1. Introduction
Glioblastoma multiforme (GBM) is a deadly brain tumor affecting many people for whom treatment options at the moment are extremely limited [
1]. GBM occurs when progenitor cells of astroglial origin transform due to anomalies in signaling mechanisms that are essential for their differentiation into more mature cells. The major complication in treating GBM patients lies in the heterogeneity of the origin of the tumor [
2,
3,
4]. Alterations in many signaling molecules, transcription factors (TFs), co-factors, and epigenetic modifying molecules have been implicated in the pathogenesis of GBM [
5,
6,
7,
8,
9]. The rapid progression and invasive properties of GBM make it very hard to treat GBM patients. Current protocols for the treatment of GBM patients involve the maximal surgical removal of the tumor tissue followed by chemotherapy with the alkylating agent temozolomide and radiotherapy. However, this treatment option is highly ineffective, and the survival rate of GBM patients post-treatment remains poor, at only 1.5 years. The tumor relapses very quickly and becomes resistant to temozolomide treatment, adding complications to the treatment strategy. Therefore, there is a huge need for the development of novel treatment options to prevent not only GBM pathogenesis but also its relapse following treatment [
10,
11,
12].
The signaling molecules of Janus-associated kinase (JAK) and TFs of signal transducer and activator of transcription (STAT) families are intricately involved in the proliferation, differentiation, and function of various cells [
13]. There are four JAK family kinases, JAK1, JAK2, JAK3, and TYK2, present in various cells and involved in the activation of the STAT TFs. Whereas JAK1 and JAK2 are universally present in all cells, JAK3’s distribution has been reported to be prevalently in hematopoietic cells. Accordingly, the phenotypes of
Jak1-/- and
Jak2-/- mice are more severe compared with
Jak3-/- mice, where gross hematopoietic anomalies were reported [
14,
15]. Cell surface receptor ligation leads to the phosphorylation of JAK proteins in the cytoplasmic tails, which subsequently leads to the recruitment of STAT proteins and their phosphorylation by JAKs. There are six STAT proteins, STAT1, STAT2, STAT3, STAT4, STAT5, and STAT6, and their cellular distribution is variable, with STAT5 being more active in hematopoietic cells and STAT3 in other cell types. JAK-STAT signaling can be activated by a host of signaling molecules such as cytokines, growth factors, inflammatory molecules, etc., which is both beneficial and harmful depending on the context [
16].
Apart from its roles in cellular proliferation, differentiation, and function, JAK-STAT signaling is a prime factor in tumorigenesis. Gain-of-function mutations in JAK and STAT molecules have been reported in both hematological and non-hematological tumors [
17,
18]. JAK-STAT involvement in GBM pathogenesis has been reported [
19,
20]. Among the JAK-STAT family members, JAK2 and STAT3’s involvement in GBM has been widely studied, and their enhanced activity has been linked to the severity of GBM [
21,
22]. Increased JAK-STAT signaling has been reported to maintain the stemness of GBM cells by upregulating and maintaining the expression of various stem cell genes such as CD44, NESTIN, PROMININ, PAX6, etc. [
23]. Thereby, it facilitates tumor relapse following surgery and chemotherapy. Therefore, the inhibition of JAK-STAT signaling in GBM remains a possible therapeutic option. As the epigenetic machinery in tumor cells is unstable and they are prone to frequent modifications, genetic intervention to downregulate JAK-STAT activity in GBM cells is not a very attractive option. Small-molecule inhibitors of JAKs or STATs are a better option to inhibit their activity in GBM and hold promise, as reported in multiple studies. Herein, we used the specific JAK3 inhibitors WHI-P131 (4-(4′-hydroxylphenyl)-amino-6,7-dimethoxyquinazoline) and PF-956980 (((3R,4R)-4-Methyl-3-(methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino)piperidin-1-yl)(pyrrolidin1-yl)methanone) to evaluate their efficacy in preventing GBM cell proliferation, inducing their differentiation, and in downregulating their stemness characteristics. Our results show that both small-molecule inhibitors of JAK3 signaling are very effective in blocking GBM cell proliferation and neurosphere formation, and in inducing differentiation into neurons and other neuronal lineage cells. Interestingly, these effects were executed by downregulating the stem cell characteristics of the GBM cells. Overall, our study demonstrates the potential therapeutic value of these JAK3 inhibitors in preventing GBM pathogenesis and relapse.
