Investigation of Tumor Heterogeneity Using Integrated Single-Cell RNA Sequence Analysis to Focus on Genes Related to Breast Cancer-, EMT-, CSC-, and Metastasis-Related Markers in Patients with HER2-Positive Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
- Luminal subtype: Cases with either ER or PgR positivity on IHC staining. ER and PgR status were considered positive using a cut off value of ≥ 1% according to the American Society of Clinical Oncology/College American Pathologists (ASCO/CAP) guideline [9]. We considered either 2+ or 3+ as positive in datasets which were described as 0+, 1+, 2+, or 3+.
- HER2 subtype: Cases with HER2 positivity. We considered 3+ or 2+ with HER2 amplification on IHC staining as positive with reference to the ASCO/CAP guideline [10]. The results of HER2 amplification is necessary in cases with unavailable HER2 results on IHC score.
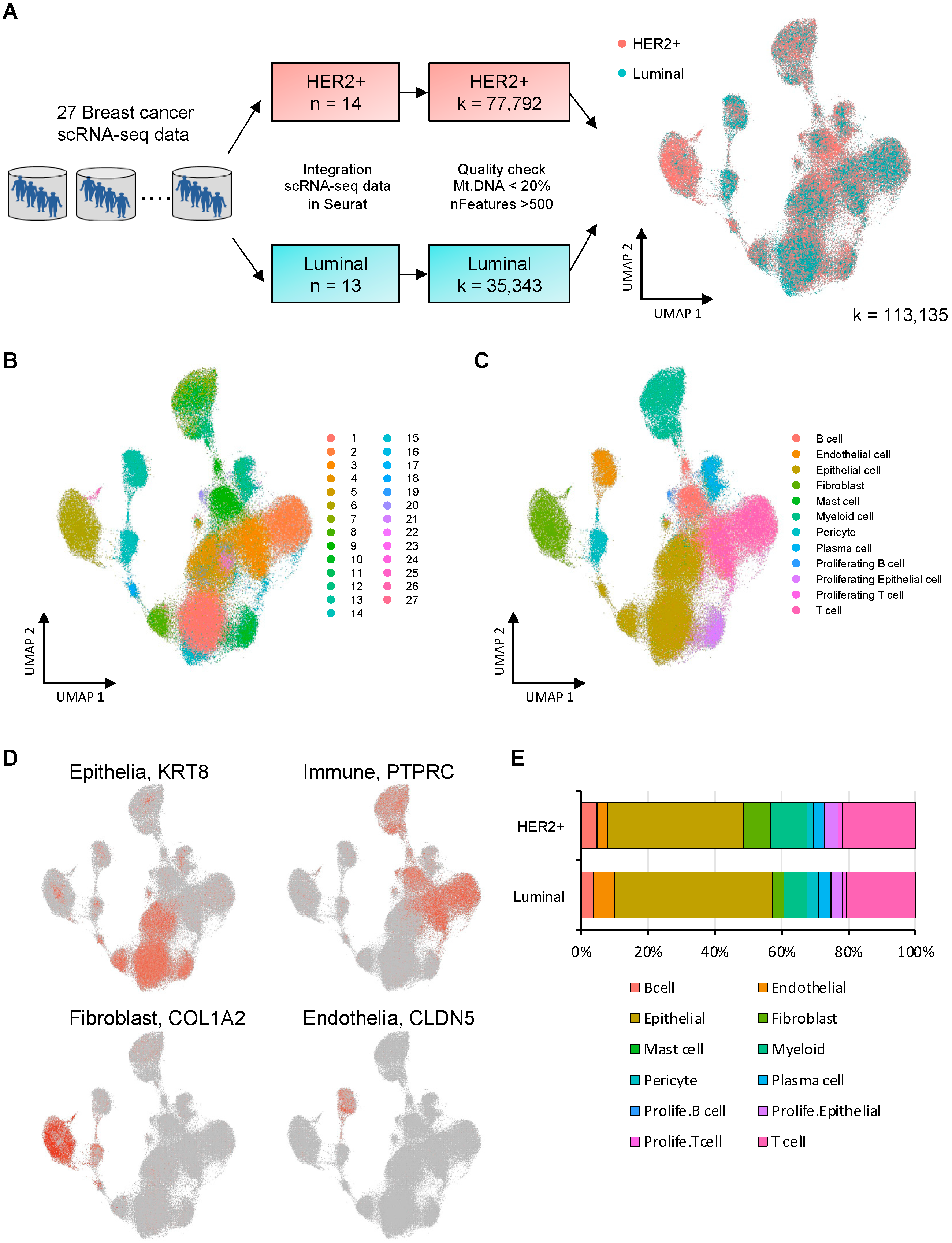
2.2. The Integration of Datasets, Data Quality Control, and the Removal of Batch Effects
2.3. Data Clustering and Cell Type Annotation
2.4. Pathway Enrichment Analysis
2.5. Data Visualization
3. Results
3.1. Integrated Single-Cell RNA-seq Data for 14 HER2 Subtypes and 13 Luminal Subtypes
3.2. The Intra-Tumor Heterogeneity of the Epithelial Population in the HER2 and Luminal Subtype
3.3. The Expression Levels of Typical Breast Cancer-, CSC-, EMT-, and Metastasis-Related Markers across Patients
3.4. The Expression Levels of Typical Breast Cancer-, CSC-, EMT-, and Metastasis-Related Markers across the Epithelial Clusters
3.5. Comparison of Gene Expression between the ERBB2-High Group and ERBB2-Low Group
3.6. Comparison of the Gene Expression Levels of Typical Breast-Cancer-Related Markers on Each Subtype at Single-Cell Levels: Luminal-HER2 Subtype, Pure-HER2 Subtype, and Luminal Subtype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, X.; Yang, H.; Yu, X.; Qin, J.-J. Drug-resistant HER2-positive breast cancer: Molecular mechanisms and overcoming strategies. Front. Pharmacol. 2022, 13, 1012552. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, E.; Shastry, M.; Shiller, S.M.; Ren, R. Targeting HER2 heterogeneity in breast cancer. Cancer Treat. Rev. 2021, 100, 102286. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Nitta, H.; Wei, L.; Banks, P.M.; Portier, B.; Parwani, A.V.; Li, Z. HER2 intratumoral heterogeneity is independently associated with incomplete response to anti-HER2 neoadjuvant chemotherapy in HER2-positive breast carcinoma. Breast Cancer Res. Treat. 2017, 166, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Filho, O.M.; Viale, G.; Stein, S.; Trippa, L.; Yardley, D.A.; Mayer, I.A.; Abramson, V.G.; Arteaga, C.L.; Spring, L.M.; Waks, A.G.; et al. Impact of HER2 Heterogeneity on Treatment Response of Early-Stage HER2-Positive Breast Cancer: Phase II Neoadjuvant Clinical Trial of T-DM1 Combined with Pertuzumab. Cancer Discov. 2021, 11, 2474–2487. [Google Scholar] [CrossRef] [PubMed]
