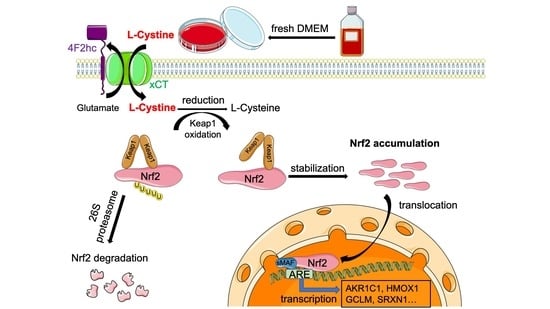
Fresh Medium or L-Cystine as an Effective Nrf2 Inducer for Cytoprotection in Cell Culture
Abstract
:1. Introduction
2. Materials and Methods
3. Results
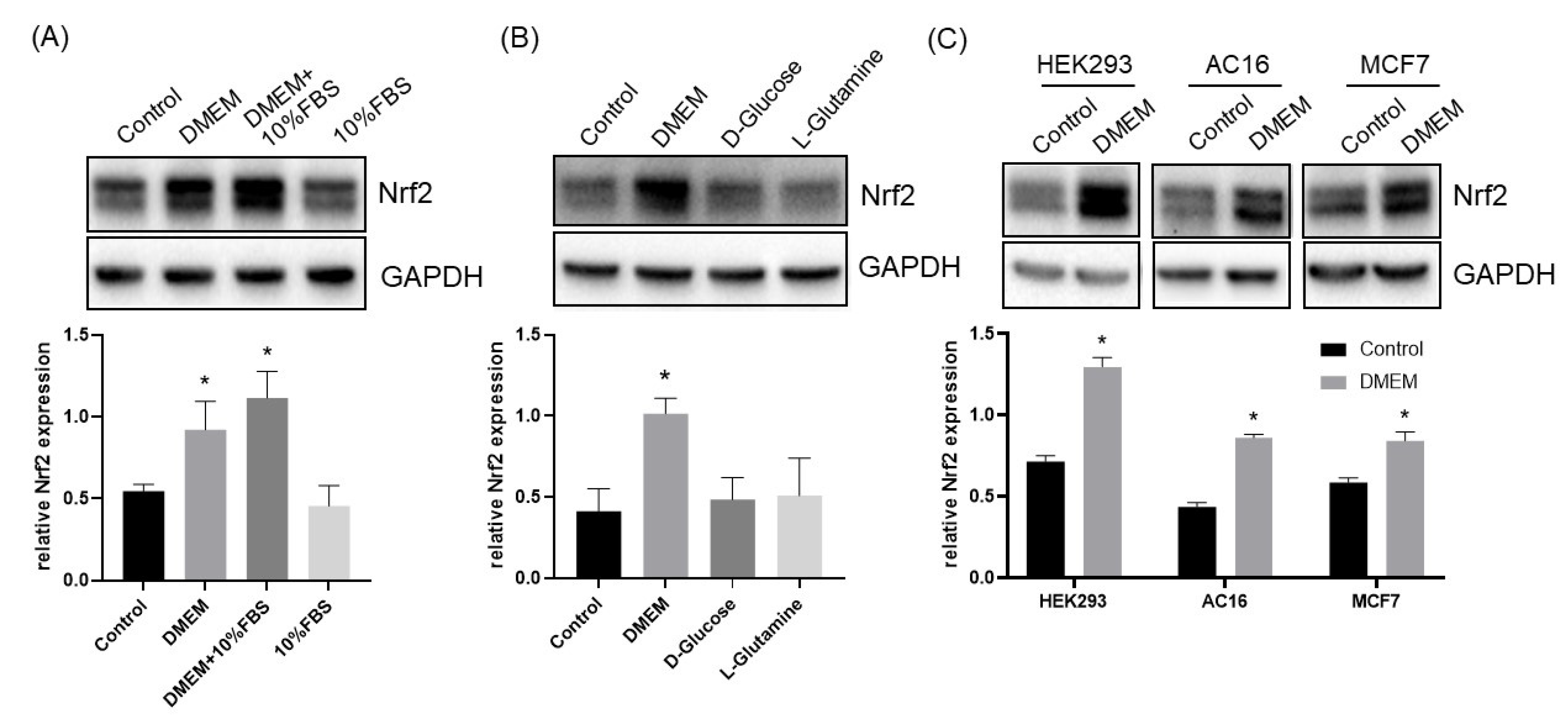
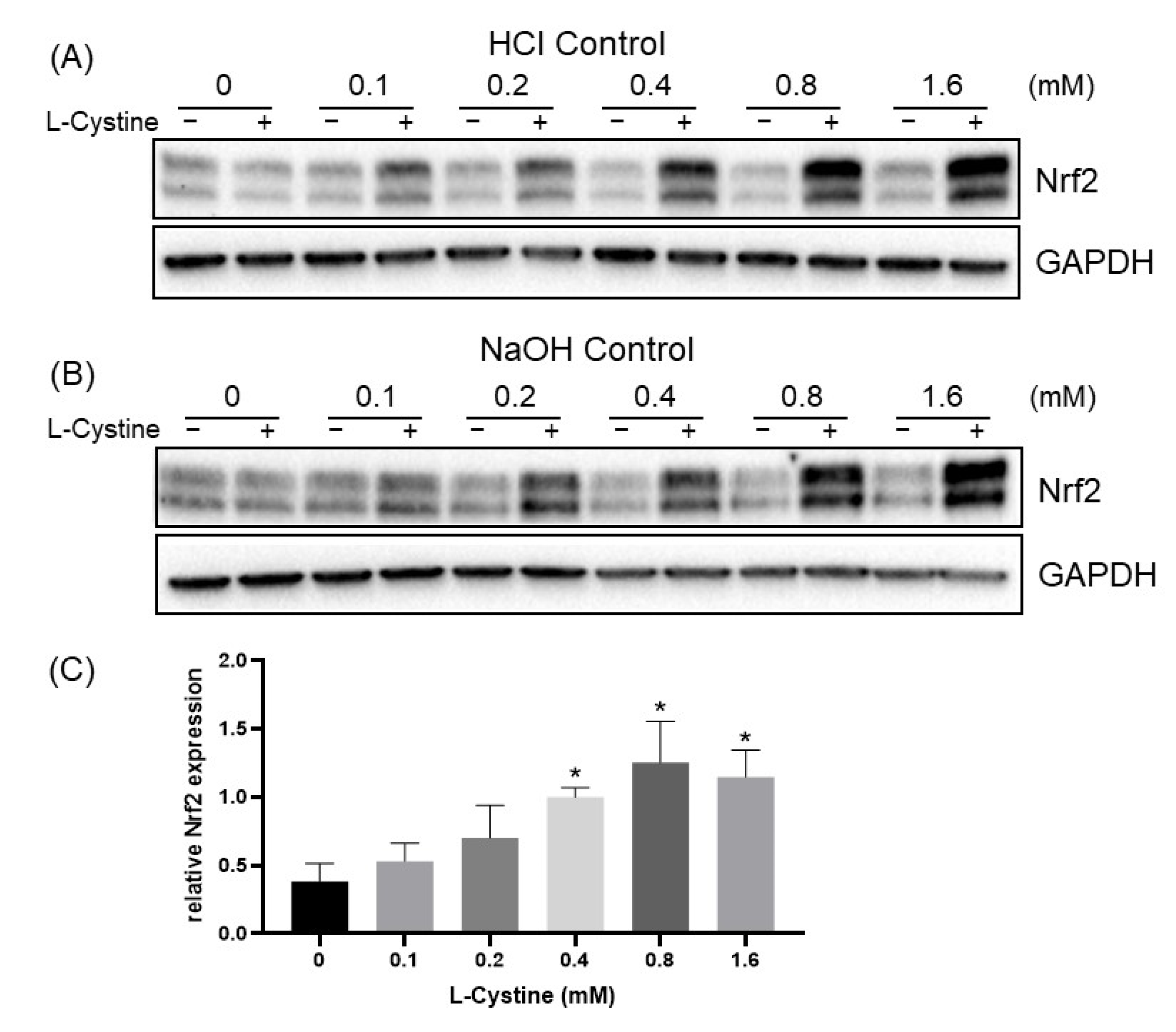
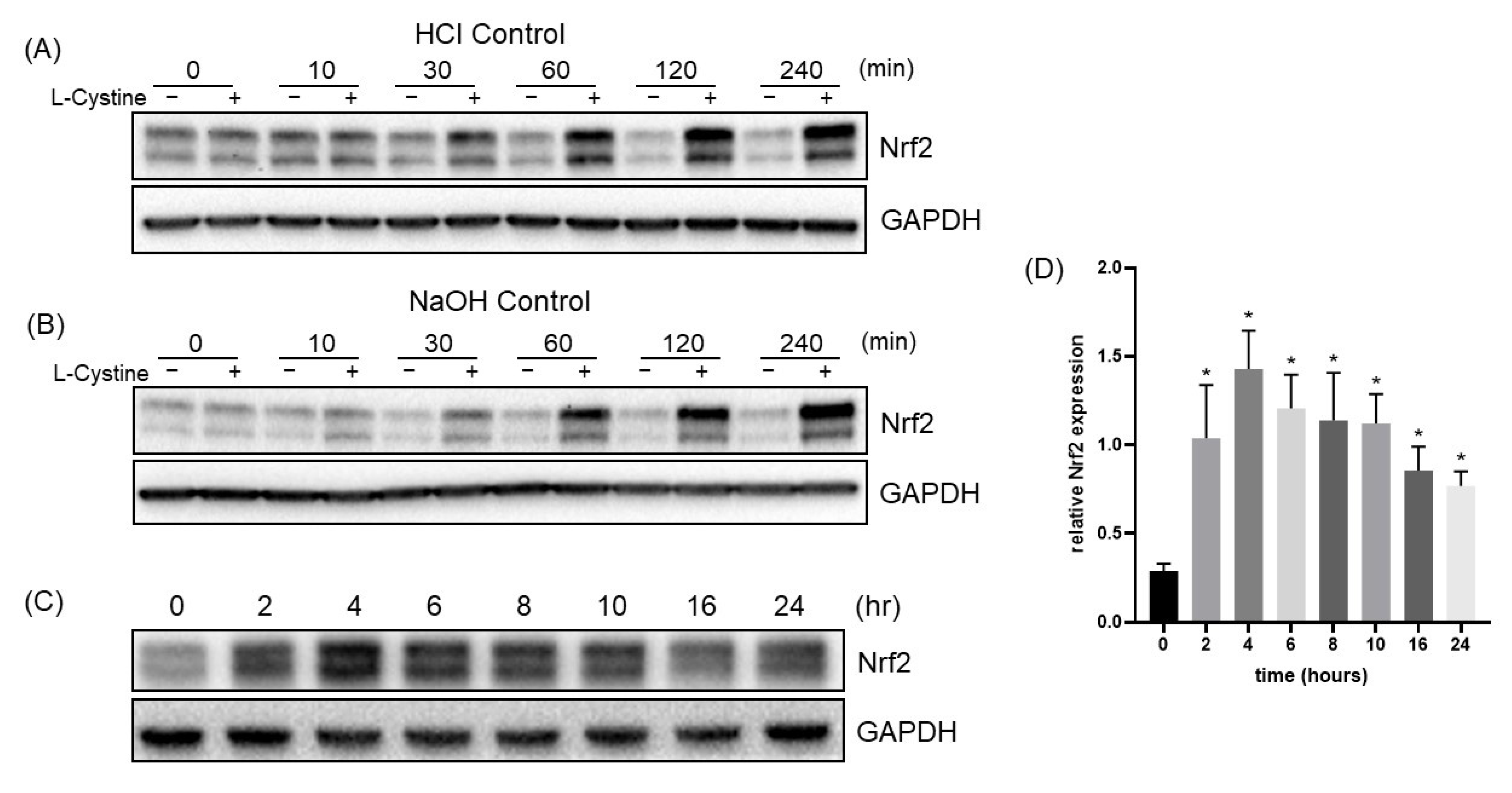
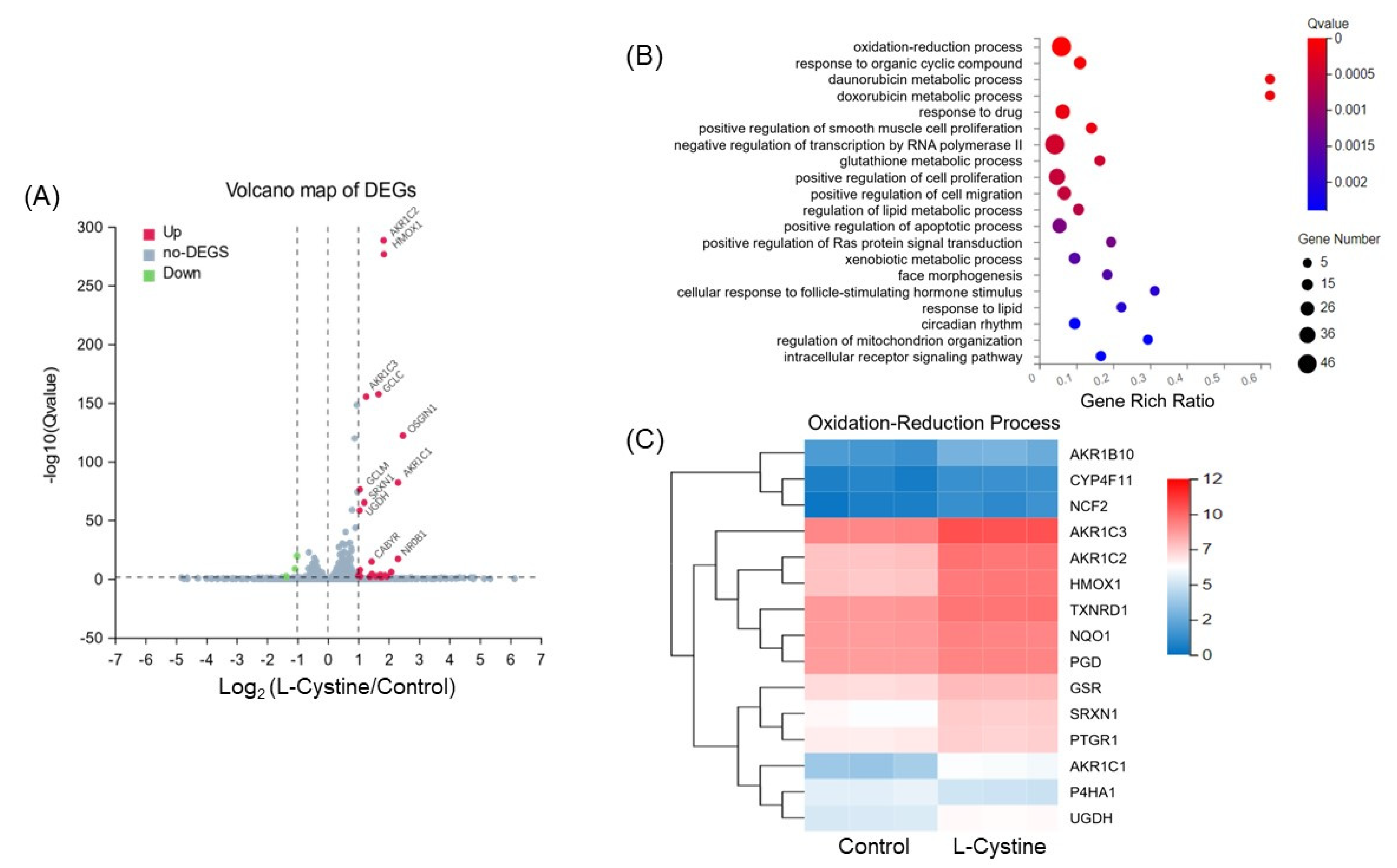
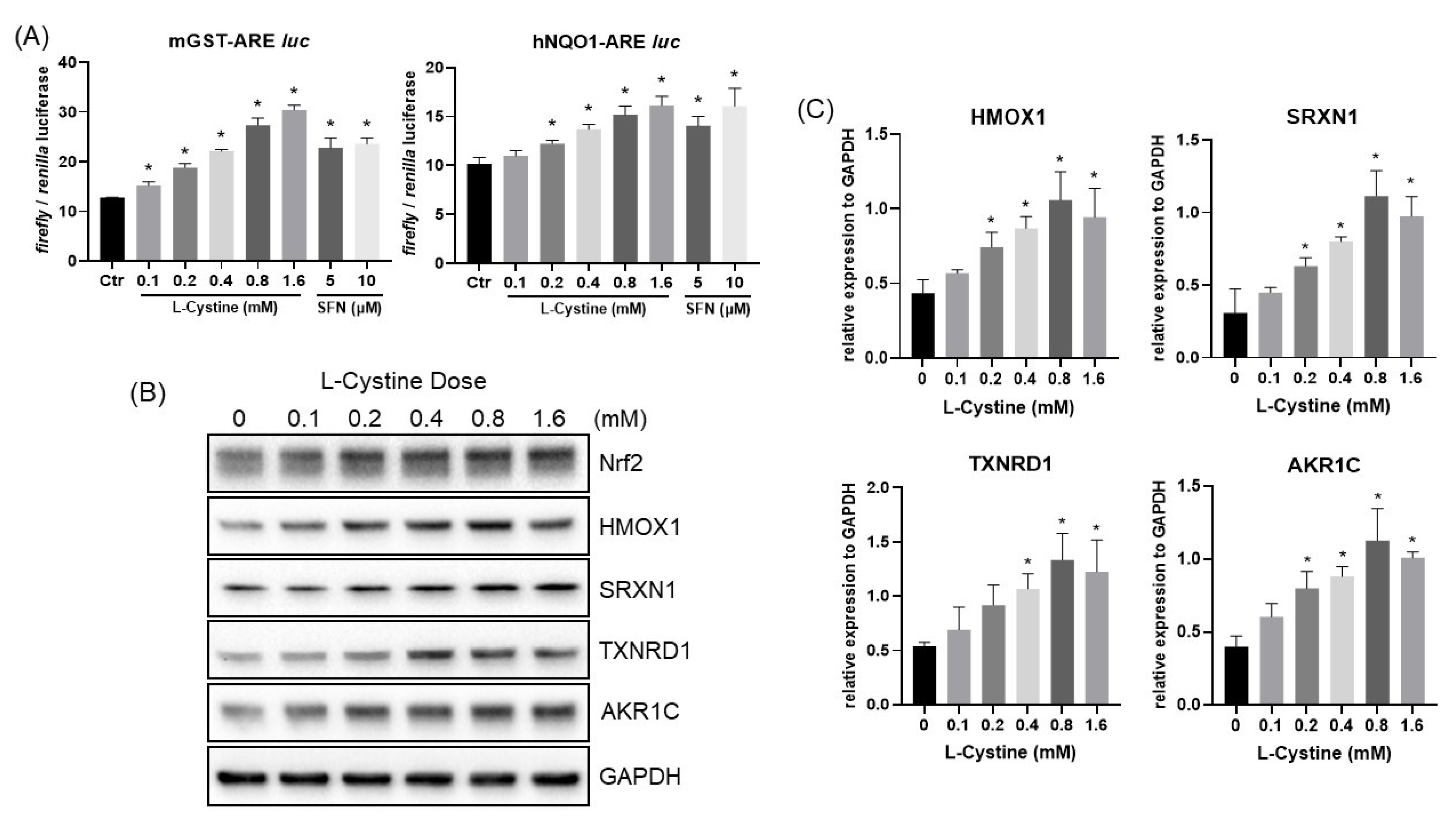
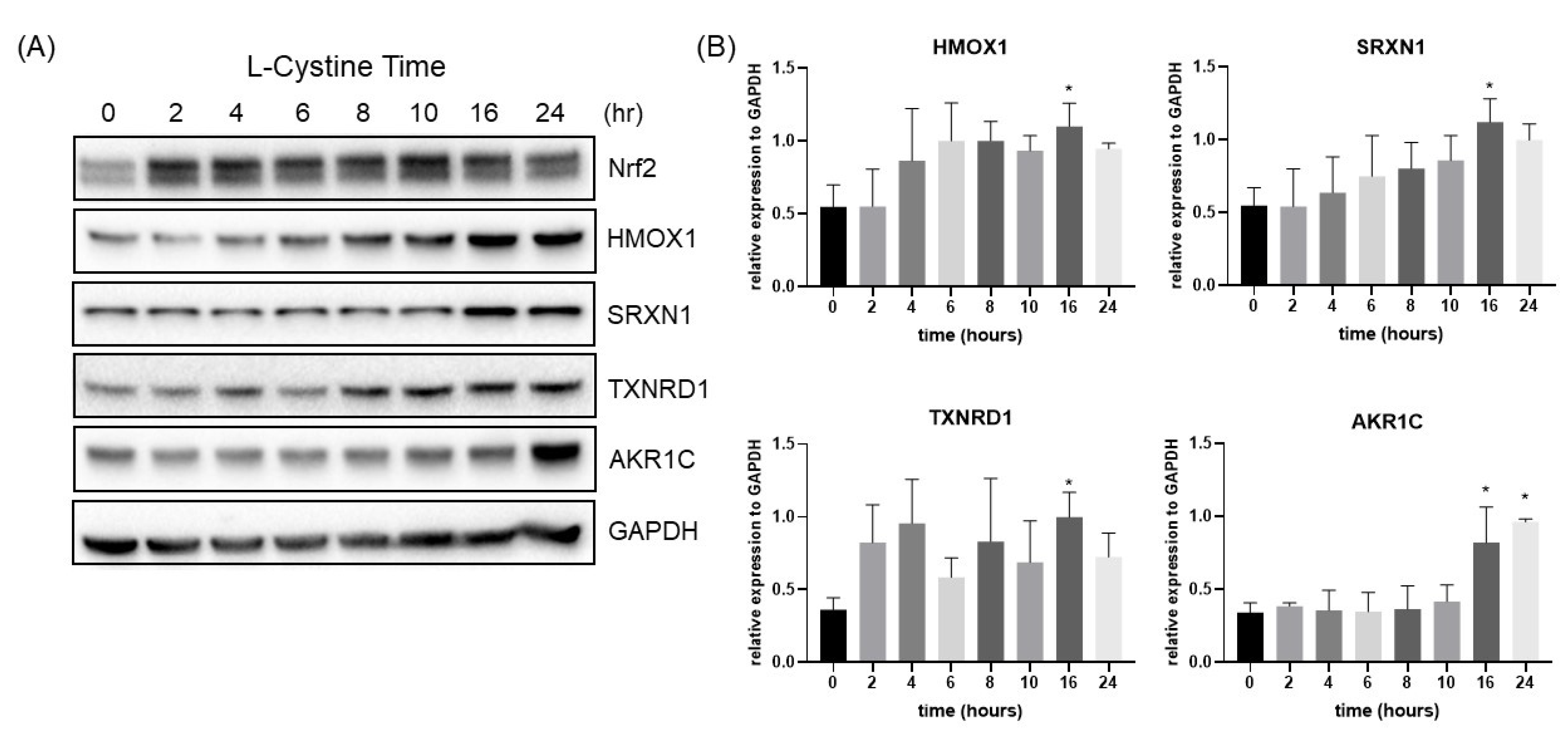
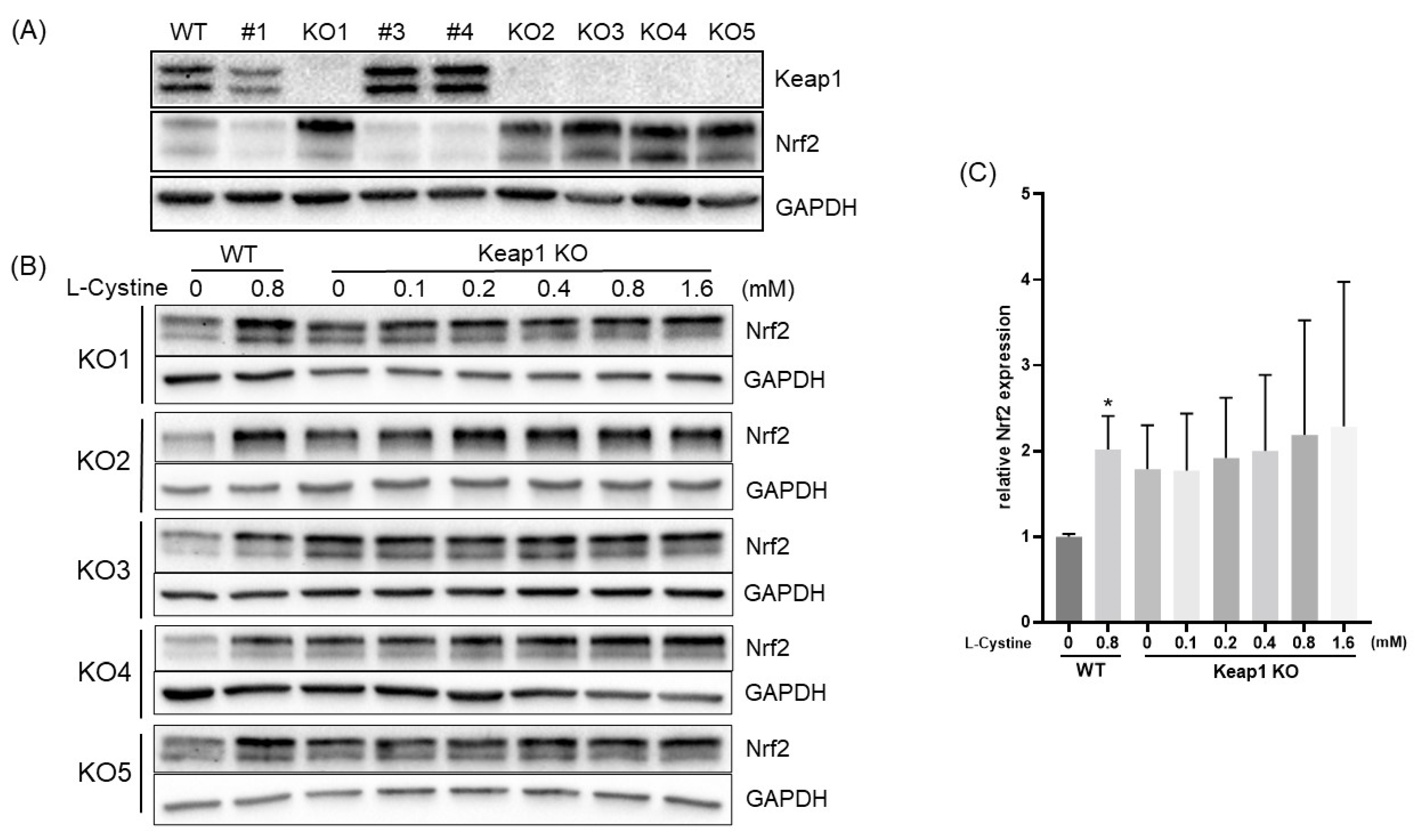
3.1. L-Cystine Induces Nrf2 Protein Elevation and Activates Nrf2 Transcription Factor
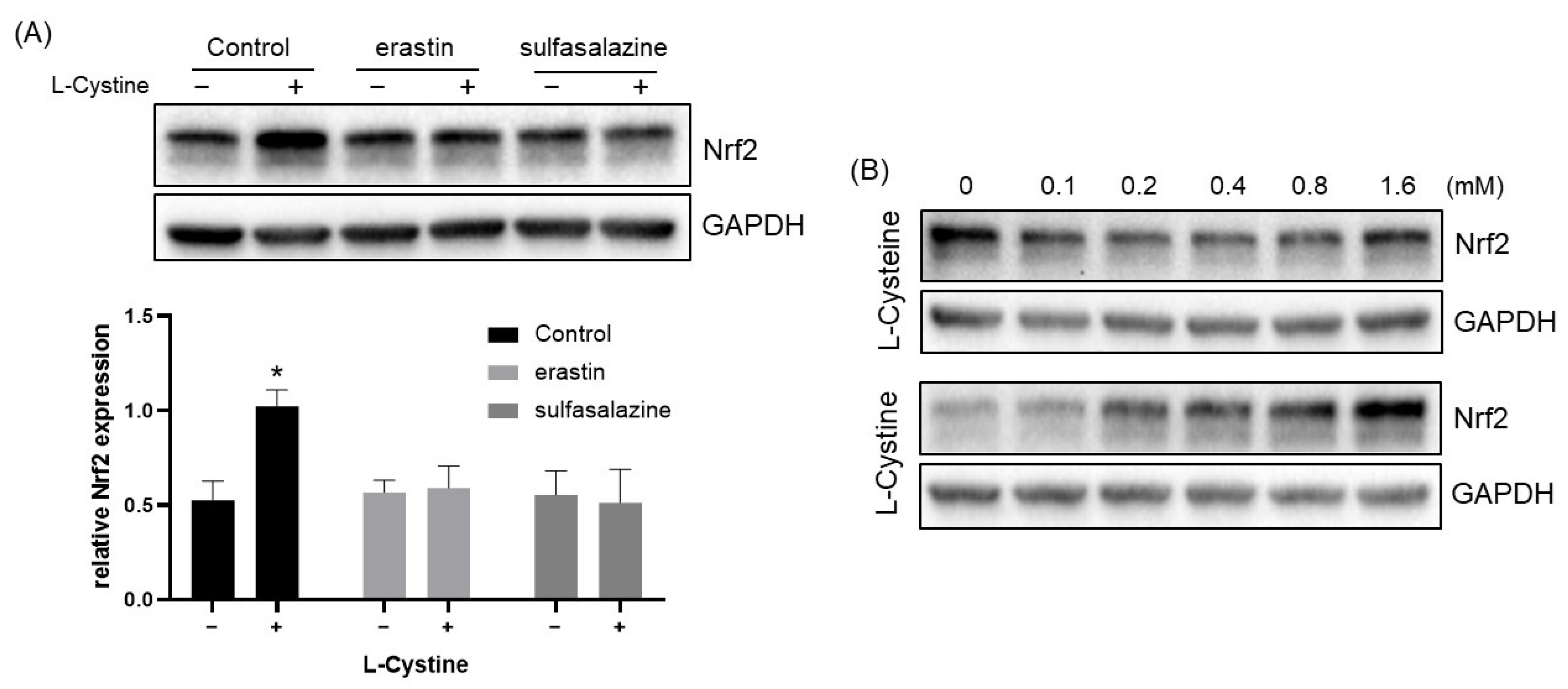
3.2. Mechanism of L-Cystine Induced Nrf2 Protein Elevation
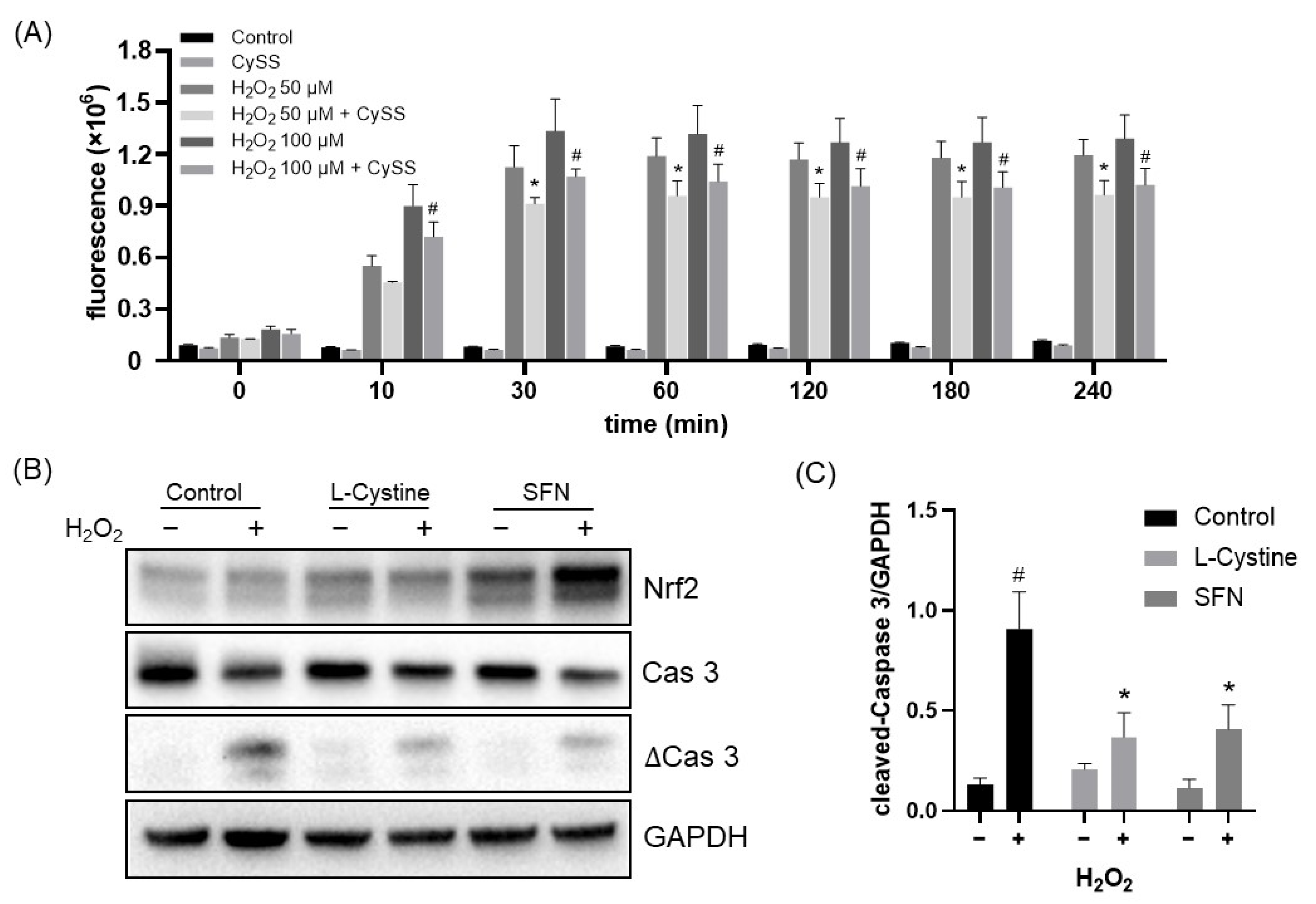
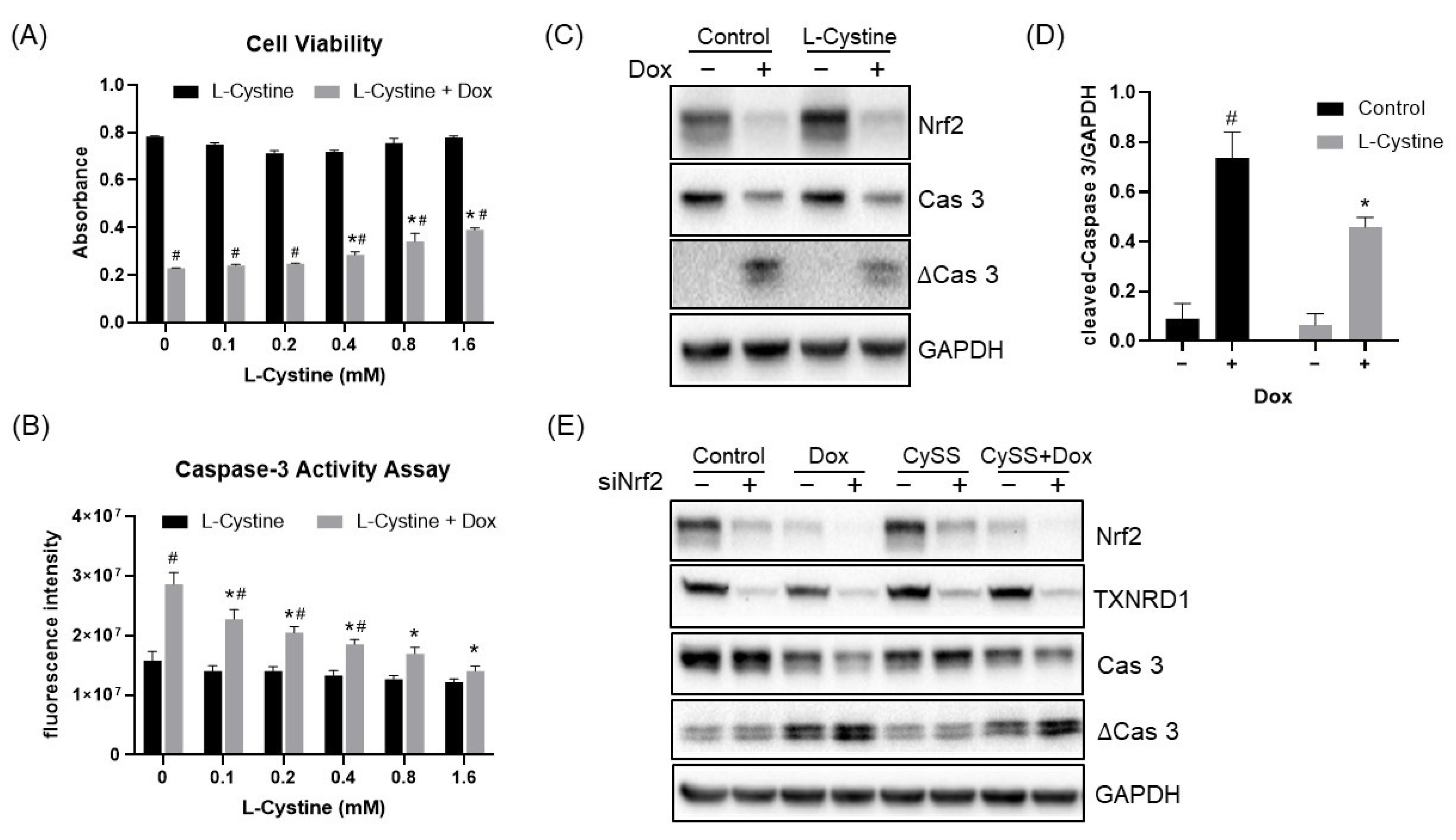
3.3. L-Cystine Protects against ROS Generation and Cell Death
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKR1C1 | Aldo-keto Reductase 1 family member C1 |
| CySS | L-Cystine |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| GCLC | Glutamate-cysteine Ligase catalytic subunit |
| GCLM | Glutamate-cysteine Ligase regulatory subunit |
| HMOX1 | Heme Oxygenase 1 |
| NQO1 | NAD(P)H Quinone Oxidoreductase 1 |
| OSGIN1 | Oxidative Stress-Induced Growth Inhibitor 1 |
| PVDF | polyvinylidene fluoride membrane |
| SRXN1 | Sulfiredoxin 1 |
| b-TrCP | b-transducin repeat-containing protein |
