Sedentary Behavior Impacts on the Epigenome and Transcriptome: Lessons from Muscle Inactivation in Drosophila Larvae
Abstract
:1. Introduction
2. Methods
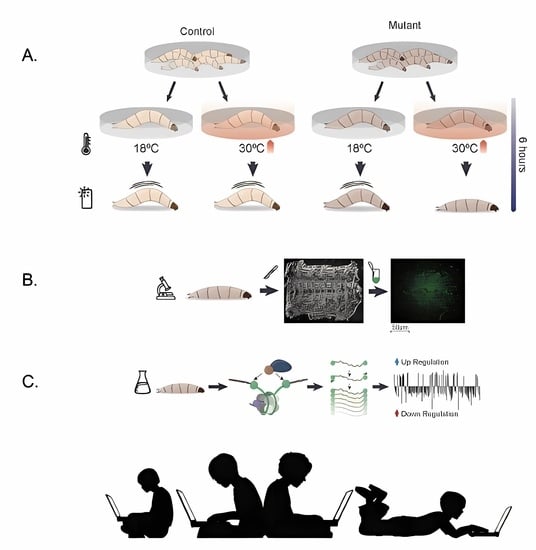
2.1. Fly Stocks and Husbandry
2.2. Induction of Larvae Immobilization
2.3. Larval Locomotion Assay
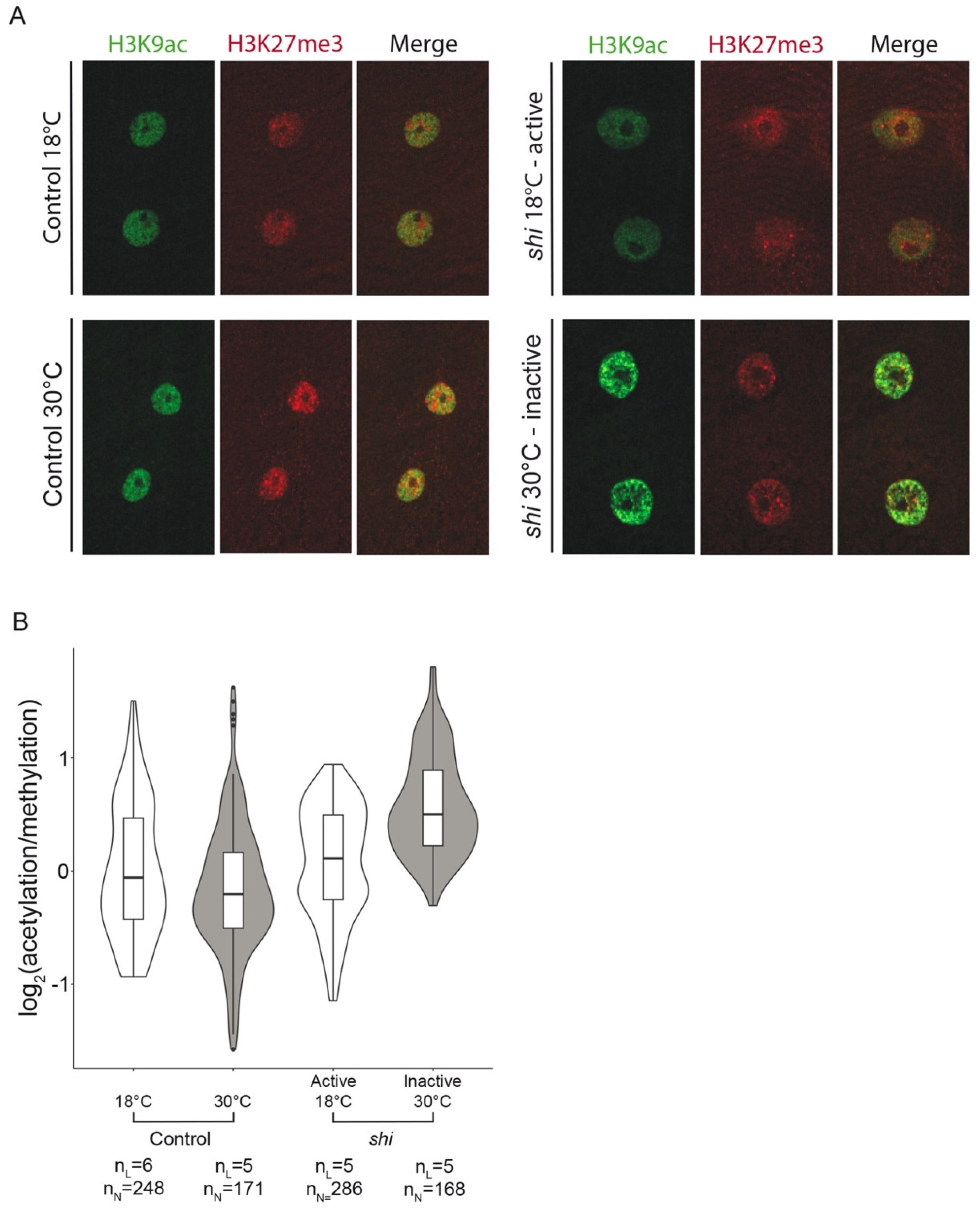
2.4. Antibody Labeling of Myonuclear Epigenetic Marks in Larvae
2.5. Microscopy, Image Acquisition, and Analysis
2.6. Targeted DamID
2.7. Statistical Analysis
3. Results
3.1. Inhibition of Larval Muscle Contractions in Shibire Mutants
3.2. Epigenetic Landscape Analysis in Myonuclei
3.3. Altered Genome Pol II Binding Profile to DNA in the Inactive Muscles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tremblay, M.S.; Aubert, S.; Barnes, J.D.; Saunders, T.J.; Carson, V.; Latimer-Cheung, A.E.; Chastin, S.F.M.; Altenburg, T.M.; Chinapaw, M.J.M.; on behalf of SBRN Terminology Consensus Project Participants. Sedentary Behavior Research Network (SBRN)—Terminology Consensus Project process and outcome. Int. J. Behav. Nutr. Phys. Act. 2017, 14, 75. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.B. Sedentary lifestyle and risk of obesity and type 2 diabetes. Lipids 2003, 38, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Hooker, S.P.; Diaz, K.M.; Blair, S.N.; Colabianchi, N.; Hutto, B.; McDonnell, M.N.; Vena, J.E.; Howard, V.J. Association of Accelerometer-Measured Sedentary Time and Physical Activity with Risk of Stroke among US Adults. JAMA Netw. Open 2022, 5, e2215385. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Chen, P.J.; Xiao, W.H. Mechanism of increased risk of insulin resistance in aging skeletal muscle. Diabetol. Metab. Syndr. 2020, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Brener, A.; Lebenthal, Y.; Shtamler, A.; Levy, S.; Stein, R.; Fattal-Valevski, A.; Sagi, L. The endocrine manifestations of spinal muscular atrophy, a real-life observational study. Neuromuscul. Disord. 2020, 30, 270–276. [Google Scholar] [CrossRef]
- Hamburg, N.M.; McMackin, C.J.; Huang, A.L.; Shenouda, S.M.; Widlansky, M.E.; Schulz, E.; Gokce, N.; Ruderman, N.B.; Keaney, J.F.; Vita, J.A.; et al. Physical inactivity rapidly induces insulin resistance and microvascular dysfunction in healthy volunteers. Arter. Thromb. Vasc. Biol. 2007, 27, 2650–2656. [Google Scholar] [CrossRef]
- Plaza-Diaz, J.; Izquierdo, D.; Torres-Martos, Á.; Baig, A.T.; Aguilera, C.M.; Ruiz-Ojeda, F.J. Impact of Physical Activity and Exercise on the Epigenome in Skeletal Muscle and Effects on Systemic Metabolism. Biomedicines 2022, 10, 126. [Google Scholar] [CrossRef]