- Muus, C.; Luecken, M.D.; Eraslan, G.; Sikkema, L.; Waghray, A.; Heimberg, G.; Kobayashi, Y.; Vaishnav, E.D.; Subrama-nian, A.; Smillie, C.; et al. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat. Med. 2021, 27, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Y.; Wang, C.; Yang, Y.; Ni, Y.; Wang, Z.; Song, T.; Yao, M.; Liu, Z.; Chao, N.; et al. Integrated single-cell RNA sequencing analysis reveals distinct cellular and transcriptional modules associated with survival in lung cancer. Signal Transduct. Target. Ther. 2022, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Z.-X.; Chen, Y.-X.; Wu, H.-X.; Yin, L.; Zhao, Q.; Luo, H.-Y.; Zeng, Z.-L.; Qiu, M.-Z.; Xu, R.-H. Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response. Genome Med. 2022, 14, 45. [Google Scholar] [CrossRef]
- Pegram, M.; Jackisch, C.; Johnston, S.R.D. Estrogen/HER2 receptor crosstalk in breast cancer: Combination therapies to improve outcomes for patients with hormone receptor-positive/HER2-positive breast cancer. NPJ Breast Cancer 2023, 9, 45. [Google Scholar] [CrossRef]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and progesterone receptor testing in breast cancer: AS-CO/CAP guideline update. J. Clin. Oncol. 2020, 38, 1346–1366. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef]
- Pal, B.; Chen, Y.; Vaillant, F.; Capaldo, B.D.; Joyce, R.; Song, X.; Bryant, V.L.; Penington, J.S.; Di Stefano, L.; Tubau Ribera, N.; et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 2021, 40, e107333. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wang, R.; Xie, H.; Hu, L.; Wang, C.; Xu, J.; Zhu, C.; Liu, Y.; Gao, F.; Li, X.; et al. Single-cell RNA sequencing reveals cell heterogeneity and transcriptome profile of breast cancer lymph node metastasis. Oncogenesis 2021, 10, 66. [Google Scholar] [CrossRef] [PubMed]
- Tokura, M.; Nakayama, J.; Prieto-Vila, M.; Shiino, S.; Yoshida, M.; Yamamoto, T.; Watanabe, N.; Takayama, S.; Suzuki, Y.; Okamoto, K.; et al. Single-Cell Transcriptome Profiling Reveals Intratumoral Heterogeneity and Molecular Features of Ductal Carcinoma In Situ. Cancer Res. 2022, 82, 3236–3248. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e1821. [Google Scholar] [CrossRef] [PubMed]
- Xi, N.M.; Li, J.J. Benchmarking Computational Doublet-Detection Methods for Single-Cell RNA Sequencing Data. Cell Syst. 2020, 12, 176–194.e6. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e4. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Tran, H.T.N.; Ang, K.S.; Chevrier, M.; Zhang, X.; Lee, N.Y.S.; Goh, M.; Chen, J. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020, 21, 12. [Google Scholar] [CrossRef]