2. Materials and Methods
2.1. Cell Lines
Human GBM cell lines U87 and U251 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS), 100 units of penicillin and streptomycin, 1% non-essential amino acids, 1% HEPES, and 0.05 mM of 2-mercaptoethanol. The cells were maintained in a 37 °C incubator with 5% CO2 and were regularly checked for contamination. To avoid continuous prolonged culture, a new vial from the frozen stock was revitalized every six months and used for experiments. Mycoplasma contamination was regularly checked using a PCR-based detection kit (Sigma Aldrich, St Louis, MI, USA).
2.2. Proliferation and Differentiation Assays
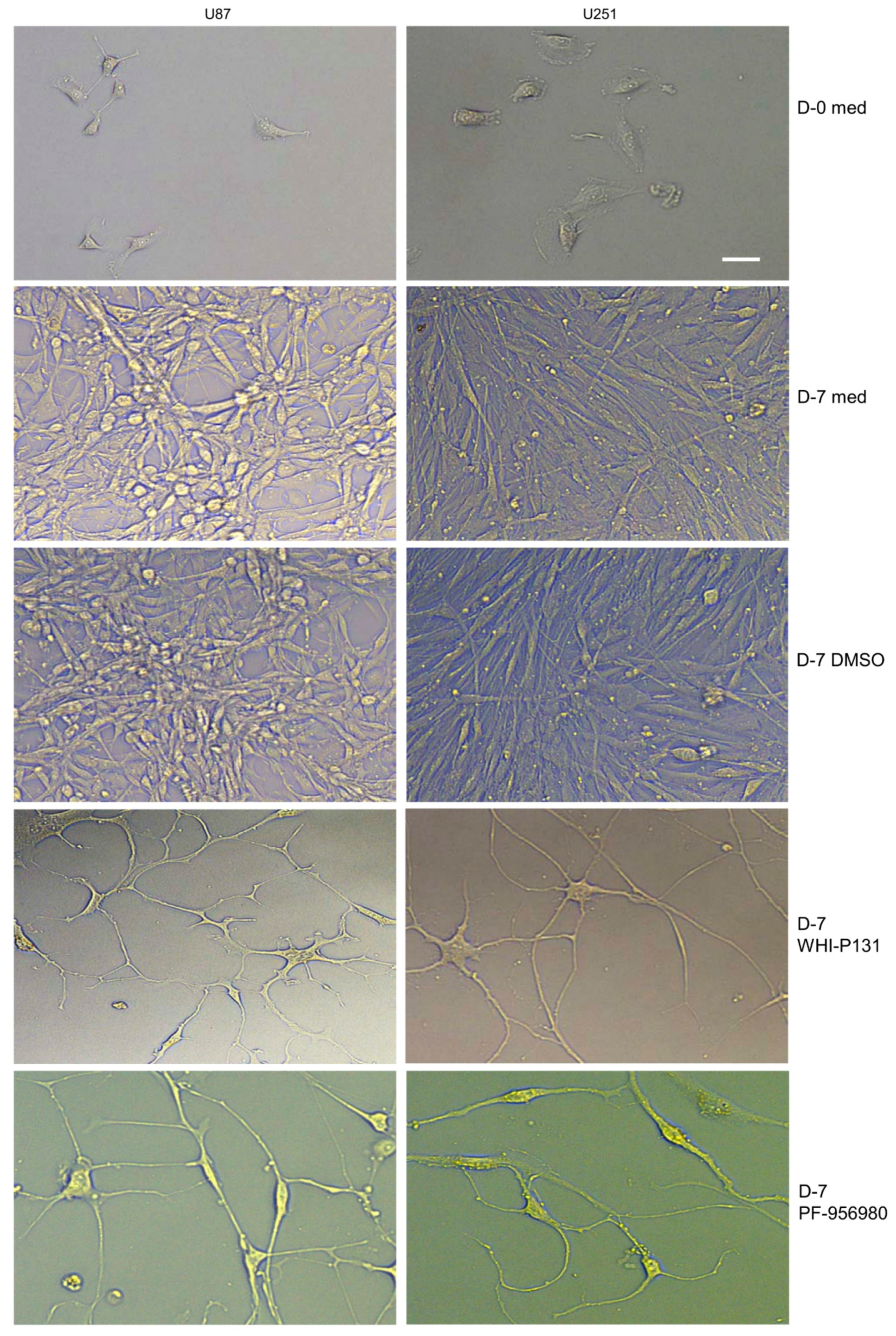
Single-cell suspensions of 5000 U87 or U251 cells were cultured in 1 mL of complete DMEM in each well of a 12-well plate. Cells were cultured in the presence or absence of 50 μM of WHI-P131 or 250 μM of PF-956980 for 7 or 14 days. An equal volume of DMSO as that of the drugs was added to the control cells. Afterward, the cells were collected, and the total number was counted according to the trypan blue exclusion method to check the effects of the drugs on GBM cell proliferation. Cellular differentiation into neuronal lineage cells in JAK3-inhibitor-treated wells and control cells was recorded on day 7 of culture using a Leica IM8 microscope.
2.3. CFSE Staining
A total of 1 × 10
6 U87 or U251 cells were stained with carboxyfluoresceine succinimidyl ester (CFSE) as described previously [
