Long-Term Memory Formation in Drosophila Depends on the 3′UTR of CPEB Gene orb2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fly Strains
2.2. Locomotion Assays
2.3. Behavioral Assay
2.4. Antibodies
2.5. RNA Immunoprecipitation
2.6. RNA Isolation, Reverse Transcription, qPCR
2.7. Next-Generation Sequencing Data Analysis
2.8. Crude Synaptosome Preparation
2.9. Semiquantitative Western Blot Analysis
2.10. Whole Mount Immunostaining
3. Results
3.1. Deletion of orb2 3’UTR Does Not Affect Locomotion
3.2. Long-Term, but Not Short- or Mid-Term, Memory Formation Is Disrupted in the orb2R Mutant
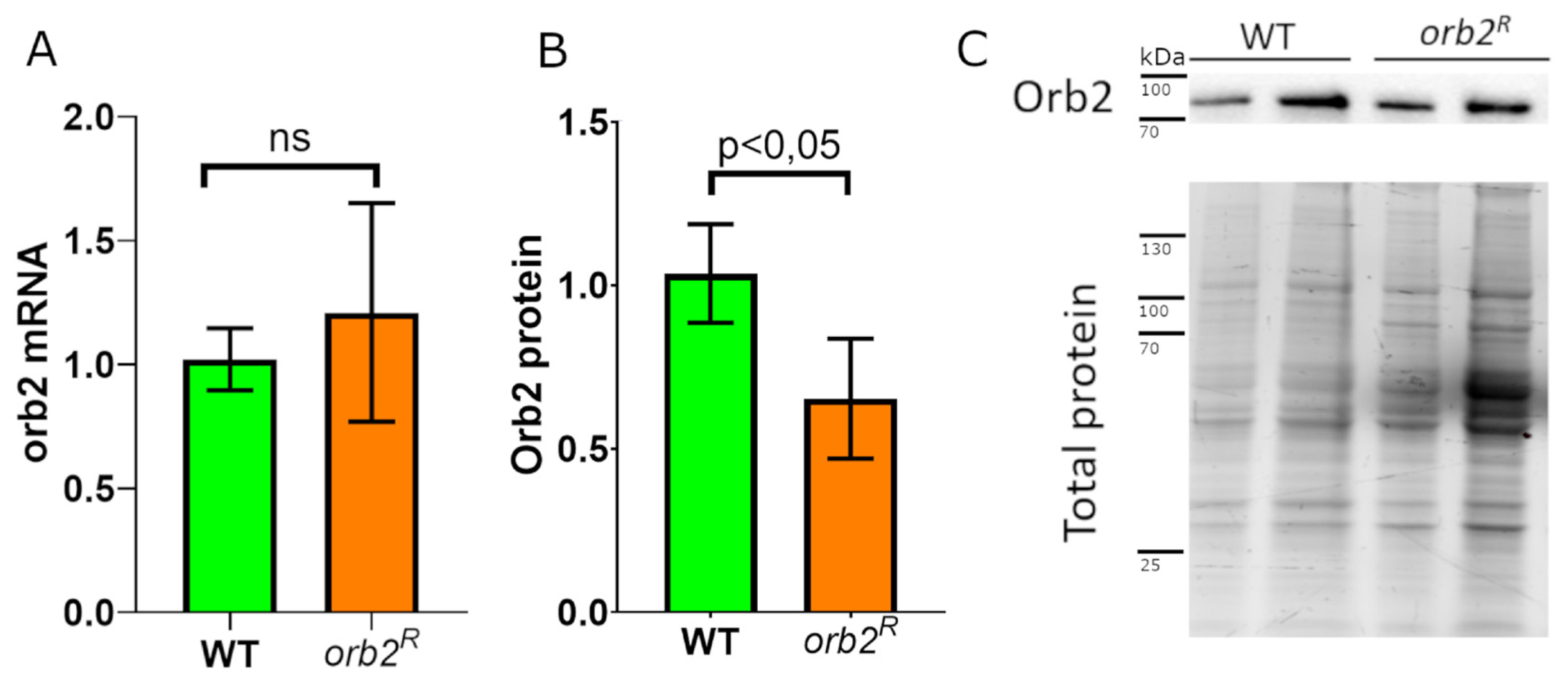
3.3. Orb2 Protein Levels in the Brain Are Altered in orb2R
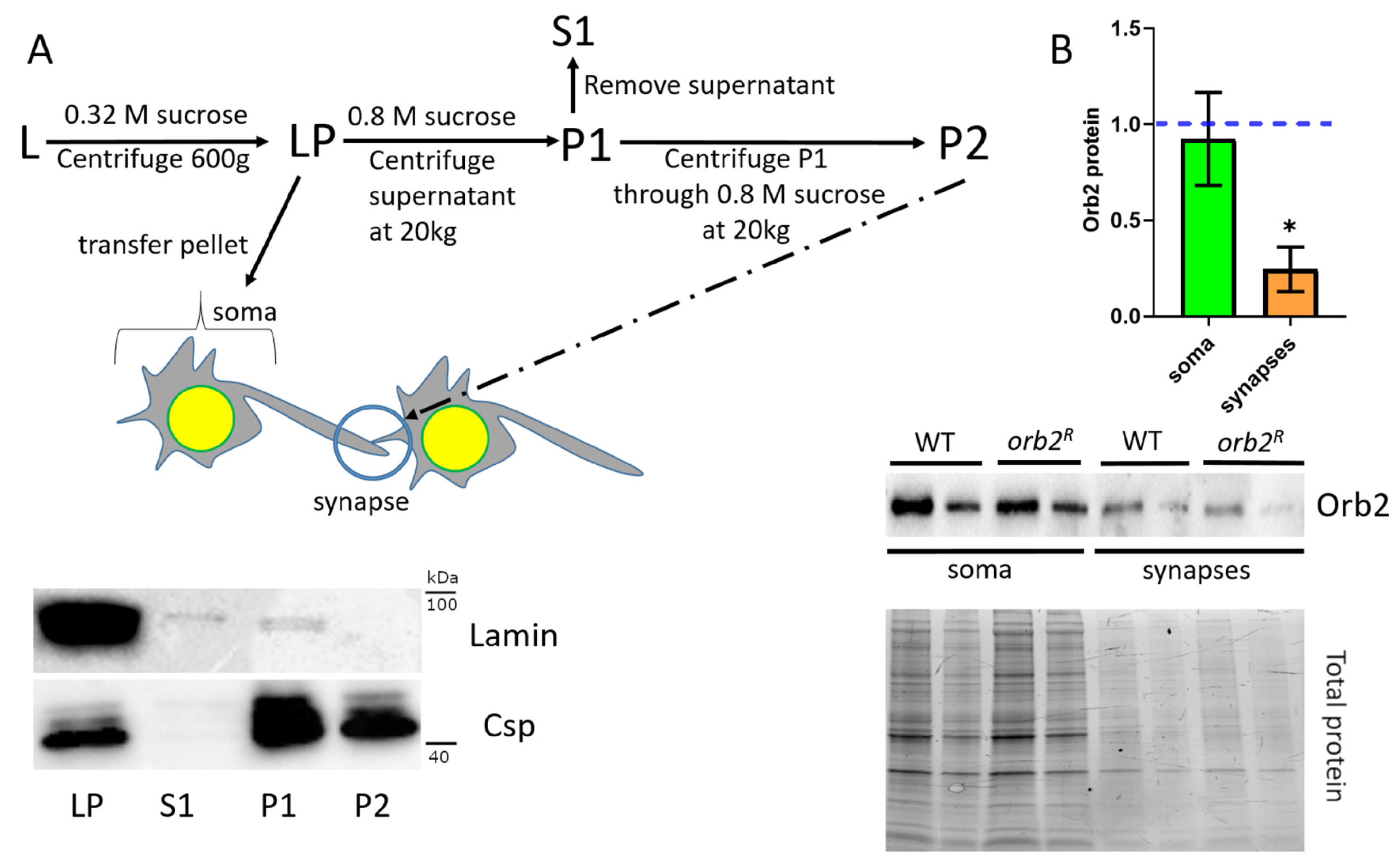
3.4. Orb2 Accumulation in Synaptic Fractions Is Affected in orb2R Mutant
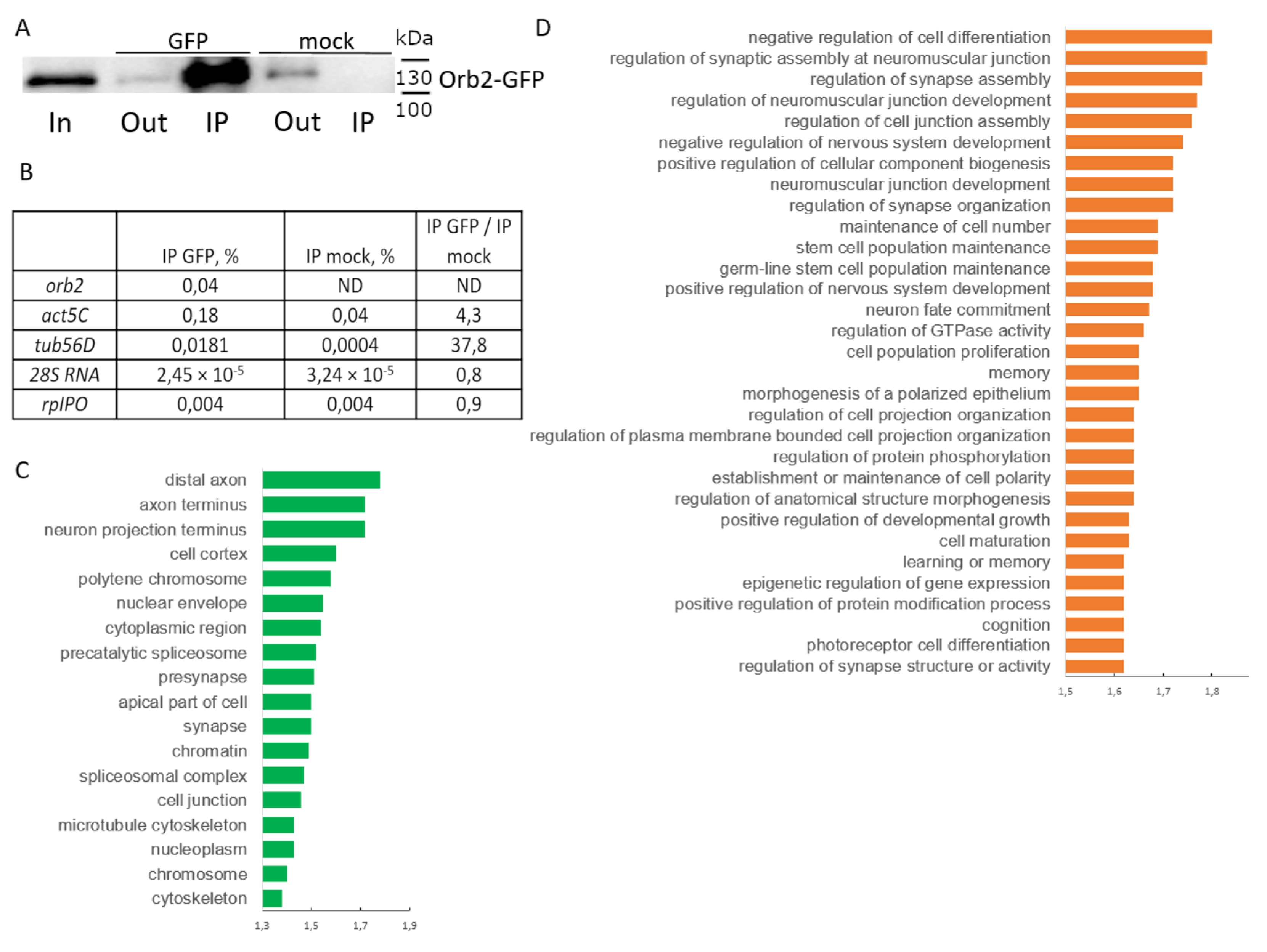
3.5. Identification of Orb2 mRNA Targets in the Nervous System
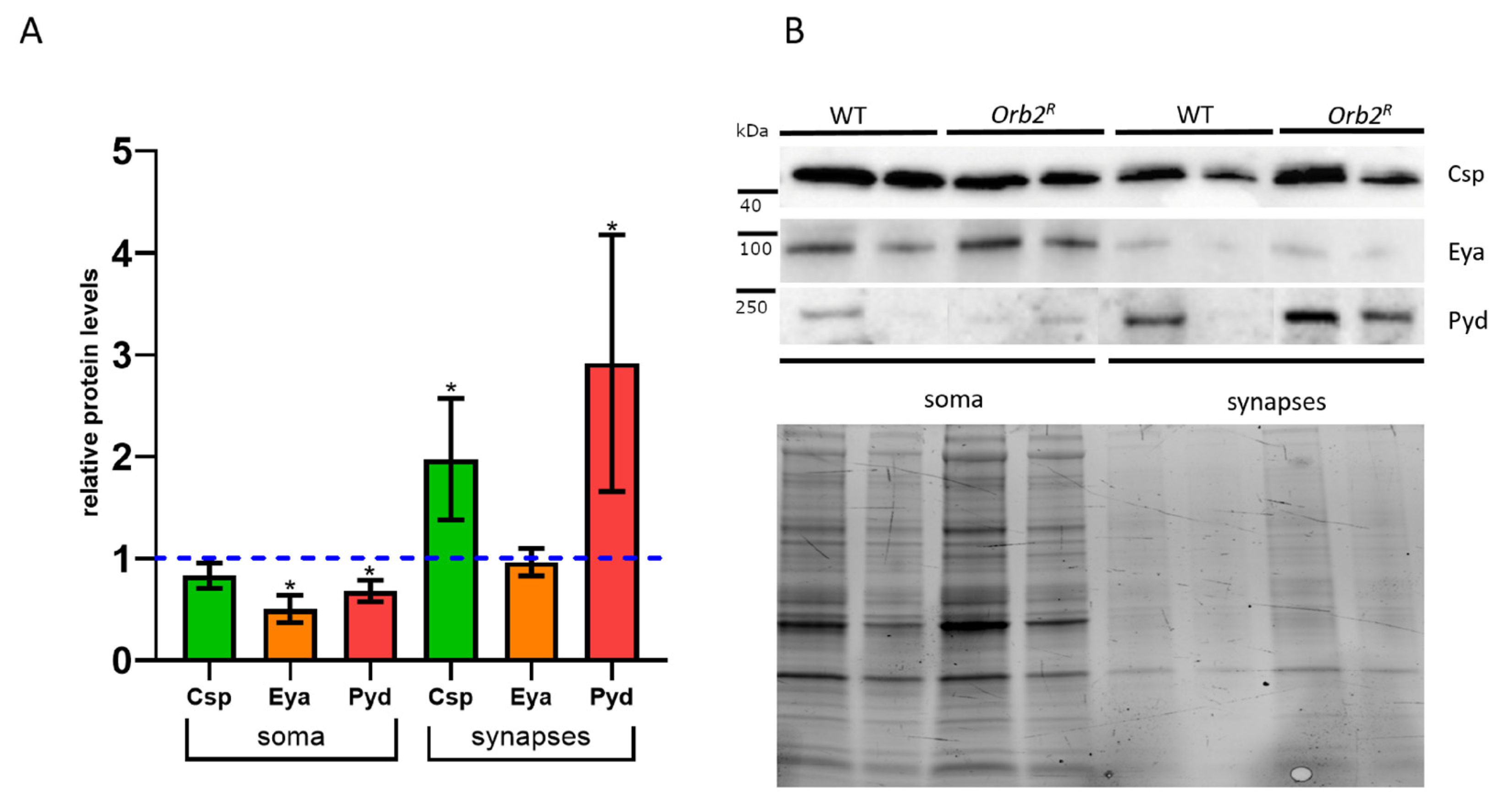
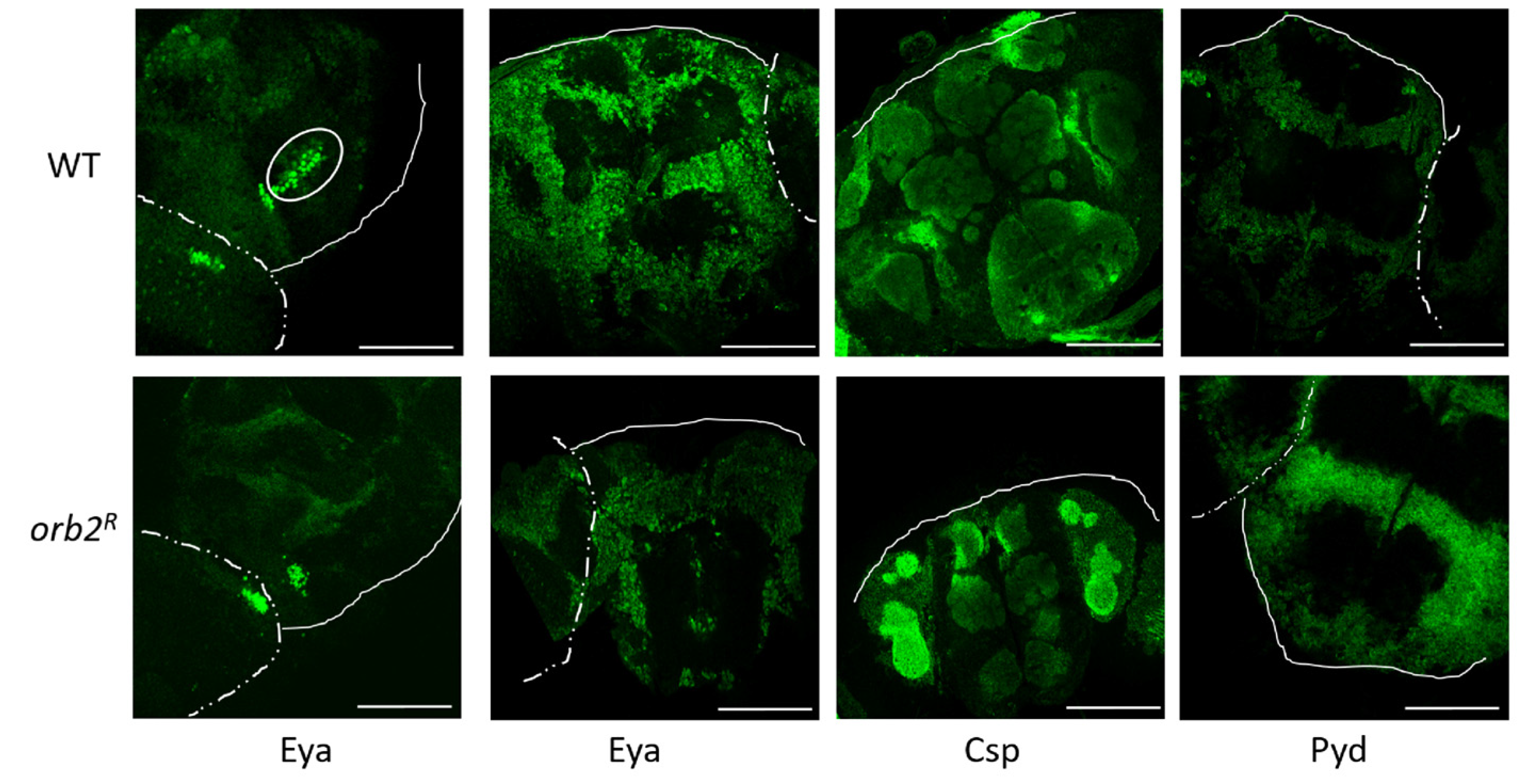
3.6. Intracellular Distribution of Proteins in Neurons Is Altered in orb2R Flies
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glanzman, D.L. Common mechanisms of synaptic plasticity in vertebrates and invertebrates. Curr. Biol. 2010, 20, R31–R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandel, E.R.; Dudai, Y.; Mayford, M.R. The molecular and systems biology of memory. Cell 2014, 157, 163–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelleher, R.J., 3rd; Govindarajan, A.; Tonegawa, S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron 2004, 44, 59–73. [Google Scholar] [CrossRef] [Green Version]
- Josselyn, S.A.; Tonegawa, S. Memory engrams: Recalling the past and imagining the future. Science 2020, 367, eaaw4325. [Google Scholar] [CrossRef]
- Doyle, M.; Kiebler, M.A. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011, 30, 3540–3552. [Google Scholar] [CrossRef] [Green Version]
- Sudhakaran, I.P.; Ramaswami, M. Long-term memory consolidation: The role of RNA-binding proteins with prion-like domains. RNA Biol. 2017, 14, 568–586. [Google Scholar] [CrossRef]
- Keleman, K.; Kruttner, S.; Alenius, M.; Dickson, B.J. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 2007, 10, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Kruttner, S.; Traunmuller, L.; Dag, U.; Jandrasits, K.; Stepien, B.; Iyer, N.; Fradkin, L.G.; Noordermeer, J.N.; Mensh, B.D.; Keleman, K. Synaptic Orb2A Bridges Memory Acquisition and Late Memory Consolidation in Drosophila. Cell Rep. 2015, 11, 1953–1965. [Google Scholar] [CrossRef] [Green Version]
- McGrew, L.L.; Richter, J.D. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: Characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990, 9, 3743–3751. [Google Scholar] [CrossRef]
- Si, K.; Choi, Y.B.; White-Grindley, E.; Majumdar, A.; Kandel, E.R. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 2010, 140, 421–435. [Google Scholar] [CrossRef]
- Bowler, J.T.; Sawaya, M.R.; Boyer, D.R.; Cascio, D.; Bali, M.; Eisenberg, D.S. Micro-electron diffraction structure of the aggregation-driving N terminus of Drosophila neuronal protein Orb2A reveals amyloid-like β-sheets. J. Biol. Chem. 2022, 298, 102396. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, M.; Lasko, P.; Richter, J.D. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 2014, 30, 393–415. [Google Scholar] [CrossRef] [PubMed]