- Kumar, T.; Nee, K.; Wei, R.; He, S.; Nguyen, Q.H.; Bai, S.; Blake, K.; Pein, M.; Gong, Y.; Sei, E.; et al. A spatially resolved single-cell genomic atlas of the adult human breast. Nature 2023, 620, 181–191. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.A.; Bhakta, N.R.; Kessenbrock, K.; Prummel, K.D.; Yu, Y.; Takai, K.; Zhou, A.; Eyob, H.; Balakrishnan, S.; Wang, C.-Y.; et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Dave, B.; Mittal, V.; Tan, N.M.; Chang, J.C. Epithelial-mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res. 2012, 14, 202. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, S.K.; Cabrera, R.; Mao, S.P.; Christin, J.R.; Wu, B.; Guo, W.; Bravo-Cordero, J.J.; Condeelis, J.S.; Segall, J.E.; Hodgson, L. Rac3 regulates breast cancer invasion and metastasis by controlling adhesion and matrix degradation. J. Cell Biol. 2017, 216, 4331–4349. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Lu, Y.B.; He, B.F.; Xie, T.; Yan, C.L.; Liu, T.F.; Wu, S.L.; Yeh, Y.Y.; Li, Z.Y.; Huang, W.; et al. Rab32 promotes glioblastoma mi-gration and invasion via regulation of ERK/Drp1-mediated mitochondrial fission. Cell Death Dis. 2023, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Drasin, D.J.; Robin, T.P.; Ford, H.L. Breast cancer epithelial-to-mesenchymal transition: Examining the functional consequences of plasticity. Breast Cancer Res. 2011, 13, 226. [Google Scholar] [CrossRef]
- Ingthorsson, S.; Andersen, K.; Hilmarsdottir, B.; Maelandsmo, G.M.; Magnusson, M.K.; Gudjonsson, T. HER2 induced EMT and tumorigenicity in breast epithelial progenitor cells is inhibited by coexpression of EGFR. Oncogene 2015, 35, 4244–4255. [Google Scholar] [CrossRef]
- Farabaugh, S.M.; Boone, D.N.; Lee, A.V. Role of IGF1R in Breast Cancer Subtypes, Stemness, and Lineage Differentiation. Front. Endocrinol. 2015, 6, 59. [Google Scholar] [CrossRef]
- Schettini, F.; Pascual, T.; Conte, B.; Chic, N.; Brasó-Maristany, F.; Galván, P.; Martínez, O.; Adamo, B.; Vidal, M.; Muñoz, M.; et al. HER2-enriched subtype and pathological complete response in HER2-positive breast cancer: A systematic review and meta-analysis. Cancer Treat. Rev. 2020, 84, 101965. [Google Scholar] [CrossRef]
- Carey, L.A.; Berry, D.A.; Cirrincione, C.T.; Barry, W.T.; Pitcher, B.N.; Harris, L.N.; Ollila, D.W.; Krop, I.E.; Henry, N.L.; Weckstein, D.J.; et al. Molecular Heterogeneity and Response to Neoadjuvant Human Epidermal Growth Factor Receptor 2 Targeting in CALGB 40601, a Randomized Phase III Trial of Paclitaxel Plus Trastuzumab With or Without Lapatinib. J. Clin. Oncol. 2016, 34, 542–549. [Google Scholar] [CrossRef]
