; Morelli, M. Neuroprotective and anti-inflammatory effects of the adenosine A(2A) receptor antagonist ST1535 in a MPTP mouse model of Parkinson’s disease. Synapse 2011, 65, 181–188. [Google Scholar] [CrossRef]
- Minghetti, L.; Greco, A.; Potenza, R.L.; Pezzola, A.; Blum, D.; Bantubungi, K.; Popoli, P. Effects of the adenosine A2A receptor antagonist SCH 58621 on cyclooxygenase-2 expression, glial activation, and brain-derived neurotrophic factor availability in a rat model of striatal neurodegeneration. J. Neuropathol. Exp. Neurol. 2007, 66, 363–371. [Google Scholar] [CrossRef]
- Saura, J.; Angulo, E.; Ejarque, A.; Casado, V.; Tusell, J.M.; Moratalla, R.; Chen, J.-F.F.; Schwarzschild, M.A.; Lluis, C.; Franco, R.; et al. Adenosine A2A receptor stimulation potentiates nitric oxide release by activated microglia. J. Neurochem. 2005, 95, 919–929. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jhun, B.S.; Oh, Y.T.; Lee, J.H.; Choe, W.; Baik, H.H.; Ha, J.; Yoon, K.S.; Kim, S.S.; Kang, I. Activation of adenosine A3 receptor suppresses lipopolysaccharide-induced TNF-alpha production through inhibition of PI 3-kinase/Akt and NF-kappaB activation in murine BV2 microglial cells. Neurosci. Lett. 2006, 396, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hammarberg, C.; Schulte, G.; Fredholm, B.B. Evidence for functional adenosine A3 receptors in microglia cells. J. Neurochem. 2003, 86, 1051–1054. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.P.; Christopoulos, A.; Davenport, A.P.; Kelly, E.; Mathie, A.; Peters, J.A.; Veale, E.L.; Armstrong, J.F.; Faccenda, E.; Harding, S.D.; et al. The concise guide to pharmacology 2019/20: G protein-coupled receptors. Br. J. Pharmacol. 2019, 176, S21–S141. [Google Scholar] [CrossRef] [PubMed]