- Lantz, V.; Chang, J.S.; Horabin, J.I.; Bopp, D.; Schedl, P. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 1994, 8, 598–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barr, J.; Gilmutdinov, R.; Wang, L.; Shidlovskii, Y.; Schedl, P. The Drosophila CPEB Protein Orb Specifies Oocyte Fate by a 3’UTR-Dependent Autoregulatory Loop. Genetics 2019, 213, 1431–1446. [Google Scholar] [CrossRef]
- Barr, J.; Charania, S.; Gilmutdinov, R.; Yakovlev, K.; Shidlovskii, Y.; Schedl, P. The CPEB translational regulator, Orb, functions together with Par proteins to polarize the Drosophila oocyte. PLoS Genet. 2019, 15, e1008012. [Google Scholar] [CrossRef]
- Pai, T.P.; Chen, C.C.; Lin, H.H.; Chin, A.L.; Lai, J.S.; Lee, P.T.; Tully, T.; Chiang, A.S. Drosophila ORB protein in two mushroom body output neurons is necessary for long-term memory formation. Proc. Natl. Acad. Sci. USA 2013, 110, 7898–7903. [Google Scholar] [CrossRef] [Green Version]
- Hafer, N.; Xu, S.; Bhat, K.M.; Schedl, P. The Drosophila CPEB protein Orb2 has a novel expression pattern and is important for asymmetric cell division and nervous system function. Genetics 2011, 189, 907–921. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Hafer, N.; Agunwamba, B.; Schedl, P. The CPEB protein Orb2 has multiple functions during spermatogenesis in Drosophila melanogaster. PLoS Genet. 2012, 8, e1003079. [Google Scholar] [CrossRef] [Green Version]
- Kruttner, S.; Stepien, B.; Noordermeer, J.N.; Mommaas, M.A.; Mechtler, K.; Dickson, B.J.; Keleman, K. Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron 2012, 76, 383–395. [Google Scholar] [CrossRef] [Green Version]
- Hervas, R.; Li, L.; Majumdar, A.; Fernandez-Ramirez Mdel, C.; Unruh, J.R.; Slaughter, B.D.; Galera-Prat, A.; Santana, E.; Suzuki, M.; Nagai, Y.; et al. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLoS Biol. 2016, 14, e1002361. [Google Scholar] [CrossRef]
- Khan, M.R.; Li, L.; Perez-Sanchez, C.; Saraf, A.; Florens, L.; Slaughter, B.D.; Unruh, J.R.; Si, K. Amyloidogenic Oligomerization Transforms Drosophila Orb2 from a Translation Repressor to an Activator. Cell 2015, 163, 1468–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanfilippo, P.; Wen, J.; Lai, E.C. Landscape and evolution of tissue-specific alternative polyadenylation across Drosophila species. Genome Biol. 2017, 18, 229. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Chen, Y.C.; Gillen, A.E.; Taliaferro, J.M.; Deplancke, B.; Li, H.; Lai, E.C. Diverse cell-specific patterns of alternative polyadenylation in Drosophila. Nat. Commun. 2022, 13, 5372. [Google Scholar] [CrossRef] [PubMed]
- Bae, B.; Miura, P. Emerging Roles for 3’ UTRs in Neurons. Int. J. Mol. Sci. 2020, 21, 3413. [Google Scholar] [CrossRef]
- Barkoff, A.F.; Dickson, K.S.; Gray, N.K.; Wickens, M. Translational control of cyclin B1 mRNA during meiotic maturation: Coordinated repression and cytoplasmic polyadenylation. Dev. Biol. 2000, 220, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Gilmutdinov, R.; Kozlov, E.N.; Yakovlev, K.V.; Olenina, L.V.; Kotov, A.A.; Barr, J.; Zhukova, M.; Schedl, P.; Shidlovskii, Y.V. The 3’UTR of the Drosophila CPEB translation factor gene orb2 plays a crucial role in spermatogenesis. Development 2021, 148, dev198788. [Google Scholar] [CrossRef]