- Nuciforo, P.; Pascual, T.; Cortés, J.; Llombart-Cussac, A.; Fasani, R.; Paré, L.; Oliveira, M.; Galvan, P.; Martínez, N.; Bermejo, B.; et al. A predictive model of pathologic response based on tumor cellularity and tumor-infiltrating lymphocytes (CelTIL) in HER2-positive breast cancer treated with chemo-free dual HER2 blockade. Ann. Oncol. 2017, 29, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Campbell, C.; Savas, P.; Nuciforo, P.; Aura, C.; de Azambuja, E.; Eidtmann, H.; Ellis, C.E.; Baselga, J.; et al. Tumor-Infiltrating Lymphocytes and Associations With Pathological Complete Response and Event-Free Survival in HER2-Positive Early-Stage Breast Cancer Treated With Lapatinib and Trastuzumab: A Secondary Analysis of the NeoALTTO Trial. JAMA Oncol. 2015, 1, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, Y.; Olopade, O.I. MYC and Breast Cancer. Genes Cancer 2010, 1, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Risom, T.; Wang, X.; Liang, J.; Zhang, X.; Pelz, C.; Campbell, L.G.; Eng, J.; Chin, K.; Farrington, C.; Narla, G.; et al. Dereg-ulating MYC in a model of HER2+ breast cancer mimics human intertumoral heterogeneity. J. Clin. Investig. 2020, 130, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, P.; Hamilton, E.; Tolaney, S.M.; Cortes, J.; Morganti, S.; Ferraro, E.; Marra, A.; Viale, G.; Trapani, D.; Cardoso, F.; et al. HER2-Low Breast Cancer: Pathological and Clinical Landscape. J. Clin. Oncol. 2020, 38, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Jacot, W.; Yamashita, T.; Sohn, J.; Vidal, M.; Tokunaga, E.; Tsurutani, J.; Ueno, N.T.; Prat, A.; Chae, Y.S.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N. Engl. J. Med. 2022, 387, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Schettini, F.; Chic, N.; Brasó-Maristany, F.; Paré, L.; Pascual, T.; Conte, B.; Martínez-Sáez, O.; Adamo, B.; Vidal, M.; Barnadas, E.; et al. Clinical, pathological, and PAM50 gene expression features of HER2-low breast cancer. NPJ Breast Cancer 2021, 7, 1. [Google Scholar] [CrossRef]
- Ke, M.; Elshenawy, B.; Sheldon, H.; Arora, A.; Buffa, F.M. Single cell RNA-sequencing: A powerful yet still challenging technology to study cellular heterogeneity. BioEssays 2022, 44, e2200084. [Google Scholar] [CrossRef]
- Schiffer, M. Going single but not solo with podocytes: Potentials, limitations, and pitfalls of single-cell analysis. Kidney Int. 2017, 92, 1038–1041. [Google Scholar] [CrossRef]
- Fogazzi, V.; Kapahnke, M.; Cataldo, A.; Plantamura, I.; Tagliabue, E.; Di Cosimo, S.; Cosentino, G.; Iorio, M.V. The Role of MicroRNAs in HER2-Positive Breast Cancer: Where We Are and Future Prospective. Cancers 2022, 14, 5326. [Google Scholar] [CrossRef]
- Wu, X. Expressions of miR-21 and miR-210 in Breast Cancer and Their Predictive Values for Prognosis. Iran. J. Public Health 2020, 49, 21–29. [Google Scholar] [CrossRef]