- Lillo, A.; Martínez-Pinilla, E.; Reyes-Resina, I.; Navarro, G.; Franco, R. Adenosine A2a and A3 receptors are able to interact with each other. A further piece in the puzzle of adenosine receptor-mediated signaling. Int. J. Mol. Sci. 2020, 21, 5070. [Google Scholar] [CrossRef]
- Newell, E.A.; Exo, J.L.; Verrier, J.D.; Jackson, T.C.; Gillespie, D.G.; Janesko-Feldman, K.; Kochanek, P.M.; Jackson, E.K. 2′,3′-cAMP, 3′-AMP, 2′-AMP and adenosine inhibit TNF-α and CXCL10 production from activated primary murine microglia via A2A receptors. Brain Res. 2015, 1594, 27–35. [Google Scholar] [CrossRef]
- Pulido-Salgado, M.; Vidal-Taboada, J.M.; Garcia Diaz-Barriga, G.; Serratosa, J.; Valente, T.; Castillo, P.; Matalonga, J.; Straccia, M.; Canals, J.M.; Valledor, A.; et al. Myeloid C/EBPβ deficiency reshapes microglial gene expression and is protective in experimental autoimmune encephalomyelitis. J. Neuroinflamm. 2017, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Saura, J.; Tusell, J.M.; Serratosa, J. High-Yield Isolation of Murine Microglia by Mild Trypsinization. Glia 2003, 44, 183–189. [Google Scholar] [CrossRef]
- Lillo, A.; Serrano-Marín, J.; Lillo, J.; Raïch, I.; Navarro, G.; Franco, R. Gene regulation in activated microglia by adenosine A3 receptor agonists. A transcriptomics study. Purinergic Signal. 2023; in press. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Cid, E.; Marquez-Galera, A.; Valero, M.; Gal, B.; Medeiros, D.C.; Navarron, C.M.; Ballesteros-Esteban, L.; Reig-Viader, R.; Morales, A.V.; Fernandez-Lamo, I.; et al. Sublayer- and cell-type-specific neurodegenerative transcriptional trajectories in hippocampal sclerosis. Cell Rep. 2021, 35, 109229. [Google Scholar] [CrossRef] [PubMed]