| TXNRD1 | thioredoxin reductase 1 |
References
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef] [Green Version]
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, S.; Levonen, A.L.; Kensler, T.W.; et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019, 18, 295–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.M. Nrf2 for cardiac protection: Pharmacological options against oxidative stress. Trends Pharm. Sci. 2021, 42, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.M. Nrf2 for protection against oxidant generation and mitochondrial damage in cardiac injury. Free Radic. Biol. Med. 2022, 179, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef] [Green Version]
- Raghunath, A.; Sundarraj, K.; Nagarajan, R.; Arfuso, F.; Bian, J.; Kumar, A.P.; Sethi, G.; Perumal, E. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018, 17, 297–314. [Google Scholar] [CrossRef]
- Ma, Q.; He, X. Molecular basis of electrophilic and oxidative defense: Promises and perils of Nrf2. Pharmacol. Rev. 2012, 64, 1055–1081. [Google Scholar] [CrossRef] [Green Version]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhao, F.; Gao, B.; Tan, C.; Yagishita, N.; Nakajima, T.; Wong, P.K.; Chapman, E.; Fang, D.; Zhang, D.D. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014, 28, 708–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purdom-Dickinson, S.E.; Sheveleva, E.V.; Sun, H.; Chen, Q.M. Translational Control of Nrf2 Protein in Activation of Antioxidant Response Element by Oxidants. Mol. Pharm. 2007, 72, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dinh, T.N.; Kappeler, K.; Tsaprailis, G.; Chen, Q.M. La autoantigen mediates oxidant induced de novo Nrf2 protein translation. Mol. Cell. Proteom. 2012, 11, M111.015032. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Zhang, J.; Strom, J.; Lee, S.; Chen, Q.M. Myocardial ischemic reperfusion induces de novo Nrf2 protein translation. Biochim. Biophys. Acta 2014, 1842, 1638–1647. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Zhang, J.; Strom, J.; Yang, D.; Dinh, T.N.; Kappeler, K.; Chen, Q.M. G-Quadruplex in the NRF2 mRNA 5′ Untranslated Region Regulates De Novo NRF2 Protein Translation under Oxidative Stress. Mol. Cell. Biol. 2017, 37, e00122-16. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Qu, H.; Zhang, J.; Thongkum, A.; Dinh, T.N.; Kappeler, K.V.; Chen, Q.M. Far Upstream Binding Protein 1 (FUBP1) participates in translational regulation of Nrf2 protein under oxidative stress. Redox Biol. 2021, 41, 101906. [Google Scholar] [CrossRef]
- Li, W.; Thakor, N.; Xu, E.Y.; Huang, Y.; Chen, C.; Yu, R.; Holcik, M.; Kong, A.N. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 2010, 38, 778–788. [Google Scholar] [CrossRef] [Green Version]