- Yang, F.; Li, Y.; Xu, L.; Zhu, Y.; Gao, H.; Zhen, L.; Fang, L. miR-17 as a diagnostic biomarker regulates cell proliferation in breast cancer. OncoTargets Ther. 2017, ume 10, 543–550. [Google Scholar] [CrossRef]
- Peng, J.; Yuan, C.; Wu, Z.; Wang, Y.; Yin, W.; Lin, Y.; Zhou, L.; Lu, J. Upregulation of microRNA-1 inhibits proliferation and metastasis of breast cancer. Mol. Med. Rep. 2020, 22, 454–464. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, L.; Wu, Z.; Wang, M.; Jin, F.; Wang, N.; Hu, X.; Liu, Z.; Zhang, C.-Y.; Zen, K.; et al. miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer 2016, 16, 826. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, W.; Tang, T.; Wang, Y.; Yin, X.; Chen, Y.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. miR-29a contributes to breast cancer cells epithelial–mesenchymal transition, migration, and invasion via down-regulating histone H4K20 trimethylation through directly targeting SUV420H2. Cell Death Dis. 2019, 10, 176. [Google Scholar] [CrossRef]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self re-newal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef]
- Thammaiah, C.K.; Jayaram, S. Role of let-7 family microRNA in breast cancer. Non-Coding RNA Res. 2016, 1, 77–82. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiino, S.; Tokura, M.; Nakayama, J.; Yoshida, M.; Suto, A.; Yamamoto, Y. Investigation of Tumor Heterogeneity Using Integrated Single-Cell RNA Sequence Analysis to Focus on Genes Related to Breast Cancer-, EMT-, CSC-, and Metastasis-Related Markers in Patients with HER2-Positive Breast Cancer. Cells 2023, 12, 2286. https://doi.org/10.3390/cells12182286
Shiino S, Tokura M, Nakayama J, Yoshida M, Suto A, Yamamoto Y. Investigation of Tumor Heterogeneity Using Integrated Single-Cell RNA Sequence Analysis to Focus on Genes Related to Breast Cancer-, EMT-, CSC-, and Metastasis-Related Markers in Patients with HER2-Positive Breast Cancer. Cells. 2023; 12(18):2286. https://doi.org/10.3390/cells12182286
Chicago/Turabian StyleShiino, Sho, Momoko Tokura, Jun Nakayama, Masayuki Yoshida, Akihiko Suto, and Yusuke Yamamoto. 2023. "Investigation of Tumor Heterogeneity Using Integrated Single-Cell RNA Sequence Analysis to Focus on Genes Related to Breast Cancer-, EMT-, CSC-, and Metastasis-Related Markers in Patients with HER2-Positive Breast Cancer" Cells 12, no. 18: 2286. https://doi.org/10.3390/cells12182286
APA StyleShiino, S., Tokura, M., Nakayama, J., Yoshida, M., Suto, A., & Yamamoto, Y. (2023). Investigation of Tumor Heterogeneity Using Integrated Single-Cell RNA Sequence Analysis to Focus on Genes Related to Breast Cancer-, EMT-, CSC-, and Metastasis-Related Markers in Patients with HER2-Positive Breast Cancer. Cells, 12(18), 2286. https://doi.org/10.3390/cells12182286