- Kamyshev, N.G.; Iliadi, K.G.; Bragina, J.V. Drosophila conditioned courtship: Two ways of testing memory. Learn. Mem. 1999, 6, 1–20. [Google Scholar] [CrossRef]
- Trapnell, C., L. Pachter, and S.L. Salzberg, TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Anders, S., P. T. Pyl, and W. Huber, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef] [PubMed]
- 31. Lyne, R.; Smith, R.; Rutherford, K.; Wakeling, M.; Varley, A.; Guillier, F.; Janssens, H.; Ji, W.; Mclaren, P.; North, P.; et al. FlyMine: An integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007, 8, R129. [Google Scholar] [CrossRef] [Green Version]
- Kamat, P.K.; Kalani, A.; Tyagi, N. Method and validation of synaptosomal preparation for isolation of synaptic membrane proteins from rat brain. MethodsX 2014, 1, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Savvateeva, E.; Popov, A.; Kamyshev, N.; Bragina, J.; Heisenberg, M.; Senitz, D.; Kornhuber, J.; Riederer, P. Age-dependent memory loss, synaptic pathology and altered brain plasticity in the Drosophila mutant cardinal accumulating 3-hydroxykynurenine. J. Neural Transm 2000, 107, 581–601. [Google Scholar] [CrossRef] [PubMed]
- Stepien, B.K.; Oppitz, C.; Gerlach, D.; Dag, U.; Novatchkova, M.; Kruttner, S.; Stark, A.; Keleman, K. RNA-binding profiles of Drosophila CPEB proteins Orb and Orb2. Proc. Natl. Acad. Sci. USA 2016, 113, E7030–E7038. [Google Scholar] [CrossRef] [Green Version]
- Mastushita-Sakai, T.; White-Grindley, E.; Samuelson, J.; Seidel, C.; Si, K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc. Natl. Acad. Sci. USA 2010, 107, 11987–11992. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Tyagi, S.; Schedl, P. Spermatid cyst polarization in Drosophila depends upon apkc and the CPEB family translational regulator orb2. PLoS Genet. 2014, 10, e1004380. [Google Scholar] [CrossRef]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011, 471, 473–479. [Google Scholar] [CrossRef] [Green Version]
- Dawson-Scully, K.; Bronk, P.; Atwood, H.L.; Zinsmaier, K.E. Cysteine-string protein increases the calcium sensitivity of neurotransmitter exocytosis in Drosophila. J. Neurosci. 2000, 20, 6039–6047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moressis, A.; Friedrich, A.R.; Pavlopoulos, E.; Davis, R.L.; Skoulakis, E.M. A dual role for the adaptor protein DRK in Drosophila olfactory learning and memory. J. Neurosci. 2009, 29, 2611–2625. [Google Scholar] [CrossRef] [Green Version]
- Eddison, M.; Belay, A.T.; Sokolowski, M.B.; Heberlein, U. A genetic screen for olfactory habituation mutations in Drosophila: Analysis of novel foraging alleles and an underlying neural circuit. PLoS ONE 2012, 7, e51684. [Google Scholar] [CrossRef]
- Zinsmaier, K.E.; Hofbauer, A.; Heimbeck, G.; Pflugfelder, G.O.; Buchner, S.; Buchner, E. A cysteine-string protein is expressed in retina and brain of Drosophila. J. Neurogenet. 1990, 7, 15–29. [Google Scholar] [CrossRef] [PubMed]
- White-Grindley, E.; Li, L.; Mohammad Khan, R.; Ren, F.; Saraf, A.; Florens, L.; Si, K. Contribution of Orb2A stability in regulated amyloid-like oligomerization of Drosophila Orb2. PLoS Biol. 2014, 12, e1001786. [Google Scholar] [CrossRef] [Green Version]