- Robledinos-Anton, N.; Fernandez-Gines, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxid. Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef] [Green Version]
- Staurengo-Ferrari, L.; Badaro-Garcia, S.; Hohmann, M.S.N.; Manchope, M.F.; Zaninelli, T.H.; Casagrande, R.; Verri, W.A., Jr. Contribution of Nrf2 Modulation to the Mechanism of Action of Analgesic and Anti-inflammatory Drugs in Pre-clinical and Clinical Stages. Front. Pharmacol. 2018, 9, 1536. [Google Scholar] [CrossRef]
- Jeong, W.S.; Jun, M.; Kong, A.N. Nrf2: A potential molecular target for cancer chemoprevention by natural compounds. Antioxid. Redox Signal. 2006, 8, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.M.; Maltagliati, A.J. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genom. 2018, 50, 77–97. [Google Scholar] [CrossRef] [Green Version]
- Ilatovskaya, D.V.; Pavlov, T.S.; Levchenko, V.; Staruschenko, A. ROS production as a common mechanism of ENaC regulation by EGF, insulin, and IGF-1. Am. J. Physiol. Cell Physiol. 2013, 304, C102–C111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazaki, K.; Matsuno, Y.; Yoshida, K.; Sherpa, M.; Nakajima, M.; Matsuyama, M.; Kiwamoto, T.; Morishima, Y.; Ishii, Y.; Hizawa, N. ROS-Nrf2 pathway mediates the development of TGF-beta1-induced epithelial-mesenchymal transition through the activation of Notch signaling. Eur. J. Cell Biol. 2021, 100, 151181. [Google Scholar] [CrossRef]
