CREB1 Is Involved in miR-134-5p-Mediated Endometrial Stromal Cell Proliferation, Apoptosis, and Autophagy
Abstract
:1. Introduction
2. Results
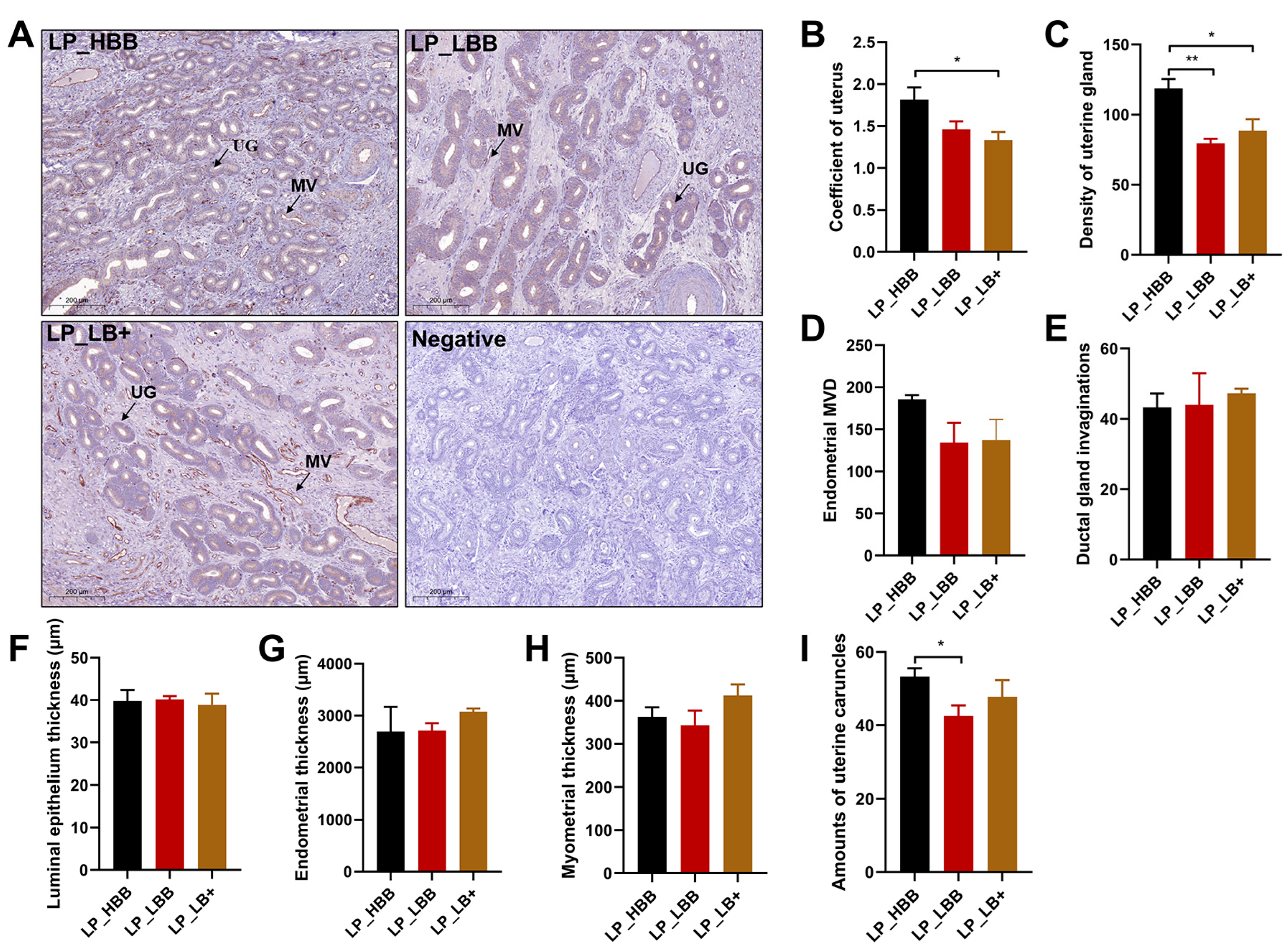