- Holt, C.E.; Martin, K.C.; Schuman, E.M. Local translation in neurons: Visualization and function. Nat. Struct. Mol. Biol. 2019, 26, 557–566. [Google Scholar] [CrossRef]
- Das, S.; Singer, R.H.; Yoon, Y.J. The travels of mRNAs in neurons: Do they know where they are going? Curr. Opin. Neurobiol. 2019, 57, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Smibert, P.; Miura, P.; Westholm, J.O.; Shenker, S.; May, G.; Duff, M.O.; Zhang, D.; Eads, B.D.; Carlson, J.; Brown, J.B.; et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012, 1, 277–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, Y.; Ihara, K.; Masuda, T.; Yamamoto, T.; Iwata, I.; Takahashi, A.; Awata, H.; Nakamura, N.; Takakura, M.; Suzuki, Y.; et al. Shifting transcriptional machinery is required for long-term memory maintenance and modification in Drosophila mushroom bodies. Nat. Commun. 2016, 7, 13471. [Google Scholar] [CrossRef] [Green Version]
- Mariano, V.; Achsel, T.; Bagni, C.; Kanellopoulos, A.K. Modelling Learning and Memory in Drosophila to Understand Intellectual Disabilities. Neuroscience 2020, 445, 12–30. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Truglio, M.; Scott, M.J.; Fitzsimons, H.L. Long-Term Memory in Drosophila Is Influenced by Histone Deacetylase HDAC4 Interacting with SUMO-Conjugating Enzyme Ubc9. Genetics 2016, 203, 1249–1264. [Google Scholar] [CrossRef] [Green Version]
- Walkinshaw, E.; Gai, Y.; Farkas, C.; Richter, D.; Nicholas, E.; Keleman, K.; Davis, R.L. Identification of genes that promote or inhibit olfactory memory formation in Drosophila. Genetics 2015, 199, 1173–1182. [Google Scholar] [CrossRef] [Green Version]
- Berger, K.H.; Kong, E.C.; Dubnau, J.; Tully, T.; Moore, M.S.; Heberlein, U. Ethanol sensitivity and tolerance in long-term memory mutants of Drosophila melanogaster. Alcohol Clin. Exp. Res. 2008, 32, 895–908. [Google Scholar] [CrossRef]
- Chia, W.; Somers, W.G.; Wang, H. Drosophila neuroblast asymmetric divisions: Cell cycle regulators, asymmetric protein localization, and tumorigenesis. J. Cell Biol. 2008, 180, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Kelsom, C.; Lu, W. Uncovering the link between malfunctions in Drosophila neuroblast asymmetric cell division and tumorigenesis. Cell Biosci. 2012, 2, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Wang, H.; Groth, C. Drosophila neuroblasts as a new model for the study of stem cell self-renewal and tumour formation. Biosci. Rep. 2014, 34, e00125. [Google Scholar] [CrossRef]
- Kang, K.H.; Reichert, H. Control of neural stem cell self-renewal and differentiation in Drosophila. Cell Tissue Res. 2015, 359, 33–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brody, T.; Stivers, C.; Nagle, J.; Odenwald, W.F. Identification of novel Drosophila neural precursor genes using a differential embryonic head cDNA screen. Mech. Dev. 2002, 113, 41–59. [Google Scholar] [CrossRef]
- Ding, W.Y.; Huang, J.; Wang, H. Waking up quiescent neural stem cells: Molecular mechanisms and implications in neurodevelopmental disorders. PLoS Genet. 2020, 16, e1008653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumuller, R.A.; Richter, C.; Fischer, A.; Novatchkova, M.; Neumuller, K.G.; Knoblich, J.A. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 2011, 8, 580–593. [Google Scholar] [CrossRef] [Green Version]