- Zhuang, J.; Jiang, T.; Lu, D.; Luo, Y.; Zheng, C.; Feng, J.; Yang, D.; Chen, C.; Yan, X. NADPH oxidase 4 mediates reactive oxygen species induction of CD146 dimerization in VEGF signal transduction. Free Radic. Biol. Med. 2010, 49, 227–236. [Google Scholar] [CrossRef]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.M.; Alexander, D.; Sun, H.; Xie, L.; Lin, Y.; Terrand, J.; Morrissy, S.; Purdom, S. Corticosteroids inhibit cell death induced by doxorubicin in cardiomyocytes: Induction of antiapoptosis, antioxidant, and detoxification genes. Mol. Pharmacol. 2005, 67, 1861–1873. [Google Scholar] [CrossRef]
- Brennan, M.S.; Matos, M.F.; Richter, K.E.; Li, B.; Scannevin, R.H. The NRF2 transcriptional target, OSGIN1, contributes to monomethyl fumarate-mediated cytoprotection in human astrocytes. Sci. Rep. 2017, 7, 42054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewerenz, J.; Hewett, S.J.; Huang, Y.; Lambros, M.; Gout, P.W.; Kalivas, P.W.; Massie, A.; Smolders, I.; Methner, A.; Pergande, M.; et al. The cystine/glutamate antiporter system x(c)(-) in health and disease: From molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 2013, 18, 