- Bult, C.J.; Eppig, J.T.; Blake, J.A.; Kadin, J.A.; Richardson, J.E.; Anagnostopoulos, A.; Baldarelli, R.M.; Beal, J.S.; Bello, S.M.; Berghout, J.; et al. Mouse genome database 2016. Nucleic Acids Res. 2016, 44, D840–D847. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sekine, M.; Seng, S.; Avraham, S.; Avraham, H.K. BRCA1 interacts with Smad3 and regulates Smad3-mediated TGF-beta signaling during oxidative stress responses. PLoS ONE 2009, 4, e7091. [Google Scholar] [CrossRef]
- Xu, L.; Wu, M.; Hu, J.; Zhai, Z.; Shu, H. Identification of downstream genes up-regulated by the tumor necrosis factor family member TALL-1. J. Leukoc. Biol. 2002, 72, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.; Roccato, E.; Raho, G.; Pagliardini, S.; Pierotti, M.A.; Greco, A. The TFG protein, involved in oncogenic rearrangements, interacts with TANK and NEMO, two proteins involved in the NF-kappaB pathway. J. Cell. Physiol. 2006, 208, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Cui, L. Resveratrol suppresses melanoma by inhibiting NF-κB/miR-221 and inducing TFG expression. Arch. Dermatol. Res. 2017, 309, 823–831. [Google Scholar] [CrossRef]
- Kamato, D.; Burch, M.L.; Piva, T.J.; Rezaei, H.B.; Rostam, M.A.; Xu, S.; Zheng, W.; Little, P.J.; Osman, N. Transforming growth factor-β signalling: Role and consequences of Smad linker region phosphorylation. Cell. Signal. 2013, 25, 2017–2024. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qi, Y.; Qi, J.; Wu, J.; Lin, F.; Cui, X.; Ge, J.; Liu, Z. Activin A is a novel chemoattractant for migration of microglial BV2 cells. J. Neuroimmunol. 2022, 371, 577929. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.A.; Kim, Y.J.; Jeon, Y.J. The diverse repertoire of ISG15: More intricate than initially thought. Exp. Mol. Med. 2022, 54, 1779–1792. [Google Scholar] [CrossRef] [PubMed]