- Southall, T.D.; Brand, A.H. Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J. 2009, 28, 3799–3807. [Google Scholar] [CrossRef] [Green Version]
- Gabilondo, H.; Losada-Perez, M.; del Saz, D.; Molina, I.; Leon, Y.; Canal, I.; Torroja, L.; Benito-Sipos, J. A targeted genetic screen identifies crucial players in the specification of the Drosophila abdominal Capaergic neurons. Mech. Dev. 2011, 128, 208–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Q.; Brbic, M.; Horns, F.; Kolluru, S.S.; Jones, R.C.; Li, J.; Reddy, A.R.; Xie, A.; Kohani, S.; Li, Z.; et al. Temporal evolution of single-cell transcriptomes of Drosophila olfactory projection neurons. Elife 2021, 10, e63450. [Google Scholar] [CrossRef]
- Bayraktar, O.A.; Doe, C.Q. Combinatorial temporal patterning in progenitors expands neural diversity. Nature 2013, 498, 449–455. [Google Scholar] [CrossRef] [Green Version]
- Bivik, C.; Bahrampour, S.; Ulvklo, C.; Nilsson, P.; Angel, A.; Fransson, F.; Lundin, E.; Renhorn, J.; Thor, S. Novel Genes Involved in Controlling Specification of Drosophila FMRFamide Neuropeptide Cells. Genetics 2015, 200, 1229–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrero, P.; Estacio-Gomez, A.; Moris-Sanz, M.; Alvarez-Rivero, J.; Diaz-Benjumea, F.J. Origin and specification of the brain leucokinergic neurons of Drosophila: Similarities to and differences from abdominal leucokinergic neurons. Dev. Dyn. 2014, 243, 402–414. [Google Scholar] [CrossRef]
- Ma, D.; Przybylski, D.; Abruzzi, K.C.; Schlichting, M.; Li, Q.; Long, X.; Rosbash, M. A transcriptomic taxonomy of Drosophila circadian neurons around the clock. Elife 2021, 10, e63056. [Google Scholar] [CrossRef] [PubMed]
- Liebl, F.L.; Featherstone, D.E. Genes involved in Drosophila glutamate receptor expression and localization. BMC Neurosci. 2005, 6, 44. [Google Scholar] [CrossRef] [Green Version]
- Michki, N.S.; Li, Y.; Sanjasaz, K.; Zhao, Y.; Shen, F.Y.; Walker, L.A.; Cao, W.; Lee, C.Y.; Cai, D. The molecular landscape of neural differentiation in the developing Drosophila brain revealed by targeted scRNA-seq and multi-informatic analysis. Cell Rep. 2021, 35, 109039. [Google Scholar] [CrossRef]
- Apitz, H.; Salecker, I. A challenge of numbers and diversity: Neurogenesis in the Drosophila optic lobe. J. Neurogenet 2014, 28, 233–249. [Google Scholar] [CrossRef]
- Mira, H.; Morante, J. Neurogenesis From Embryo to Adult—Lessons From Flies and Mice. Front. Cell Dev. Biol. 2020, 8, 533. [Google Scholar] [CrossRef] [PubMed]
- Yasugi, T.; Nishimura, T. Temporal regulation of the generation of neuronal diversity in Drosophila. Dev. Growth Differ. 2016, 58, 73–87. [Google Scholar] [CrossRef]
- Kearney, J.B.; Wheeler, S.R.; Estes, P.; Parente, B.; Crews, S.T. Gene expression profiling of the developing Drosophila CNS midline cells. Dev. Biol. 2004, 275, 473–492. [Google Scholar] [CrossRef]
- Karlin, S.; Burge, C. Trinucleotide repeats and long homopeptides in genes and proteins associated with nervous system disease and development. Proc. Natl. Acad. Sci. USA 1996, 93, 1560–1565. [Google Scholar] [CrossRef] [Green Version]