522–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, D.; Kharkar, P.S.; Gandhi, N.S.; Kaur, E.; Dutt, S.; Nandave, M. Novel analogs of sulfasalazine as system xc (-) antiporter inhibitors: Insights from the molecular modeling studies. Drug Dev. Res. 2019, 80, 758–777. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Mann, G.E. Redox status in mammalian cells and stem cells during culture in vitro: Critical roles of Nrf2 and cystine transporter activity in the maintenance of redox balance. Redox Biol. 2014, 2, 786–794. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.P.; Go, Y.M.; Anderson, C.L.; Ziegler, T.R.; Kinkade, J.M., Jr.; Kirlin, W.G. Cysteine/cystine couple is a newly recognized node in the circuitry for biologic redox signaling and control. FASEB J. 2004, 18, 1246–1248. [Google Scholar] [CrossRef] [Green Version]
- Knickelbein, R.G.; Seres, T.; Lam, G.; Johnston, R.B., Jr.; Warshaw, J.B. Characterization of multiple cysteine and cystine transporters in rat alveolar type II cells. Am. J. Physiol. 1997, 273, L1147–L1155. [Google Scholar] [CrossRef]
- Ghezzi, P. Oxidoreduction of protein thiols in redox regulation. Biochem. Soc. Trans. 2005, 33, 1378–1381. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.D.; Lo, S.C.; Cross, J.V.; Templeton, D.J.; Hannink, M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004, 24, 10941–10953. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Eggler, A.L.; Mesecar, A.D.; van Breemen, R.B. Modification of keap1 cysteine residues by sulforaphane. Chem. Res. Toxicol. 2011, 24, 515–521. [Google Scholar] [CrossRef] [Green Version]
- Liby, K.; Hock, T.; Yore, M.M.; Suh, N.; Place, A.E.; Risingsong, R.; Williams, C.R.; Royce, D.B.; Honda, T.; Honda, Y.; et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005, 65, 4789–4798. [Google Scholar] [CrossRef]
- Suzuki, T.; Muramatsu, A.; Saito, R.; Iso, T.; Shibata, T.; Kuwata, K.; Kawaguchi, S.I.; Iwawaki, T.; Adachi, S.; Suda, H.; et al. Molecular Mechanism of Cellular Oxidative Stress Sensing by Keap1. Cell Rep. 2019, 28, 746–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Zeeuw, D.; Akizawa, T.; Audhya, P.; Bakris, G.L.; Chin, M.; Christ-Schmidt, H.; Goldsberry, A.; Houser, M.; Krauth, M.; Lambers Heerspink, H.J.; et al. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N. Engl. J. Med. 2013, 369, 2492–2503. [Google Scholar] [CrossRef] [Green Version]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Sun, Z. Overexpression of membranal SLC3A2 regulates the proliferation of oral squamous cancer cells and affects the prognosis of oral cancer patients. J. Oral Pathol. Med. 2021, 50, 371–377. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Lautenbacher, L.; Samaras, P.; Muller, J.; Grafberger, A.; Shraideh, M.; Rank, J.; Fuchs, S.T.; Schmidt, T.K.; The, M.; Dallago, C.; et al. ProteomicsDB: Toward a FAIR open-source resource for life-science research. Nucleic Acids Res. 2022, 50, D1541–D1552. [Google Scholar] [CrossRef]
- Samaras, P.; Schmidt, T.; Frejno, M.; Gessulat, S.; Reinecke, M.; Jarzab, A.; Zecha, J.; Mergner, J.; Giansanti, P.; Ehrlich, H.C.; et al. ProteomicsDB: A multi-omics and multi-organism resource for life science research. Nucleic Acids Res. 2020, 48, D1153–D1163. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, W.; Chen, Q.M. Fresh Medium or L-Cystine as an Effective Nrf2 Inducer for Cytoprotection in Cell Culture. Cells 2023, 12, 291. https://doi.org/10.3390/cells12020291
Dai W, Chen QM. Fresh Medium or L-Cystine as an Effective Nrf2 Inducer for Cytoprotection in Cell Culture. Cells. 2023; 12(2):291. https://doi.org/10.3390/cells12020291
Chicago/Turabian StyleDai, Wujing, and Qin M. Chen. 2023. "Fresh Medium or L-Cystine as an Effective Nrf2 Inducer for Cytoprotection in Cell Culture" Cells 12, no. 2: 291. https://doi.org/10.3390/cells12020291
APA StyleDai, W., & Chen, Q. M. (2023). Fresh Medium or L-Cystine as an Effective Nrf2 Inducer for Cytoprotection in Cell Culture. Cells, 12(2), 291. https://doi.org/10.3390/cells12020291