Mesothelial Cells Exhibit Characteristics of Perivascular Cells in an In Vitro Angiogenesis Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Basic Cell Culture
2.2. Lentiviral Infections
2.3. Flow Cytometry
2.4. Cell Culture Experiments
2.4.1. GFP-MC Stimulation
2.4.2. Culture of GFP-MCs on NHDFs
2.4.3. Basic Angiogenesis Assay
2.4.4. Cardiac Angiogenesis Assay Using HCMECs and HCFs
2.4.5. GFP-MCs in the Cardiac Spheroid Model
2.5. Boyden Chamber Assay
2.6. Immunofluorescence
2.7. Live Cell Imaging of GFP-MCs in Angiogenesis Co-Culture
2.8. Image Analysis
2.9. qRT-PCR
2.10. Statistics and Data Analysis
3. Results
3.1. Mesothelial Cells Showed Specific Alignment with Endothelial Tubes in an In Vitro Angiogenesis Assay
3.2. Mesothelial Cells Positively Affected Branch Point Formation
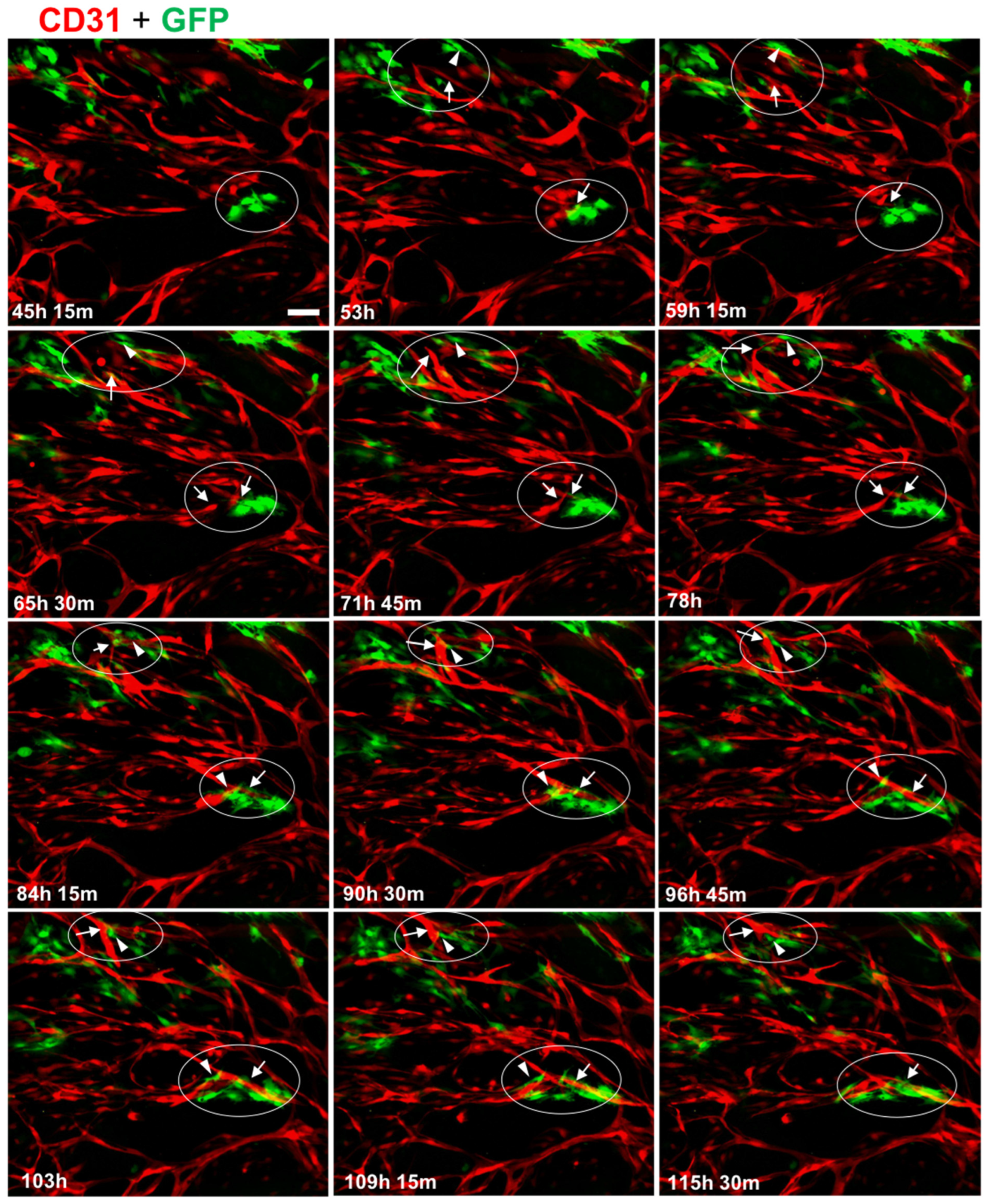
3.3. MC Interacted with and Supported Endothelial Tube Formation
3.4. Mesothelial Cells Upregulated Expression of the Pericyte Marker Ng2 When Co-Cultured in the Angiogenesis Assay
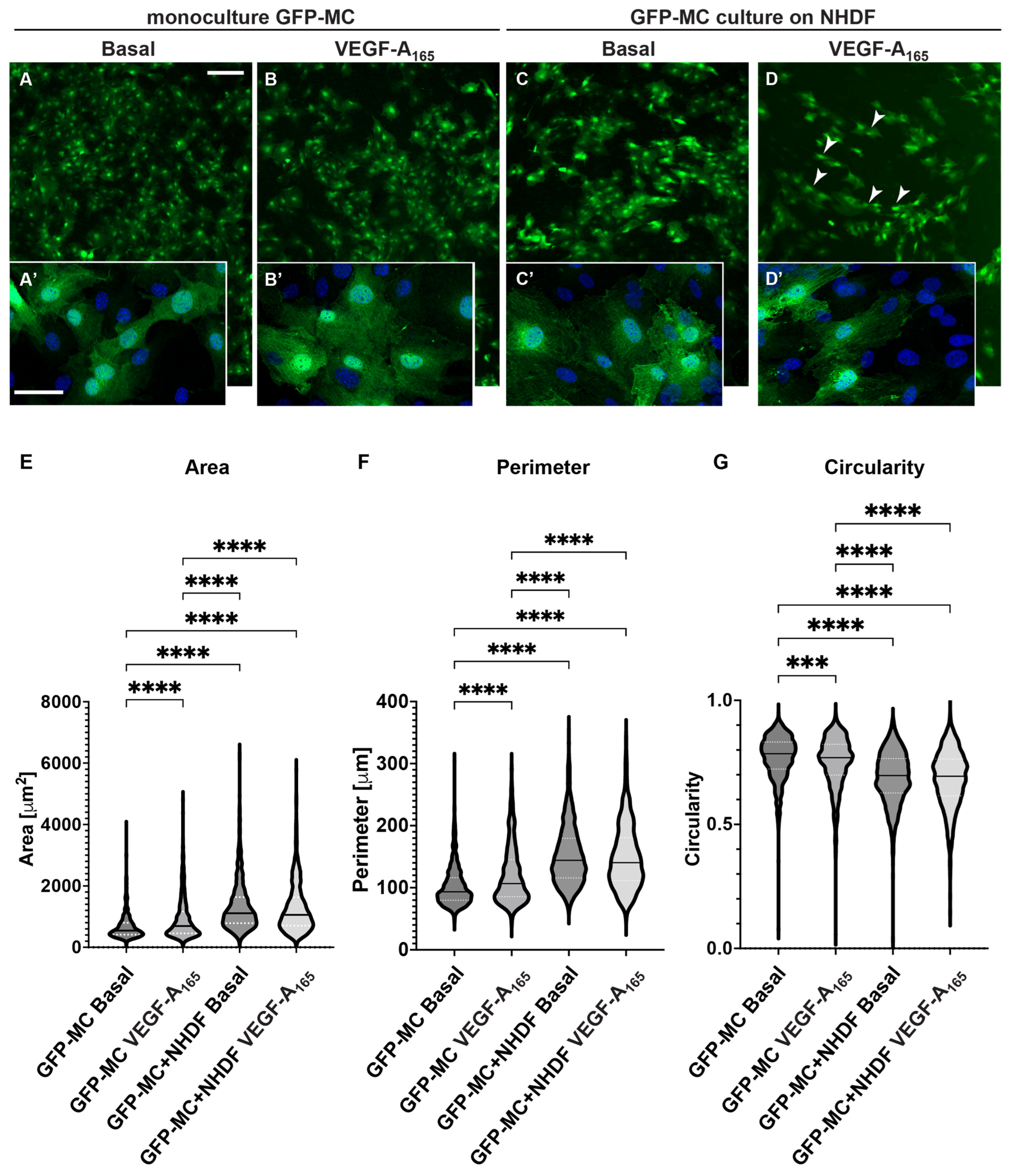
3.5. Mesothelial Cells Changed Cell Shape in Response to Culture on Fibroblast Feeder Cells and with VEGF
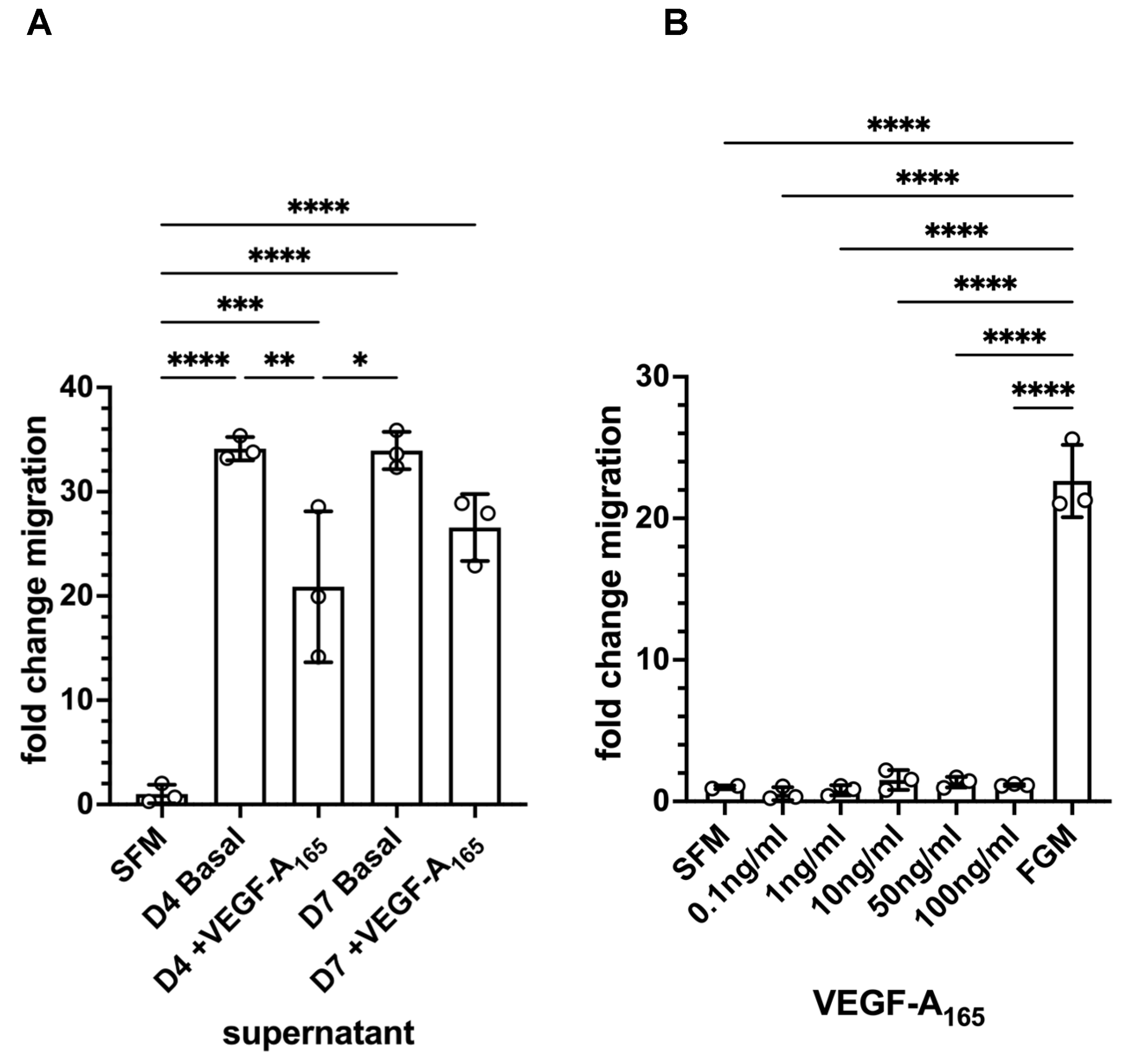
3.6. Mesothelial Cells Showed Migratory Behaviour in Response to Supernatant from HDMEC–NHDF Co-Culture but Not VEGF
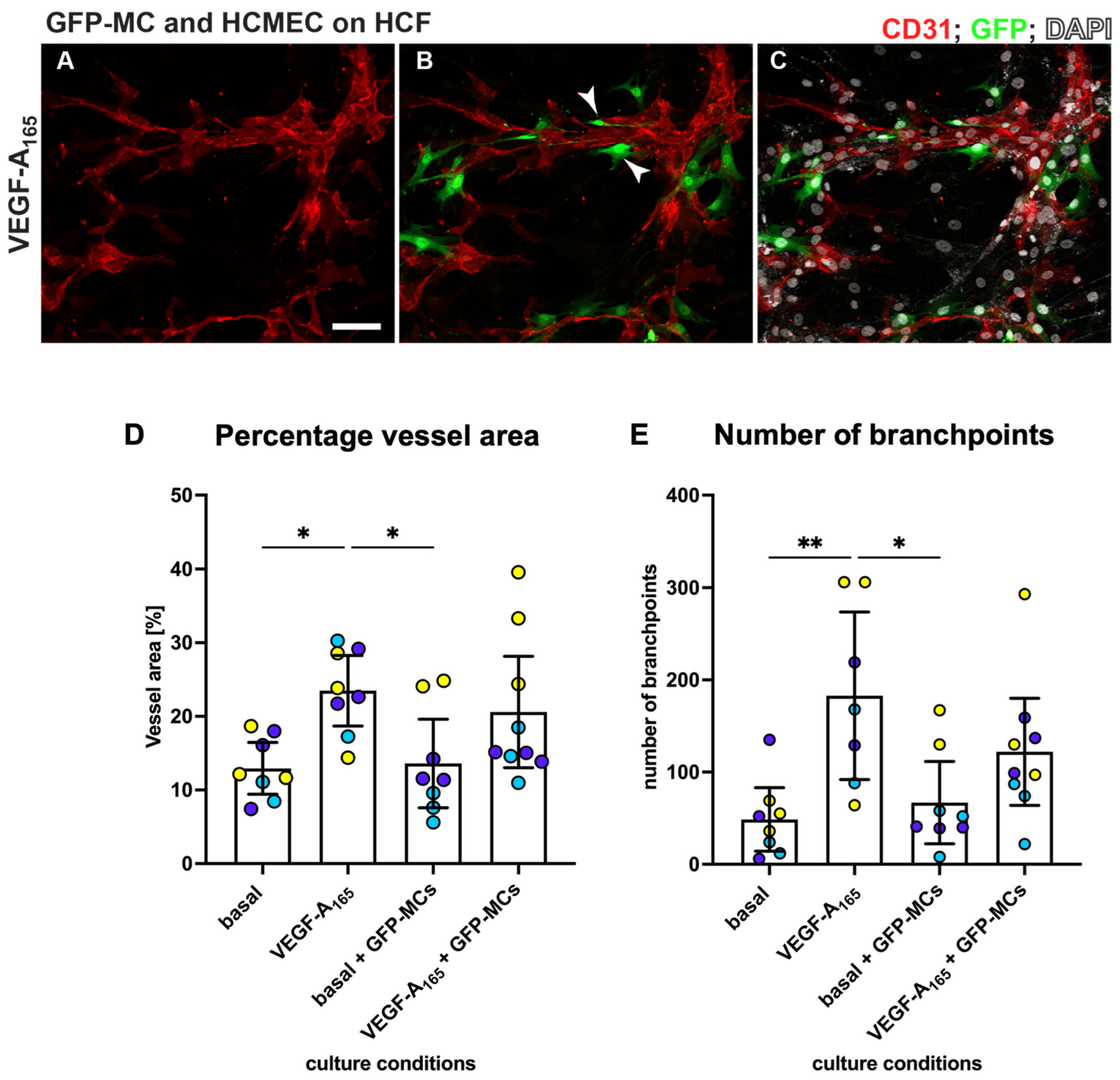
3.7. Mesothelial Cells Aligned and Supported Tube Formation in a Cardiac Angiogenesis Assay
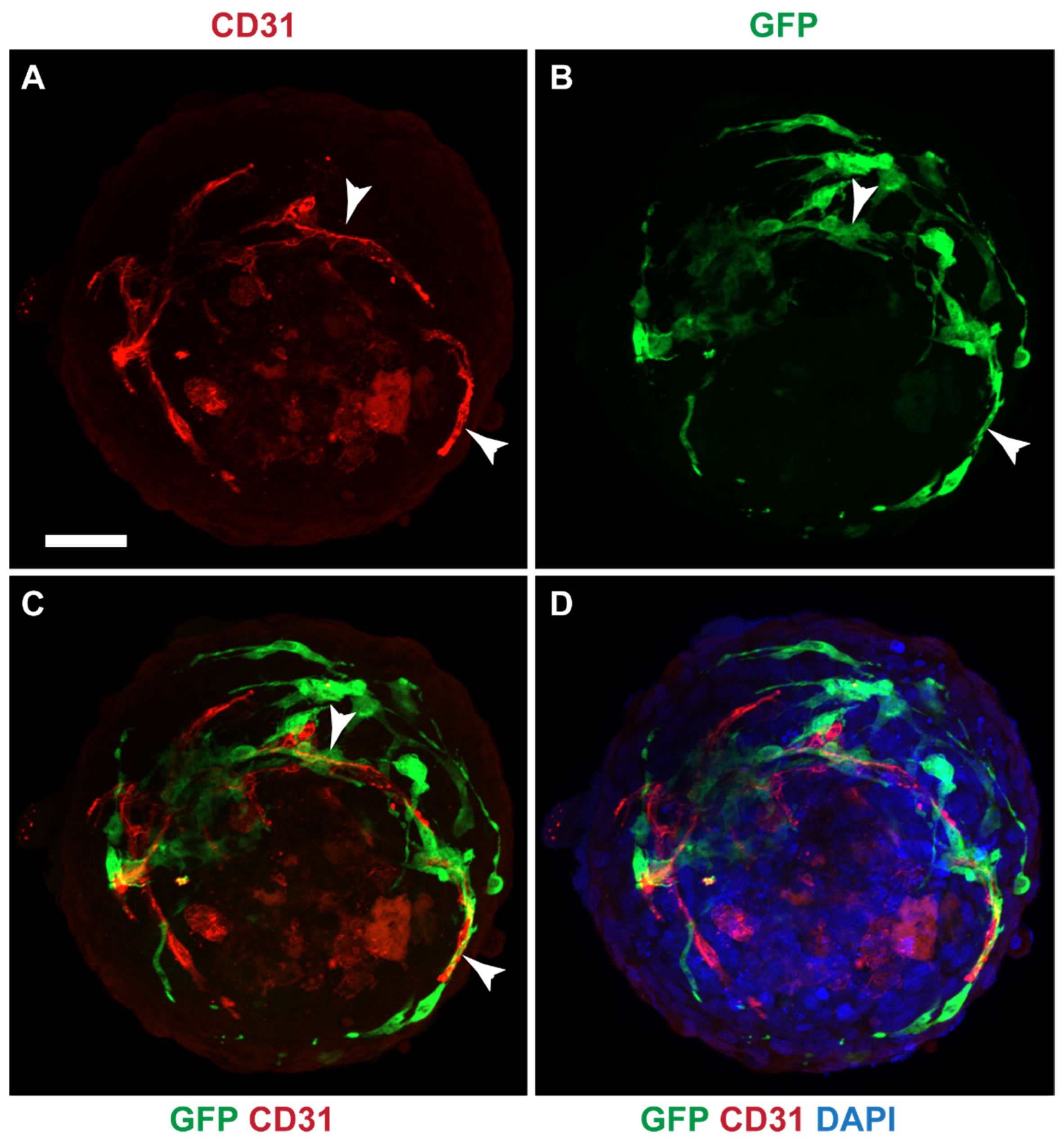