- Zwarts, L.; Vanden Broeck, L.; Cappuyns, E.; Ayroles, J.F.; Magwire, M.M.; Vulsteke, V.; Clements, J.; Mackay, T.F.; Callaerts, P. The genetic basis of natural variation in mushroom body size in Drosophila melanogaster. Nat. Commun. 2015, 6, 10115. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, A.I.; Rovescalli, A.C.; Pozzi, P.; Yoo, S.; Mozer, B.; Li, H.P.; Yu, S.H.; Higashida, H.; Guo, V.; Spencer, M.; et al. Genes required for Drosophila nervous system development identified by RNA interference. Proc. Natl. Acad. Sci. USA 2004, 101, 16216–16221. [Google Scholar] [CrossRef] [Green Version]
- Ackley, B.D.; Jin, Y. Genetic analysis of synaptic target recognition and assembly. Trends Neurosci. 2004, 27, 540–547. [Google Scholar] [CrossRef]
- Pazos Obregon, F.; Papalardo, C.; Castro, S.; Guerberoff, G.; Cantera, R. Putative synaptic genes defined from a Drosophila whole body developmental transcriptome by a machine learning approach. BMC Genom. 2015, 16, 694. [Google Scholar] [CrossRef] [Green Version]
- Harris, K.P.; Littleton, J.T. Transmission, Development, and Plasticity of Synapses. Genetics 2015, 201, 345–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.S.; Nolte, H.; Jacob, W.; Ziegler, A.B.; Putz, S.; Grosjean, Y.; Szczepanowska, K.; Trifunovic, A.; Braun, T.; Heumann, H.; et al. Human R1441C LRRK2 regulates the synaptic vesicle proteome and phosphoproteome in a Drosophila model of Parkinson’s disease. Hum. Mol. Genet. 2016, 25, 5365–5382. [Google Scholar] [CrossRef] [Green Version]
- Haussmann, I.U.; White, K.; Soller, M. Erect wing regulates synaptic growth in Drosophila by integration of multiple signaling pathways. Genome Biol. 2008, 9, R73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Suzuki, T. Activity-Dependent Synaptic Plasticity in Drosophila melanogaster. Front. Physiol. 2020, 11, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olesnicky, E.C.; Killian, D.J.; Garcia, E.; Morton, M.C.; Rathjen, A.R.; Sola, I.E.; Gavis, E.R. Extensive use of RNA-binding proteins in Drosophila sensory neuron dendrite morphogenesis. G3 2014, 4, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Usui, T.; Satoh, D.; Moriyama, S.; Shimono, K.; Itoh, T.; Shirahige, K.; Uemura, T. Sensory-neuron subtype-specific transcriptional programs controlling dendrite morphogenesis: Genome-wide analysis of Abrupt and Knot/Collier. Dev. Cell 2013, 27, 530–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrish, J.Z.; Kim, M.D.; Jan, L.Y.; Jan, Y.N. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006, 20, 820–835. [Google Scholar] [CrossRef]
- Berger, J.; Senti, K.A.; Senti, G.; Newsome, T.P.; Asling, B.; Dickson, B.J.; Suzuki, T. Systematic identification of genes that regulate neuronal wiring in the Drosophila visual system. PLoS Genet. 2008, 4, e1000085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mindorff, E.N.; O’Keefe, D.D.; Labbe, A.; Yang, J.P.; Ou, Y.; Yoshikawa, S.; van Meyel, D.J. A gain-of-function screen for genes that influence axon guidance identifies the NF-kappaB protein dorsal and reveals a requirement for the kinase Pelle in Drosophila photoreceptor axon targeting. Genetics 2007, 176, 2247–2263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraut, R.; Menon, K.; Zinn, K. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 2001, 11, 417–430. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Schuldiner, O. Axon and dendrite pruning in Drosophila. Curr. Opin. Neurobiol. 2014, 27, 192–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirilly, D.; Wong, J.J.; Lim, E.K.; Wang, Y.; Zhang, H.; Wang, C.; Liao, Q.; Wang, H.; Liou, Y.C.; Wang, H.; et al. Intrinsic epigenetic factors cooperate with the steroid hormone ecdysone to govern dendrite pruning in Drosophila. Neuron 2011, 72, 86–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alyagor, I.; Berkun, V.; Keren-Shaul, H.; Marmor-Kollet, N.; David, E.; Mayseless, O.; Issman-Zecharya, N.; Amit, I.; Schuldiner, O. Combining Developmental and Perturbation-Seq Uncovers Transcriptional Modules Orchestrating Neuronal Remodeling. Dev. Cell 2018, 47, 38–52.e36. [Google Scholar] [CrossRef] [Green Version]
- Hoopfer, E.D.; Penton, A.; Watts, R.J.; Luo, L. Genomic analysis of Drosophila neuronal remodeling: A role for the RNA-binding protein Boule as a negative regulator of axon pruning. J. Neurosci. 2008, 28, 6092–6103. [Google Scholar] [CrossRef] [Green Version]
- Chew, L.Y.; Zhang, H.; He, J.; Yu, F. The Nrf2-Keap1 pathway is activated by steroid hormone signaling to govern neuronal remodeling. Cell Rep. 2021, 36, 109466. [Google Scholar] [CrossRef]
- Chen, D.; Qu, C.; Bjorum, S.M.; Beckingham, K.M.; Hewes, R.S. Neuronal remodeling during metamorphosis is regulated by the alan shepard (shep) gene in Drosophila melanogaster. Genetics 2014, 197, 1267–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, W.P.; Mateos, A.; Koch, M.; Klussman, S.; Yang, C.; Lu, N.; Kumar, S.; Limpert, S.; Gopferich, M.; Zschaetzsch, M.; et al. Regulation of Adult CNS Axonal Regeneration by the Post-transcriptional Regulator Cpeb1. Front. Mol. Neurosci. 2017, 10, 445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecuyer, E.; Yoshida, H.; Parthasarathy, N.; Alm, C.; Babak, T.; Cerovina, T.; Hughes, T.R.; Tomancak, P.; Krause, H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007, 131, 174–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Sartain, C.V.; Pleiss, J.A.; Wolfner, M.F. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev. Biol. 2013, 383, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Hughes, J.R.; Meireles, A.M.; Fisher, K.H.; Garcia, A.; Antrobus, P.R.; Wainman, A.; Zitzmann, N.; Deane, C.; Ohkura, H.; Wakefield, J.G. A microtubule interactome: Complexes with roles in cell cycle and mitosis. PLoS Biol. 2008, 6, e98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, K.H.; Deane, C.M.; Wakefield, J.G. The functional domain grouping of microtubule associated proteins. Commun. Integr. Biol. 2008, 1, 47–50. [Google Scholar] [CrossRef] [Green Version]
- Prokop, A.; Beaven, R.; Qu, Y.; Sanchez-Soriano, N. Using fly genetics to dissect the cytoskeletal machinery of neurons during axonal growth and maintenance. J. Cell Sci. 2013, 126, 2331–2341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eade, K.T.; Fancher, H.A.; Ridyard, M.S.; Allan, D.W. Developmental transcriptional networks are required to maintain neuronal subtype identity in the mature nervous system. PLoS Genet. 2012, 8, e1002501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacin, H.; Zhu, Y.; Wilson, B.A.; Skeath, J.B. Transcription factor expression uniquely identifies most postembryonic neuronal lineages in the Drosophila thoracic central nervous system. Development 2014, 141, 1011–1021. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Dumelie, J.G.; Li, X.; Cheng, M.H.; Yang, Z.; Laver, J.D.; Siddiqui, N.U.; Westwood, J.T.; Morris, Q.; Lipshitz, H.D.; et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol. 2014, 15, R4. [Google Scholar] [CrossRef]