- Zaugg, J.B.; Sahlén, P.; Andersson, R.; Alberich-Jorda, M.; de Laat, W.; Deplancke, B.; Ferrer, J.; Mandrup, S.; Natoli, G.; Plewczynski, D.; et al. Current challenges in understanding the role of enhancers in disease. Nat. Struct. Mol. Biol. 2022, 29, 1148–1158. [Google Scholar] [CrossRef]
- Nowell, J.; Blunt, E.; Edison, P. Incretin and insulin signaling as novel therapeutic targets for Alzheimer’s and Parkinson’s disease. Mol. Psychiatry 2022, 28, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Citron, B.A.; Dennis, J.S.; Zeitlin, R.S.; Echeverria, V. Transcription factor Sp1 dysregulation in Alzheimer’s disease. J. Neurosci. Res. 2008, 86, 2499–2504. [Google Scholar] [CrossRef]
- Novikova, G.; Kapoor, M.; Tcw, J.; Abud, E.M.; Efthymiou, A.G.; Chen, S.X.; Cheng, H.; Fullard, J.F.; Bendl, J.; Liu, Y.; et al. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat. Commun. 2021, 12, 1610. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.H.; Yu, Q.; Deng, Z.L.; Yang, K.; Ye, Y.; Ge, M.X.; Yan, D.; Wang, H.T.; Chen, X.Q.; Yang, L.Q.; et al. ETS1 acts as a regulator of human healthy aging via decreasing ribosomal activity. Sci. Adv. 2022, 8, 2017. [Google Scholar] [CrossRef]
- Jantaratnotai, N.; Ling, A.; Cheng, J.; Schwab, C.; Mcgeer, P.L.; Mclarnon, J.G. Upregulation and Expression Patterns of the Angiogenic Transcription Factor Ets-1 in Alzheimer’s Disease Brain. J. Alzheimer’s Dis. 2013, 37, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Zhan, Y.; He, H.; Maynard, E.; Balschi, J.A.; Oettgen, P. Ets-1 is a