3.8. Mesothelial Cells Integrated in a Cardiac 3D Spheroid Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mutsaers, S.E. Mesothelial cells: Their structure, function and role in serosal repair. Respirology 2002, 7, 171–191. [Google Scholar] [CrossRef] [PubMed]
- Mutsaers, S.E.; Birnie, K.; Lansley, S.; Herrick, S.E.; Lim, C.B.; Prele, C.M. Mesothelial cells in tissue repair and fibrosis. Front. Pharmacol. 2015, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Buechler, M.B.; Kim, K.W.; Onufer, E.J.; Williams, J.W.; Little, C.C.; Dominguez, C.X.; Li, Q.; Sandoval, W.; Cooper, J.E.; Harris, C.A.; et al. A Stromal Niche Defined by Expression of the Transcription Factor WT1 Mediates Programming and Homeostasis of Cavity-Resident Macrophages. Immunity 2019, 51, 119–130.e5. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Bader, D.M.; Wilm, B. Serosal mesothelium retains vasculogenic potential (Developmental Dynamics 236, (2973–2979)). Dev. Dyn. 2008, 237, 525. [Google Scholar] [CrossRef]
- Que, J.; Wilm, B.; Hasegawa, H.; Wang, F.; Bader, D.; Hogan, B.L. Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc. Natl. Acad. Sci. USA 2008, 105, 16626–16630. [Google Scholar] [CrossRef]
- Wilm, B.; Ipenberg, A.; Hastie, N.D.; Burch, J.B.E.; Bader, D.M. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 2005, 132, 5317–5328. [Google Scholar] [CrossRef]
- Shao, Y.; Saredy, J.; Yang, W.Y.; Sun, Y.; Lu, Y.; Saaoud, F.; Drummer, C.t.; Johnson, C.; Xu, K.; Jiang, X.; et al. Vascular Endothelial Cells and Innate Immunity. Arterioscler. Thromb. Vasc. Biol. 2020, 40, e138–e152. [Google Scholar] [CrossRef]