- Mohr, C.; Hartmann, B. Alternative splicing in Drosophila neuronal development. J. Neurogenet. 2014, 28, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Buhlman, L.M.; Krishna, G.; Jones, T.B.; Thomas, T.C. Drosophila as a model to explore secondary injury cascades after traumatic brain injury. Biomed. Pharm. 2021, 142, 112079. [Google Scholar] [CrossRef]
- Mbodj, A.; Junion, G.; Brun, C.; Furlong, E.E.; Thieffry, D. Logical modelling of Drosophila signalling pathways. Mol. Biosyst. 2013, 9, 2248–2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, Z.; Ranjan, R.; Wenniger, J.J.; Hong, S.N.; Bronk, P.; Zinsmaier, K.E. Overexpression of cysteine-string proteins in Drosophila reveals interactions with syntaxin. J. Neurosci. 1999, 19, 10270–10279. [Google Scholar] [CrossRef] [Green Version]
- Sang, Q.; Wang, G.; Morton, D.B.; Wu, H.; Xie, B. The ZO-1 protein Polychaetoid as an upstream regulator of the Hippo pathway in Drosophila. PLoS Genet. 2021, 17, e1009894. [Google Scholar] [CrossRef] [PubMed]
- Seppa, M.J.; Johnson, R.I.; Bao, S.; Cagan, R.L. Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev. Biol. 2008, 318, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Saigo, K. Transcriptional regulation of atonal required for Drosophila larval eye development by concerted action of eyes absent, sine oculis and hedgehog signaling independent of fused kinase and cubitus interruptus. Development 2000, 127, 1531–1540. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozlov, E.N.; Tokmatcheva, E.V.; Khrustaleva, A.M.; Grebenshchikov, E.S.; Deev, R.V.; Gilmutdinov, R.A.; Lebedeva, L.A.; Zhukova, M.; Savvateeva-Popova, E.V.; Schedl, P.; et al. Long-Term Memory Formation in Drosophila Depends on the 3′UTR of CPEB Gene orb2. Cells 2023, 12, 318. https://doi.org/10.3390/cells12020318
Kozlov EN, Tokmatcheva EV, Khrustaleva AM, Grebenshchikov ES, Deev RV, Gilmutdinov RA, Lebedeva LA, Zhukova M, Savvateeva-Popova EV, Schedl P, et al. Long-Term Memory Formation in Drosophila Depends on the 3′UTR of CPEB Gene orb2. Cells. 2023; 12(2):318. https://doi.org/10.3390/cells12020318
Chicago/Turabian StyleKozlov, Eugene N., Elena V. Tokmatcheva, Anastasia M. Khrustaleva, Eugene S. Grebenshchikov, Roman V. Deev, Rudolf A. Gilmutdinov, Lyubov A. Lebedeva, Mariya Zhukova, Elena V. Savvateeva-Popova, Paul Schedl, and et al. 2023. "Long-Term Memory Formation in Drosophila Depends on the 3′UTR of CPEB Gene orb2" Cells 12, no. 2: 318. https://doi.org/10.3390/cells12020318
APA StyleKozlov, E. N., Tokmatcheva, E. V., Khrustaleva, A. M., Grebenshchikov, E. S., Deev, R. V., Gilmutdinov, R. A., Lebedeva, L. A., Zhukova, M., Savvateeva-Popova, E. V., Schedl, P., & Shidlovskii, Y. V. (2023). Long-Term Memory Formation in Drosophila Depends on the 3′UTR of CPEB Gene orb2. Cells, 12(2), 318. https://doi.org/10.3390/cells12020318