critical transcriptional regulator of reactive oxygen species and p47phox gene expression in response to angiotensin II. Circ. Res. 2007, 101, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Ndoja, A.; Reja, R.; Lee, S.H.; Webster, J.D.; Ngu, H.; Rose, C.M.; Kirkpatrick, D.S.; Modrusan, Z.; Chen, Y.J.J.; Dugger, D.L.; et al. Ubiquitin Ligase COP1 Suppresses Neuroinflammation by Degrading c/EBPβ in Microglia. Cell 2020, 182, 1156–1169.e12. [Google Scholar] [CrossRef]
- Tan, M.; Shen, L.; Hou, Y. Epigenetic modification of BDNF mediates neuropathic pain via miR-30a-3p/EP300 axis in CCI rats. Biosci. Rep. 2020, 40, 20194442. [Google Scholar] [CrossRef]
- Blalock, E.M.; Geddes, J.W.; Chen, K.C.; Porter, N.M.; Markesbery, W.R.; Landfield, P.W. Incipient Alzheimer’s disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl. Acad. Sci. USA 2004, 101, 2173–2178. [Google Scholar] [CrossRef]
- Lösing, P.; Niturad, C.E.; Harrer, M.; Reckendorf, C.M.Z.; Schatz, T.; Sinske, D.; Lerche, H.; Maljevic, S.; Knöll, B. SRF mod-ulates seizure occurrence, activity induced gene transcription and hippocampal circuit reorganization in the mouse pilocarpine epilepsy model. Mol. Brain 2017, 10, 30. [Google Scholar] [CrossRef]
- Kasza, A.; Wyrzykowska, P.; Horwacik, I.; Tymoszuk, P.; Mizgalska, D.; Palmer, K.; Rokita, H.; Sharrocks, A.D.; Jura, J. Transcription factors Elk-1 and SRF are engaged in IL1-dependent regulation of ZC3H12A expression. BMC Mol. Biol. 2010, 11, 14. [Google Scholar] [CrossRef]
- Lee, M.S.; Kim, B.; Lee, S.M.; Cho, W.C.; Lee, W.B.; Kang, J.S.; Choi, U.Y.; Lyu, J.; Kim, Y.J. Genome-Wide Profiling of In Vivo LPS-Responsive Genes in Splenic Myeloid Cells. Mol. Cells 2013, 35, 498. [Google Scholar] [CrossRef] [PubMed]