- Trimm, E.; Red-Horse, K. Vascular endothelial cell development and diversity. Nat. Rev. Cardiol. 2023, 20, 197–210. [Google Scholar] [CrossRef]
- Majesky, M.W. Vascular Development. Arterioscler. Thromb. Vasc. Biol. 2018, 38, e17–e24. [Google Scholar] [CrossRef]
- Sumpio, B.E.; Riley, J.T.; Dardik, A. Cells in focus: Endothelial cell. Int. J. Biochem. Cell Biol. 2002, 34, 1508–1512. [Google Scholar] [CrossRef]
- Risau, W.; Flamme, I. Vasculogenesis. Annu. Rev. Cell Dev. Biol. 1995, 11, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Cazares, D.; Chavez-Dominguez, R.; Carlos-Reyes, A.; Lopez-Camarillo, C.; Hernadez de la Cruz, O.N.; Lopez-Gonzalez, J.S. Contribution of Angiogenesis to Inflammation and Cancer. Front. Oncol. 2019, 9, 1399. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P. Mechanisms of angiogenesis and arteriogenesis. Nat. Med. 2000, 6, 389–395. [Google Scholar] [CrossRef]
- Adams, R.H.; Alitalo, K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 2007, 8, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Herbert, S.P.; Stainier, D.Y. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat. Rev. Mol. Cell Biol. 2011, 12, 551–564. [Google Scholar] [CrossRef]
- Gaengel, K.; Genove, G.; Armulik, A.; Betsholtz, C. Endothelial-mural cell signaling in vascular development and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 630–638. [Google Scholar] [CrossRef]
- Geevarghese, A.; Herman, I.M. Pericyte-endothelial crosstalk: Implications and opportunities for advanced cellular therapies. Transl. Res. 2014, 163, 296–306. [Google Scholar] [CrossRef]