24]. Briefly, cells were collected via trypsinization, and a single-cell suspension was prepared. The cells were washed 2 times with PBS and stained with 2 μM of CFSE in PBS for 5 min in the dark at room temperature (RT). Following staining, the cells were washed with complete DMEM, resuspended in complete DMEM, and counted. A total of 10,000 CFSE-stained U87 or U251 cells were cultured in 1 mL of complete DMEM in each well of a 12-well plate in the presence or absence of 50 μM of WHI-P131 or 250 μM of PF-956980. Cells were imaged using a Leica IM8 microscope at D-0 and at 24 h of culture to evaluate CFSE dilution.
2.4. Cell Death Analysis
2.4.1. Annexin V/PI Staining
A total of 1 × 10
4 U87 or U251 cells were cultured in 1 mL of complete DMEM in each well of a 12-well plate in the presence or absence of 50 μM of WHI-P131 or 250 μM of PF-956980. At 48 h of culture, cell death was analyzed using annexin V and propidium iodide (PI) staining. The cells were first stained with annexin V following the manufacturer’s protocol (BD Biosciences, Franklin Lakes, NJ, USA), and subsequently, PI (0.5 μg/mL) was added to the cells, and they were analyzed immediately using the FACS Aria and FACS Diva software (
https://www.bdbiosciences.com/ja-jp/products/software/instrument-software/bd-facsdiva-software, Beckton Dickinson, Heidelberg, Germany).
2.4.2. TUNEL Assay
A total of 1 × 104 U87 or U251 cells were cultured in 1 mL of complete DMEM in each well of a 12-well plate in the presence or absence of 50 μM of WHI-P131 or 250 μM of PF-956980. At 48 h of culture, cell death was analyzed via TUNEL assay using the TUNEL Assay Kit-FITC (Abcam, Cambridge, UK) following the manufacturer’s protocol. Live and dead cells were analyzed using the FACS Aria and FACS Diva software (Beckton Dickinson, Heidelberg, Germany).