- Boddicker, R.L.; Razidlo, G.L.; Feldman, A.L. Genetic alterations affecting GTPases and T-cell receptor signaling in peripheral T-cell lymphomas. Small GTPases 2019, 10, 33–39. [Google Scholar] [CrossRef]
- He, H.; Huang, J.; Wu, S.; Jiang, S.; Liang, L.; Liu, Y.; Liu, W.; Xie, L.; Tao, Y.; Jiang, Y.; et al. The roles of GTPase-activating proteins in regulated cell death and tumor immunity. J. Hematol. Oncol. 2021, 14, 171. [Google Scholar] [CrossRef] [PubMed]
- Lieschke, S.; Zechmeister, B.; Haupt, M.; Zheng, X.; Jin, F.; Hein, K.; Weber, M.S.; Hermann, D.M.; Bähr, M.; Kilic, E.; et al. CCL11 Differentially Affects Post-Stroke Brain Injury and Neuroregeneration in Mice Depending on Age. Cells 2019, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Scabia, G.; Testa, G.; Scali, M.; Del Turco, S.; Desiato, G.; Berardi, N.; Sale, A.; Matteoli, M.; Maffei, L.; Maffei, M.; et al. Reduced ccl11/eotaxin mediates the beneficial effects of environmental stimulation on the aged hippocampus. Brain Behav. Immun. 2021, 98, 234–244. [Google Scholar] [CrossRef]
- Parajuli, B.; Horiuchi, H.; Mizuno, T.; Takeuchi, H.; Suzumura, A. CCL11 enhances excitotoxic neuronal death by producing reactive oxygen species in microglia. Glia 2015, 63, 2274–2284. [Google Scholar] [CrossRef]
- Boucsein, C.; Zacharias, R.; Färber, K.; Pavlovic, S.; Hanisch, U.K.; Kettenmann, H. Purinergic receptors on microglial cells: Functional expression in acute brain slices and modulation of microglial activation in vitro. Eur. J. Neurosci. 2003, 17, 2267–2276. [Google Scholar] [CrossRef] [PubMed]
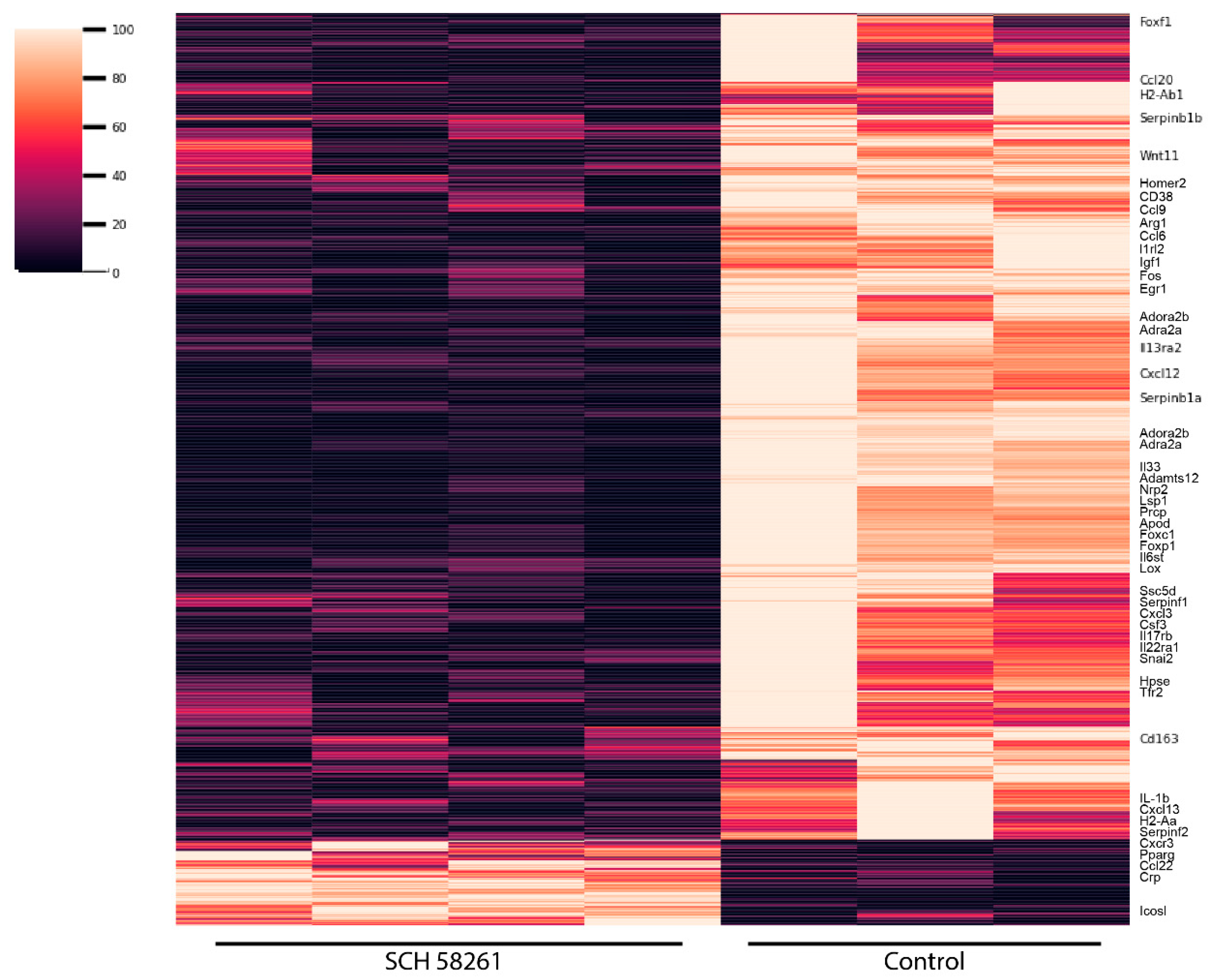
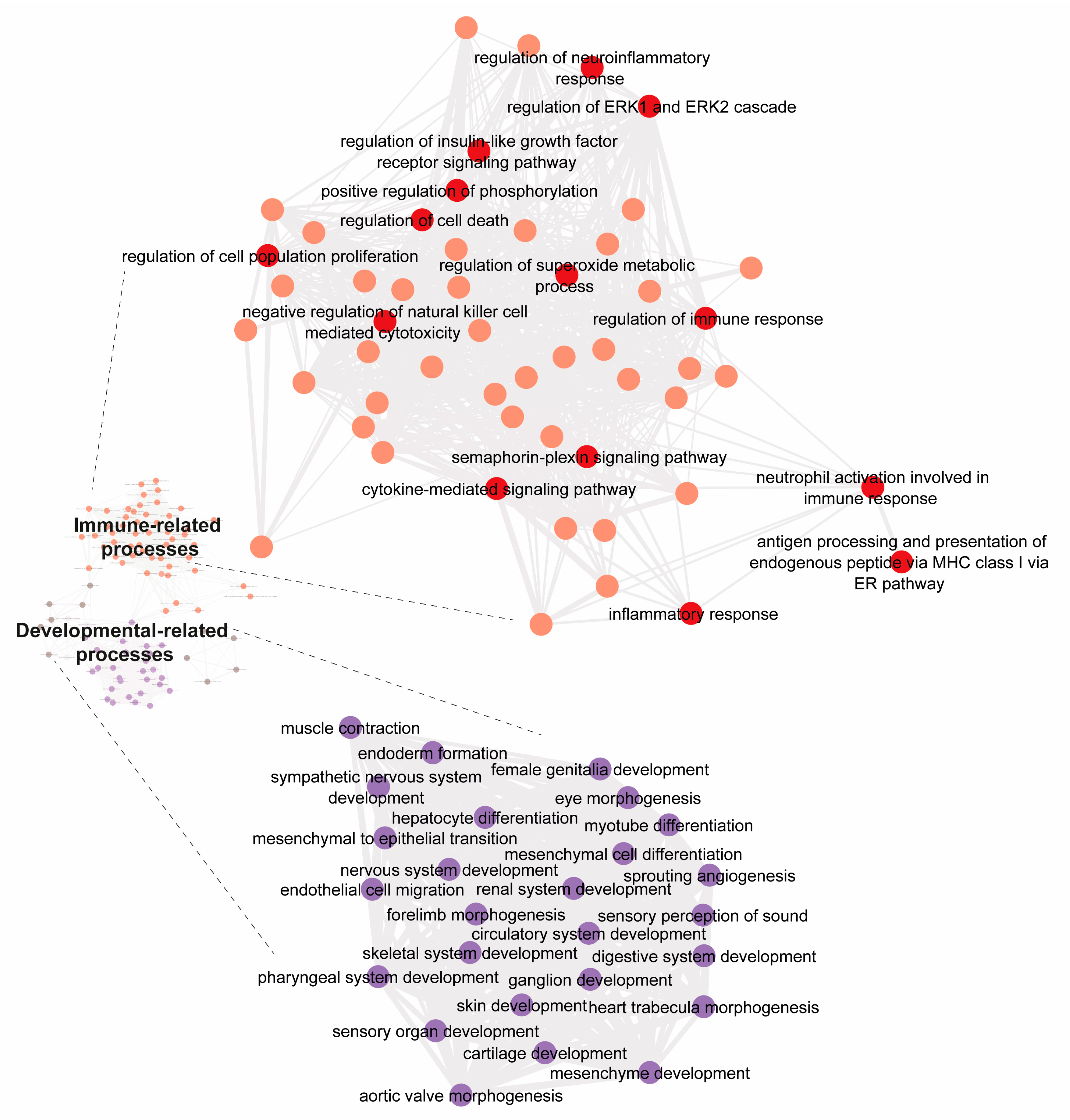
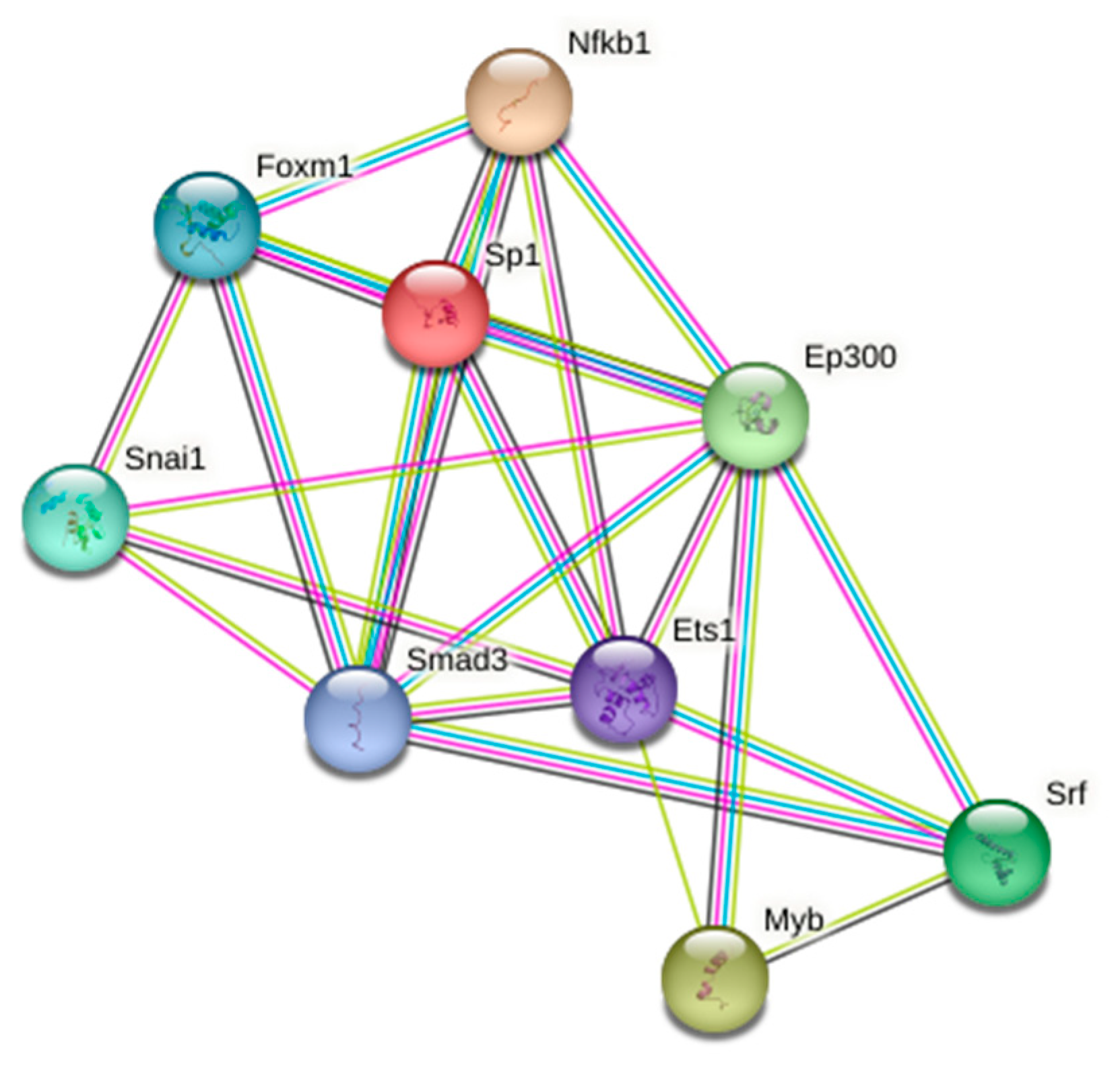