- Ding, M.; Li, H.; Zheng, L. Drosophila exercise, an emerging model bridging the fields of exercise and aging in human. Front. Cell Dev. Biol. 2022, 10, 966531. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Gomes, C.P.C.; Almeida, J.A.; Franco, O.L.; Petriz, B. Omics and the Molecular Exercise Physiology, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; Volume 96. [Google Scholar]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Zagkos, L.; Auley, M.M.; Roberts, J.; Kavallaris, N.I. Mathematical models of DNA methylation dynamics: Implications for health and ageing. J. Theor. Biol. 2019, 462, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Gensous, N.; Bacalini, M.G.; Franceschi, C.; Meskers, C.G.M.; Maier, A.B.; Garagnani, P. Age-related DNA methylation changes: Potential impact on skeletal muscle aging in humans. Front. Physiol. 2019, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.J.; Kim, K. New insights into the role of histone changes in aging. Int. J. Mol. Sci. 2020, 21, 8241. [Google Scholar] [CrossRef] [PubMed]
- Sharples, A.P.; Stewart, C.E.; Seaborne, R.A. Does skeletal muscle have an ‘epi’-memory? The role of epigenetics in nutritional programming, metabolic disease, aging and exercise. Aging Cell 2016, 15, 603–616. [Google Scholar] [CrossRef]
- Di Liegro, I. Genetic and epigenetic modulation of cell functions by physical exercise. Genes 2019, 10, 1043. [Google Scholar] [CrossRef]
- Jacques, M.; Hiam, D.; Craig, J.; Barrès, R.; Eynon, N.; Voisin, S. Epigenetic changes in healthy human skeletal muscle following exercise– a systematic review. Epigenetics 2019, 14, 633–648. [Google Scholar] [CrossRef]
- Raffin, J.; de Souto Barreto, P.; Le Traon, A.P.; Vellas, B.; Aubertin-Leheudre, M.; Rolland, Y. Sedentary behavior and the biological hallmarks of aging. Ageing Res. Rev. 2023, 83, 101807. [Google Scholar] [CrossRef]
- Sujkowski, A.; Wessells, R. Using Drosophila to Understand Biochemical and Behavioral Responses to Exercise. Exerc. Sport Sci. Rev. 2018, 46, 112–120. [Google Scholar] [CrossRef]
- Riddle, N.C. Drosophila melanogaster, a new model for exercise research. Acta Physiol. 2019, 227, e13352. [Google Scholar] [CrossRef]
- de Goede, O.M.; Nachun, D.C.; Ferraro, N.M.; Gloudemans, M.J.; Rao, A.S.; Smail, C.; Eulalio, T.Y.; Aguet, F.; Ng, B.; Xu, J.; et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 2021, 184, 2633–2648.e19. [Google Scholar] [CrossRef]
- Marshall, O.J.; Brand, A.H. Chromatin state changes during neural development revealed by in vivo cell-type specific profiling. Nat. Commun. 2017, 8, 2271. [Google Scholar] [CrossRef]
- Wang, S.; Reuveny, A.; Volk, T. Nesprin provides elastic properties to muscle nuclei by cooperating with spectraplakin and EB1. J. Cell Biol. 2015, 209, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Marshall, O.J.; Brand, A.H. Damidseq-pipeline: An automated pipeline for processing DamID sequencing datasets. Bioinformatics 2015, 31, 3371–3373. [Google Scholar] [CrossRef] [PubMed]