2.5. MTT Assay
A total of 1 × 104 U87 or U251 cells/well treated with or without 50 μM of WHI-P131 or 250 μM of PF-956980 were plated in a flat-bottom 96-well culture plate. For each condition, cells were plated in triplicate. The metabolic fitness of the control and drug-treated cells was assessed 48 h later using MTT reagent following the manufacturer’s instructions. Briefly, 10 μL of MTT reagent was added to 100 μL of a medium containing the cells and incubated at 37 °C for 4 h. Subsequently, the supernatant was removed, and 150 μL of DMSO was added to solubilize the formazan crystals. Afterward, the absorbance at 570 nm was measured using a plate reader (Molecular Devices, San Jose, CA, USA).
2.6. DNA Methylation Analysis
Genomic DNA was isolated from U87 and U251 GBM cells cultured for 24 h in the presence or absence of 50 μM of WHI-P131 following the standard protocol. The DNA concentration and quality were determined by comparing the OD 260 and 280 nm observations. An amount of 100 ng of DNA from each sample was used to determine the methylated DNA status using the MethylFlash™ Methylated DNA Quantification Kit (Epigentek, Brooklyn, NY, USA) following the manufacturer’s protocol. The methylated DNA amount was estimated at OD 450 nm using a microplate reader (Molecular Devices), and the relative quantification of methylated DNA as a percentage of total DNA was calculated following the formula 5-mC % = [(Sample OD-ME3 OD)/S]/[((ME4 OD-ME3 OD) × 2)/P] × 100%, where ME3 is the negative control, ME4 is the positive control, S is the amount of input sample DNA in ng, and P is the amount of input positive control (ME4) in ng. All DNA samples were measured in duplicate.
2.7. DNMT Activity Assay
Nuclear extracts (NEs) were prepared from 1 × 106 U87 or U251 cells cultured for 24 h in the presence or absence of 50 μM of WHI-P131, as described previously, and the protein amount was estimated using the BCA reagent. An amount of 10 μg of nuclear protein from each sample was used to determine the total DNA methyl transferase (DNMT) activity using the EpiQuik™ DNA Methyltransferase Activity/Inhibition Assay Ultra Kit (EpiGentek, Brooklyn, NY, USA) according to the manufacturer’s instructions. The total DNMT activity was estimated at OD 450 nm using a microplate reader (Molecular Devices), and the DNMT activity was calculated following the formula DNMT activity (OD/h/mg) = (No inhibitor OD-Blank OD)/[Nuclear protein amount (μg) × incubation time (h)] × 1000. All samples were measured in duplicate.
2.8. Spheroid Formation Assay
A total of 5 × 103 U87 or U251 cells/mL were cultured in a proliferation medium (DMEM-F12 supplemented with a neural stem cell supplement (2%) and 20 ng/mL each of human basic fibroblast growth factor (FGF) and human epidermal growth factor (EGF)) in the presence or absence of 50 μM of WHI-P131 or 250 μM of PF-956980 in each well of a 12-well plate. The effects of the drugs on neurosphere formation were analyzed on days 7 and 14 of culture. Images of the spheres were captured using a Leica IM8 microscope.
2.9. Immunofluorescence Microscopy
For the immunofluorescence analysis of Ki-67 and pJAK3, U87 and U251 cells were cultured in DMEM on glass coverslips in the presence or absence of 50 μM of WHI-P131 or 250 μM of PF-956980 for 24 h. Afterward, the cells were fixed in 1% formaldehyde for 10 min at room temperature (RT). The fixed cells were permeabilized with chilled acetone for 15 seconds followed by chilled methanol for 3 min. The cells were washed 3 times with PBS and incubated with a blocking solution (1% BSA and 22.52 mg/mL of glycine in PBS) for 30 min at RT. Afterward, the cells were incubated with Ki-67 (1:100 dilution) or pJAK3 (Cell Signaling Technology, Danvers, MA, USA; 1:250) in 0.1% BSA in PBS for 45 min at RT. The cells were washed 3 times in PBS and incubated with secondary anti-rabbit Alexa Flour 555 (1:250 in 0.1% BSA in PBS) for 45 min in the dark at RT. Subsequently, the cells were washed 3 times with PBS and 2 times with dH2O. A drop of Fluoromount-G containing DAPI was added to the cells, and they were mounted on a slide for observation.