- Hetheridge, C.; Mavria, G.; Mellor, H. Uses of the in vitro endothelial-fibroblast organotypic co-culture assay in angiogenesis research. Biochem. Soc. Trans. 2011, 39, 1597–1600. [Google Scholar] [CrossRef]
- Mavria, G.; Vercoulen, Y.; Yeo, M.; Paterson, H.; Karasarides, M.; Marais, R.; Bird, D.; Marshall, C.J. ERK-MAPK signaling opposes Rho-kinase to promote endothelial cell survival and sprouting during angiogenesis. Cancer Cell 2006, 9, 33–44. [Google Scholar] [CrossRef]
- Richards, M.; Mellor, H. In Vitro Coculture Assays of Angiogenesis. Methods Mol. Biol. 2016, 1430, 159–166. [Google Scholar] [CrossRef]
- Sorrell, J.M.; Baber, M.A.; Caplan, A.I. A self-assembled fibroblast-endothelial cell co-culture system that supports in vitro vasculogenesis by both human umbilical vein endothelial cells and human dermal microvascular endothelial cells. Cells Tissues Organs 2007, 186, 157–168. [Google Scholar] [CrossRef]
- Muniz-Garcia, A.; Wilm, B.; Murray, P.; Cross, M.J. Extracellular Vesicles from Human Umbilical Cord-Derived MSCs Affect Vessel Formation In Vitro and Promote VEGFR2-Mediated Cell Survival. Cells 2022, 11, 3750. [Google Scholar] [CrossRef] [PubMed]
- Sorrell, J.M.; Baber, M.A.; Caplan, A.I. Human dermal fibroblast subpopulations; differential interactions with vascular endothelial cells in coculture: Nonsoluble factors in the extracellular matrix influence interactions. Wound Repair Regen. 