- Southall, T.D.; Gold, K.S.; Egger, B.; Davidson, C.M.; Caygill, E.E.; Marshall, O.J.; Brand, A.H. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: Assaying RNA pol II occupancy in neural stem cells. Dev. Cell 2013, 26, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Ozawa, S.; Hagiwara, S. Synaptic transmission reversibly conditioned by single-gene mutation in Drosophila melanogaster. Nature 1976, 259, 489–491. [Google Scholar] [CrossRef]
- Siddiqi, O.; Benzer, S. Neurophysiological defects in temperature sensitive paralytic mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1976, 73, 3253–3257. [Google Scholar] [CrossRef]
- Krans, J.L.; Rivlin, P.K.; Hoy, R.R. Demonstrating the temperature sensitivity of synaptic transmission in a Drosophila mutant. J. Undergrad. Neurosci. Educ. 2005, 4, 27–33. [Google Scholar]
- Booth, L.N.; Brunet, A. The Aging Epigenome. Mol. Cell 2016, 62, 728–744. [Google Scholar] [CrossRef]
- Van Meter, M.; Kashyap, M.; Rezazadeh, S.; Geneva, A.J.; Morello, T.D.; Seluanov, A.; Gorbunova, V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014, 5, 5011. [Google Scholar] [CrossRef]
- Gao, W.; Liu, J.L.; Lu, X.; Yang, Q. Epigenetic regulation of energy metabolism in obesity. J. Mol. Cell Biol. 2021, 13, 480–499. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Wysocka, J. Deciphering the multi-scale, quantitative cis-regulatory code. Mol. Cell 2023, 83, 373–392. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.K.; Chan, T.F.; Bendena, W.; Hui, J.H.L. Noncoding RNA Regulation of Hormonal and Metabolic Systems in the Fruit Fly Drosophila. Metabolites 2023, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, T.; Ulfhake, B. Sarcopenia: What Is the Origin of This Aging-Induced Disorder? Front. Genet. 2021, 12, 688526. [Google Scholar] [CrossRef]
- Chen, S.; Yan, S.; Aiheti, N.; Kuribanjiang, K.; Yao, X.; Wang, Q.; Zhou, T.; Yang, L. A bi-directional Mendelian randomization study of sarcopenia-related traits and type 2 diabetes mellitus. Front. Endocrinol. 2023, 14, 1109800. [Google Scholar] [CrossRef]
- Mul, J.D.; Stanford, K.I.; Hirshman, M.F.; Goodyear, L.J. Exercise and Regulation of Carbohydrate Metabolism. Prog. Mol. Biol. Transl. Sci. 2015, 135, 17–37. [Google Scholar] [CrossRef]
- Wanders, L.; Gijbels, A.; Bakker, E.A.; Trouwborst, I.; Jardon, K.M.; Manusama, K.C.M.; Hul, G.B.; Feskens, E.J.M.; Afman, L.A.; Blaak, E.E.; et al. Physical activity and sedentary behavior show distinct associations with tissue-specific insulin sensitivity in adults with overweight. Acta Physiol. 2023, 237, e13945. [Google Scholar] [CrossRef]
- Eijkelenboom, A.; Burgering, B.M.T. FOXOs: Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013, 14, 83–97. [Google Scholar] [CrossRef]
- Demontis, F.; Perrimon, N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 2010, 143, 813–825. [Google Scholar] [CrossRef]