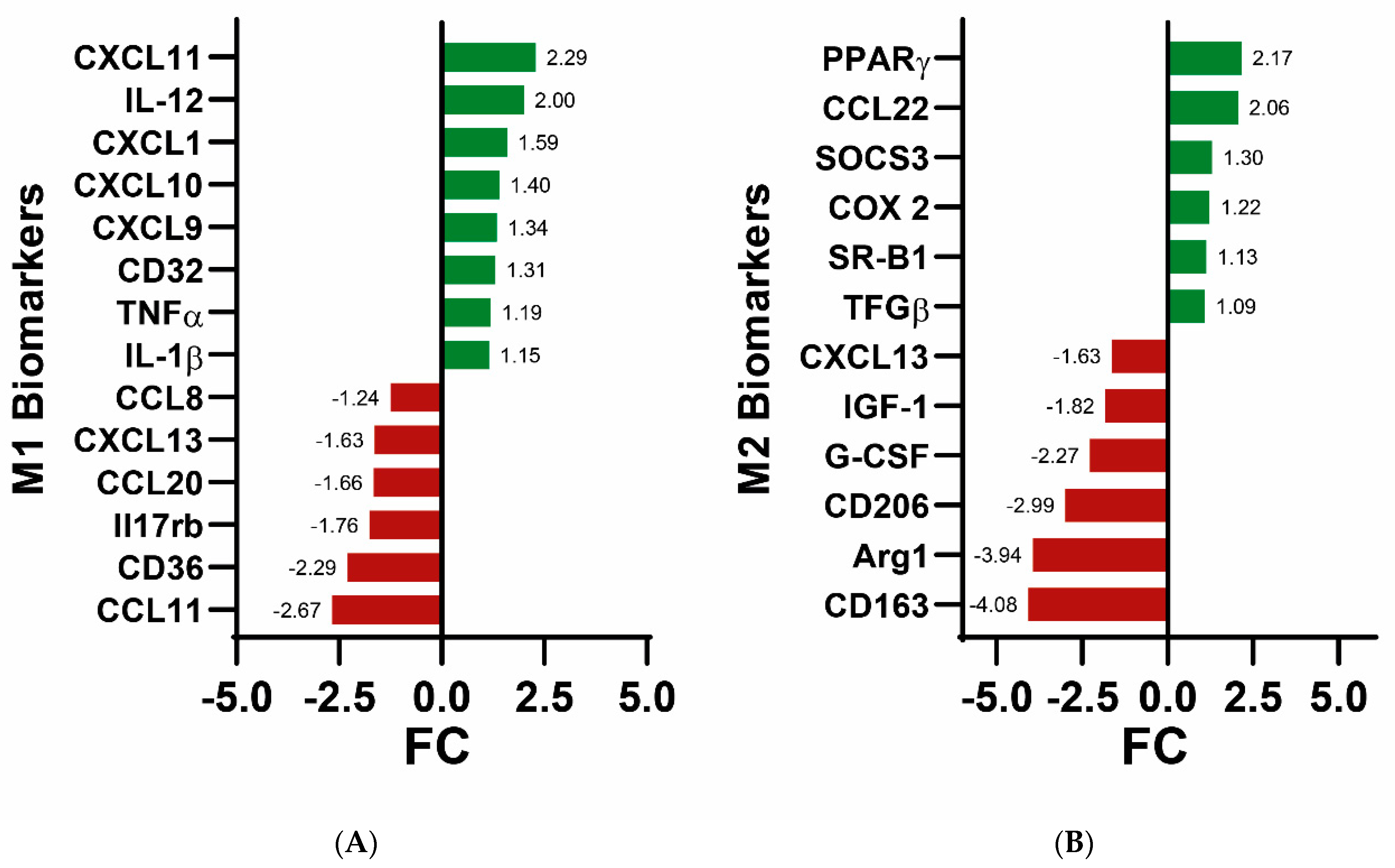

| Transcription Factor | Adjusted p-Value a | Odds Ratio | Combined Score b |
|---|---|---|---|
| SMAD3 | 4.67 × 10−3 | 4.7 | 51.69 |
| ETS1 | 1.32 × 10−2 | 3.65 | 32.03 |
| SNAI1 | 1.92 × 10−2 | 5.27 | 43.28 |
| SRF | 2.15 × 10−2 | 6.83 | 53.33 |
| SP1 | 2.15 × 10−2 | 1.93 | 15.34 |
| EP300 | 2.87 × 10−2 | 3.25 | 23.67 |
| NFKB1 | 4.06 × 10−2 | 2 | 13.68 |
| MYB | 4.86 × 10−2 | 6.38 | 42.01 |
| FOXM1 | 4.86 × 10−2 | 5.12 | 33.32 |
| Gene ID | Gene Name | FC a A2AR Antagonist | FC a A3R Agonist | FC a Dual Treatment | Ratio b |
|---|---|---|---|---|---|
| Upregulated (FC > 0) | |||||
| ENSMUSG00000116618 | AC122413.1 | 25.51 | 30.33 | 34.73 | 1.15 |
| ENSMUSG00000006638 | Abhd1 | 2.34 | 2.72 | 3.63 | 1.33 |
| ENSMUSG00000036964 | Trim17 | 1.92 | 1.97 | 2.31 | 1.17 |
| ENSMUSG00000041794 | Myrip | 1.71 | 1.88 | 2.13 | 1.13 |
| Downregulated (FC < 0) | |||||
| ENSMUSG00000040328 | Olfr56 | −12.43 | −17.88 | −34.54 | 1.93 |
| ENSMUSG00000024517 | Grp | −12.84 | −9.29 | −21.94 | 1.71 |
| ENSMUSG00000033082 | Clec1a | −5.98 | −6.70 | −12.03 | 1.79 |
| ENSMUSG00000037973 | Ccdc129 | −6.40 | −3.41 | −9.73 | 1.52 |
| ENSMUSG00000003477 | Inmt | −5.90 | −8.48 | −13.18 | 1.55 |
| ENSMUSG00000019326 | Aoc3 | −6.24 | −4.41 | −8.17 | 1.31 |
| ENSMUSG00000019932 | Kera | −5.75 | −3.34 | −7.61 | 1.32 |
| ENSMUSG00000043719 | Col6a6 | −4.55 | −4.78 | −6.38 | 1.33 |
| ENSMUSG00000033377 | Palmd | −3.18 | −2.63 | −4.34 | 1.37 |
| ENSMUSG00000088185 | Scarna2 | −2.96 | −3.86 | −6.88 | 1.78 |
| ENSMUSG00000026418 | Tnni1 | −2.94 | −3.32 | −5.52 | 1.66 |
| ENSMUSG00000047631 | Apof | −2.92 | −3.62 | −4.83 | 1.34 |
| ENSMUSG00000052854 | Nrk | −2.90 | −2.55 | −5.10 | 1.76 |