2008, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Dauleh, S.; Santeramo, I.; Fielding, C.; Ward, K.; Herrmann, A.; Murray, P.; Wilm, B. Characterisation of cultured mesothelial cells derived from the murine adult omentum. PLoS ONE 2016, 11, e0158997. [Google Scholar] [CrossRef]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder-Geleff, S.; Soleiman, A.; Kowalski, H.; Horvat, R.; Amann, G.; Kriehuber, E.; Diem, K.; Weninger, W.; Tschachler, E.; Alitalo, K.; et al. Angiosarcomas express mixed endothelial phenotypes of blood and lymphatic capillaries: Podoplanin as a specific marker for lymphatic endothelium. Am. J. Pathol. 1999, 154, 385–394. [Google Scholar] [CrossRef]
- Jiang, S.; Bailey, A.S.; Goldman, D.C.; Swain, J.R.; Wong, M.H.; Streeter, P.R.; Fleming, W.H. Hematopoietic stem cells contribute to lymphatic endothelium. PLoS ONE 2008, 3, e3812. [Google Scholar] [CrossRef]
- Kong, L.L.; Yang, N.Z.; Shi, L.H.; Zhao, G.H.; Zhou, W.; Ding, Q.; Wang, M.H.; Zhang, Y.S. The optimum marker for the detection of lymphatic vessels. Mol. Clin. Oncol. 2017, 7, 515–520. [Google Scholar] [CrossRef]
- Wilting, J.; Papoutsi, M.; Christ, B.; Nicolaides, K.H.; von Kaisenberg, C.S.; Borges, J.; Stark, G.B.; Alitalo, K.; Tomarev, S.I.; Niemeyer, C.; et al. The transcription factor Prox1 is a marker for lymphatic endothelial cells in normal and diseased human tissues. FASEB J. 2002, 16, 1271–1273. [Google Scholar] [CrossRef]