- Galves, M.; Rathi, R.; Prag, G.; Ashkenazi, A. Ubiquitin Signaling and Degradation of Aggregate-Prone Proteins. Trends Biochem. Sci. 2019, 44, 872–884. [Google Scholar] [CrossRef]
- Kurtishi, A.; Rosen, B.; Patil, K.S.; Alves, G.W.; Møller, S.G. Cellular Proteostasis in Neurodegeneration. Mol. Neurobiol. 2019, 56, 3676–3689. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, J.; Gaur, M.; Swaroop, A.; Taylor, A. Proteostasis in aging-associated ocular disease. Mol. Aspects Med. 2022, 88, 101157. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H. Ubiquitin-Specific Proteases (USPs) and Metabolic Disorders. Int. J. Mol. Sci. 2023, 24, 3219. [Google Scholar] [CrossRef]
- Li, X.; Zhou, B.; Chen, L.; Gou, L.T.; Li, H.; Fu, X.D. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 2017, 35, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Han, J.D.J. Aging-associated lncRNAs are evolutionarily conserved and participate in NFκB signaling. Nat. Aging 2021, 1, 438–453. [Google Scholar] [CrossRef]
- Ugur, B.; Chen, K.; Bellen, H.J. Drosophila tools and assays for the study of human diseases. DMM Dis. Model. Mech. 2016, 9, 235–244. [Google Scholar] [CrossRef]



| Type of Gene | Analyzed Genes | Genes Affected by
Muscle Inactivity |
|---|---|---|
| Total, n | 2010 | 121 |
| Protein coding | 1734 (86.3) | 72 (59.5) |
| lncRNA | 201 (10) | 36 (29.8) |
| antisense lncRNA | 49 (2.4) | 6 (5) |
| snoRNA | 8 (0.4) | 1 (0.8) |
| miRNA | 6 (0.3) | 3 (2.5) |
| snRNA | 7 (0.3) | 1 (0.8) |
| Pseudogene | 3 (0.2) | 1 (0.8) |
| tRNA | 2 (0.1) | 1 (0.8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brener, A.; Lorber, D.; Reuveny, A.; Toledano, H.; Porat-Kuperstein, L.; Lebenthal, Y.; Weizman, E.; Olender, T.; Volk, T. Sedentary Behavior Impacts on the Epigenome and Transcriptome: Lessons from Muscle Inactivation in Drosophila Larvae. Cells 2023, 12, 2333. https://doi.org/10.3390/cells12192333
Brener A, Lorber D, Reuveny A, Toledano H, Porat-Kuperstein L, Lebenthal Y, Weizman E, Olender T, Volk T. Sedentary Behavior Impacts on the Epigenome and Transcriptome: Lessons from Muscle Inactivation in Drosophila Larvae. Cells. 2023; 12(19):2333. https://doi.org/10.3390/cells12192333
Chicago/Turabian StyleBrener, Avivit, Dana Lorber, Adriana Reuveny, Hila Toledano, Lilach Porat-Kuperstein, Yael Lebenthal, Eviatar Weizman, Tsviya Olender, and Talila Volk. 2023. "Sedentary Behavior Impacts on the Epigenome and Transcriptome: Lessons from Muscle Inactivation in Drosophila Larvae" Cells 12, no. 19: 2333. https://doi.org/10.3390/cells12192333
APA StyleBrener, A., Lorber, D., Reuveny, A., Toledano, H., Porat-Kuperstein, L., Lebenthal, Y., Weizman, E., Olender, T., & Volk, T. (2023). Sedentary Behavior Impacts on the Epigenome and Transcriptome: Lessons from Muscle Inactivation in Drosophila Larvae. Cells, 12(19), 2333. https://doi.org/10.3390/cells12192333