| ENSMUSG00000020676 | Ccl11 | −2.67 | −3.11 | −4.41 | 1.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lillo, A.; Serrano-Marín, J.; Lillo, J.; Raïch, I.; Navarro, G.; Franco, R. Differential Gene Expression in Activated Microglia Treated with Adenosine A2A Receptor Antagonists Highlights Olfactory Receptor 56 and T-Cell Activation GTPase-Activating Protein 1 as Potential Biomarkers of the Polarization of Activated Microglia. Cells 2023, 12, 2213. https://doi.org/10.3390/cells12182213
Lillo A, Serrano-Marín J, Lillo J, Raïch I, Navarro G, Franco R. Differential Gene Expression in Activated Microglia Treated with Adenosine A2A Receptor Antagonists Highlights Olfactory Receptor 56 and T-Cell Activation GTPase-Activating Protein 1 as Potential Biomarkers of the Polarization of Activated Microglia. Cells. 2023; 12(18):2213. https://doi.org/10.3390/cells12182213
Chicago/Turabian StyleLillo, Alejandro, Joan Serrano-Marín, Jaume Lillo, Iu Raïch, Gemma Navarro, and Rafael Franco. 2023. "Differential Gene Expression in Activated Microglia Treated with Adenosine A2A Receptor Antagonists Highlights Olfactory Receptor 56 and T-Cell Activation GTPase-Activating Protein 1 as Potential Biomarkers of the Polarization of Activated Microglia" Cells 12, no. 18: 2213. https://doi.org/10.3390/cells12182213
APA StyleLillo, A., Serrano-Marín, J., Lillo, J., Raïch, I., Navarro, G., & Franco, R. (2023). Differential Gene Expression in Activated Microglia Treated with Adenosine A2A Receptor Antagonists Highlights Olfactory Receptor 56 and T-Cell Activation GTPase-Activating Protein 1 as Potential Biomarkers of the Polarization of Activated Microglia. Cells, 12(18), 2213. https://doi.org/10.3390/cells12182213