- Ravenscroft, S.M.; Pointon, A.; Williams, A.W.; Cross, M.J.; Sidaway, J.E. Cardiac Non-myocyte Cells Show Enhanced Pharmacological Function Suggestive of Contractile Maturity in Stem Cell Derived Cardiomyocyte Microtissues. Toxicol. Sci. 2016, 152, 99–112. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Zudaire, E.; Gambardella, L.; Kurcz, C.; Vermeren, S. A computational tool for quantitative analysis of vascular networks. PLoS ONE 2011, 6, e27385. [Google Scholar] [CrossRef] [PubMed]
- Stringer, C.; Wang, T.; Michaelos, M.; Pachitariu, M. Cellpose: A generalist algorithm for cellular segmentation. Nat. Methods 2021, 18, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Roberts, O.L.; Holmes, K.; Muller, J.; Cross, D.A.; Cross, M.J. ERK5 is required for VEGF-mediated survival and tubular morphogenesis of primary human microvascular endothelial cells. J. Cell Sci. 2010, 123, 3189–3200. [Google Scholar] [CrossRef]
- Maturi, V.; Enroth, S.; Heldin, C.H.; Moustakas, A. Genome-wide binding of transcription factor ZEB1 in triple-negative breast cancer cells. J. Cell Physiol. 2018, 233, 7113–7127. [Google Scholar] [CrossRef]
- Ozerdem, U.; Grako, K.A.; Dahlin-Huppe, K.; Monosov, E.; Stallcup, W.B. NG2 proteoglycan is expressed exclusively by mural cells during vascular morphogenesis. Dev. Dyn. 2001, 222, 218–227. [Google Scholar] [CrossRef]
- Davidson, M.M.; Nesti, C.; Palenzuela, L.; Walker, W.F.; Hernandez, E.; Protas, L.; Hirano, M.; Isaac, N.D. Novel cell lines derived from adult human ventricular cardiomyocytes. J. Mol. Cell Cardiol. 2005, 39, 133–147. [Google Scholar] [CrossRef]
- Seynhaeve, A.L.B.; Oostinga, D.; van Haperen, R.; Eilken, H.M.; Adams, S.; Adams, R.H.; Ten Hagen, T.L.M. Spatiotemporal endothelial cell—Pericyte association in tumors as shown by high resolution 4D intravital imaging. Sci. Rep. 2018, 8, 9596. [Google Scholar] [CrossRef]