For the immunofluorescence analysis of CD44 and NESTIN, 5 × 103 U87 or U251 cells/mL/well were cultured in 12-well plates in the presence or absence of 50 μM of WHI-P131 or 250 μM of PF-956980 for 6 days. Subsequently, the cells were processed for staining following the above protocol in the 12-well plates. Secondary anti-rabbit Alexa 555 and anti-mouse Alexa 488 were used to discriminate rabbit and mouse antibodies. DAPI (1:1000) was added to the cells in the well for 5 min at RT and washed twice before adding 250 μL of PBS to the cells. All immunofluorescence samples were imaged with a Leica IM8 fluorescence microscope.
2.10. Reverse Transcriptase PCR
U87 or U251 cells either left untreated or treated with 50 μM of WHI-P131 or 250 μM of PF-956980 for 8 days were used to synthesize cDNA using the Miltenyi Biotec μMACS™ One-step cDNA synthesis kit (130-091-902) and protocol. Semiquantitative RT-PCR was performed to reveal the levels of expression of indicated genes (ACTB: for 5′-CACACTGTGCCCATCTAC-3′, rev: 5′-TCGTAGCTCTTCTCCAGG-3′; DNMT1: for 5′-AAAACCCAGCCAACAGAG-3′, rev: 5′-GGACTGGACAGCTTGATG-3′; DNMT3B: for 5′-ACGGTTCCTGGAGTGTAA-3′, rev: 5′-TCCCCTGTTTGATCGAGT-3′; and TUBB3: for 5′-AACAGCAGCTACTTCGTG-3′, rev: 5′-GGTCGTTCATGTTGCTCT-3′) in untreated and inhibitor-treated cells.
2.11. Statistical Analysis
The data are presented as means ± sd. Statistical significance was assessed using Student’s t-test for comparisons between two groups. The p-values, wherever applicable, were calculated based on the means of the experimental replicates.
4. Discussion
GBM is a deadly tumor with a huge unmet clinical need. The therapeutic strategies available at present are grossly inadequate to have any impact on prolonging the survival of GBM patients. Although research so far has implicated several molecules to be the causative agents for GBM pathogenesis, a holistic picture to understand the pathogenesis and to apply a curative therapy to prolong the lifespan of patients is still very far away. The vigorous proliferation rate of these cells is a challenge, and several reagents have been tested to block this proliferation and kill the tumor cells. However, considering the genetic and mechanistic heterogeneity of GBM pathogenesis, a complete understanding of the growth and differentiation of GBM cells is essential to devise an efficient therapy to eliminate them.
Overactivity of JAK family kinases has been reported to play a role in various types of cancer, mostly hematological ones [
28,
29,
30]. But recently, many groups have explored their involvement in GBM pathogenesis and by using small-molecule inhibitors of JAKs or downstream TF STAT3, have observed beneficial effects in in vitro and in vivo studies [
20,
31,
32,
33]. The JAK family member that has been extensively investigated in GBM is JAK2. However, so far, nothing has progressed to a successful clinical trial without having severe side effects or toxicity. JAK3’s involvement in GBM pathogenesis has been relatively less explored. We showed that by treating U87 and U251 GBM cells with two specific JAK3 inhibitors, WHI-P131 and PF-956980, JAK3 activity could be significantly downregulated, and this has tremendous beneficial effects in preventing tumor cell proliferation and inducing cellular differentiation (
Supplementary Figure S4).