- Aguilera, A.; Aroeira, L.S.; Ramirez-Huesca, M.; Perez-Lozano, M.L.; Cirugeda, A.; Bajo, M.A.; Del Peso, G.; Valenzuela-Fernandez, A.; Sanchez-Tomero, J.A.; Lopez-Cabrera, M.; et al. Effects of rapamycin on the epithelial-to-mesenchymal transition of human peritoneal mesothelial cells. Int. J. Artif. Organs 2005, 28, 164–169. [Google Scholar] [CrossRef]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Amack, J.D. Cellular dynamics of EMT: Lessons from live in vivo imaging of embryonic development. Cell Commun. Signal. 2021, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Gupta, V.; Wang, J.; Holdway, J.E.; Wills, A.A.; Fang, Y.; Poss, K.D. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 2011, 138, 2895–2902. [Google Scholar] [CrossRef] [PubMed]
- Lepilina, A.; Coon, A.N.; Kikuchi, K.; Holdway, J.E.; Roberts, R.W.; Burns, C.G.; Poss, K.D. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006, 127, 607–619. [Google Scholar] [CrossRef]
- Chintalgattu, V.; Rees, M.L.; Culver, J.C.; Goel, A.; Jiffar, T.; Zhang, J.; Dunner, K., Jr.; Pati, S.; Bankson, J.A.; Pasqualini, R.; et al. Coronary microvascular pericytes are the cellular target of sunitinib malate-induced cardiotoxicity. Sci. Transl. Med. 2013, 5, 187ra169. [Google Scholar] [CrossRef] [PubMed]
- Ferland-McCollough, D.; Slater, S.; Richard, J.; Reni, C.; Mangialardi, G. Pericytes, an overlooked player in vascular pathobiology. Pharmacol. Ther. 2017, 171, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Marino, F.; Salerno, L.; Cianflone, E.; Molinaro, C.; Salerno, N.; De Angelis, A.; Viglietto, G.; Urbanek, K.; Torella, D. From Spheroids to Organoids: The Next Generation of Model Systems of Human Cardiac Regeneration in a Dish. Int. J. Mol. Sci. 2021, 22, 3180. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koukorava, C.; Ward, K.; Ahmed, K.; Almaghrabi, S.; Dauleh, S.; Pereira, S.M.; Taylor, A.; Haddrick, M.; Cross, M.J.; Wilm, B. Mesothelial Cells Exhibit Characteristics of Perivascular Cells in an In Vitro Angiogenesis Assay. Cells 2023, 12, 2436. https://doi.org/10.3390/cells12202436
Koukorava C, Ward K, Ahmed K, Almaghrabi S, Dauleh S, Pereira SM, Taylor A, Haddrick M, Cross MJ, Wilm B. Mesothelial Cells Exhibit Characteristics of Perivascular Cells in an In Vitro Angiogenesis Assay. Cells. 2023; 12(20):2436. https://doi.org/10.3390/cells12202436
Chicago/Turabian StyleKoukorava, Chrysa, Kelly Ward, Katie Ahmed, Shrouq Almaghrabi, Sumaya Dauleh, Sofia M. Pereira, Arthur Taylor, Malcolm Haddrick, Michael J. Cross, and Bettina Wilm. 2023. "Mesothelial Cells Exhibit Characteristics of Perivascular Cells in an In Vitro Angiogenesis Assay" Cells 12, no. 20: 2436. https://doi.org/10.3390/cells12202436
APA StyleKoukorava, C., Ward, K., Ahmed, K., Almaghrabi, S., Dauleh, S., Pereira, S. M., Taylor, A., Haddrick, M., Cross, M. J., & Wilm, B. (2023). Mesothelial Cells Exhibit Characteristics of Perivascular Cells in an In Vitro Angiogenesis Assay. Cells, 12(20), 2436. https://doi.org/10.3390/cells12202436