WHI-P131 and a related compound WHI-P154 have been previously reported to prevent GBM cell adhesion and migration [
34]. WHI-P154 alone and a conjugate of EGF receptor-WHI-P154 have also been reported to induce cytotoxicity in GBM cells [
35]. However, we did not observe any defect in cell adhesion or the induction of cytotoxicity at a range of WHI-P131 concentrations of up to 200 μM. We also tested WHI-P154 against both U87 and U251 cells and obtained similar results to those for WHI-P131 in terms of the inhibition of proliferation and induction of differentiation. PF-956980 has not been tested against GBM, but herein, we showed that it has a similar effect to that of WHI-P131 in preventing GBM pathogenesis, albeit at a higher concentration. However, even at these concentrations, they are not toxic either to the proliferating tumor cells or to the differentiated tumor cells, as we did not observe significant cell death at either early or late time points of the drug treatments. Thus, both inhibitors hold promise to be used against GBM to provide benefit to patients.
The presence of serum in culture media has been reported to induce the differentiation of neural stem cells into mature stages. However, despite the high serum amount in the culture medium and lack of neural growth factors such as EGF and FGF, these cells proliferated extensively without differentiation, showing the robustness of the tumorigenic phenotype of these cells. Both in the serum-containing differentiation medium and the EGF- and FGF-containing proliferation medium, U87 and U251 cells proliferated extensively. However, our findings show that a single treatment with either WHI-P131 or PF-956980 could not only effectively block their proliferation but also induced their differentiation into various neural lineage cells. The longest period we monitored was 75 days after a single dose of drug treatment, during which the cells remained alive and in a differentiated state. But we are sure they can be maintained in this non-tumorigenic differentiated condition as long as they are maintained with the drugs. This is an observation of huge interest as it suggests either these drugs or a more refined form of these drugs could potentially be beneficial in GBM treatment with fewer side effects.
The current treatment procedure of surgical resection followed by temozolomide therapy does not prevent tumors from developing resistance and leads to the relapse of more vigorous tumors. There are several limitations to the current treatment strategy. First of all, it is almost impossible to precisely remove all of the tumor tissue with surgery. The fact that the remaining tumor cells rapidly proliferate and re-form the tumor mass is the main roadblock to successful therapy. Our findings suggest that following the surgical removal of most of the tumor tissue, the remaining tumor cells, irrespective of whether they are cancer stem cells (CSCs) or non-stem tumor cells, could be kept in a non-proliferative, non-tumorigenic differentiated state, which might help to prolong the lifespans of GBM patients. We have shown that both WHI-P131 and PF-956980 treatment effectively act on the U87 and U251 cell populations, including CSCs, and they also downregulate the stemness-related genes.
Our observations regarding epigenetic modifications, such as increased DNMT activity, might be an important factor following JAK3 inhibition in preventing GBM cell proliferation and inducing differentiation. Methylation at the O6-methylguanine-DNA methyltransferase (MGMT) gene promoter has been detected in nearly 50% of high-grade gliomas and is being used as a good prognostic marker for effective chemotherapy. Based on our results, we can assume an enhanced MGMT methylation in JAK3-inhibitor-treated U87 and U251 cells. Whether this contributes to the observed effects of the JAK3 inhibitors in our study needs further analysis. However, this raises the interesting possibility of treating MGMT unmethylated cases of GBM with JAK3 inhibitors to make them more susceptible to chemotherapy using alkylating agents. This will potentially help many GBM patients prolong their lifespans.
In the current study, we did not look into other epigenetic changes that could be involved in the phenotype we observed following JAK3 inhibition. However, how these epigenetic changes occur, the kinetics, and the molecules involved in this process in GBM cells need further investigation. That the GBM cells remain in a differentiated state for a long time even with only a single treatment of the inhibitors holds promise for future refinement to develop more effective forms of these drugs working at a lower concentration to prevent GBM progression.