2.1. Analysis of Hu Sheep Uterine Morphology and Endocrinology in Luteal Phase
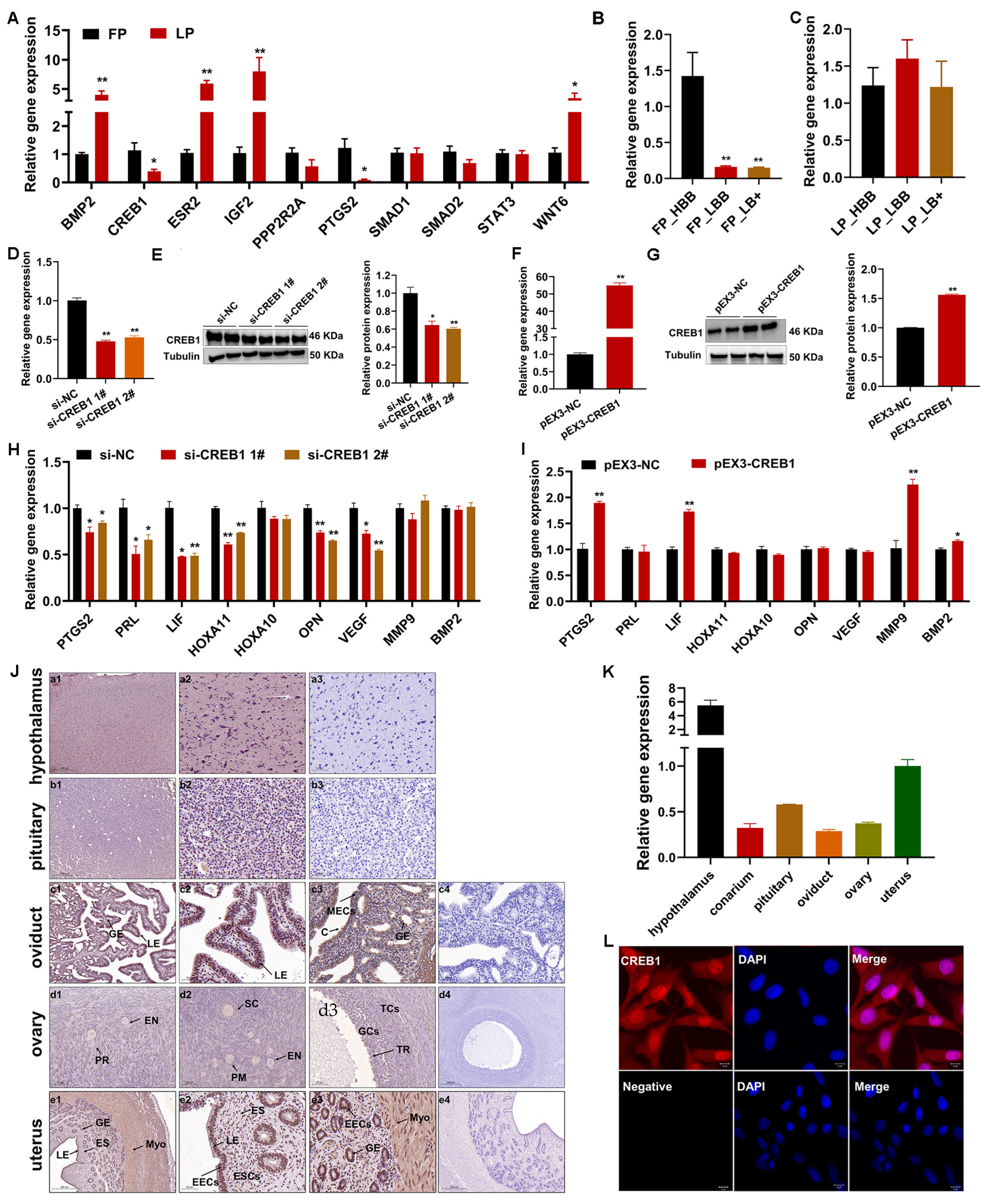
2.2. Characteristics of CREB1 during Estrous Cycle of Sheep
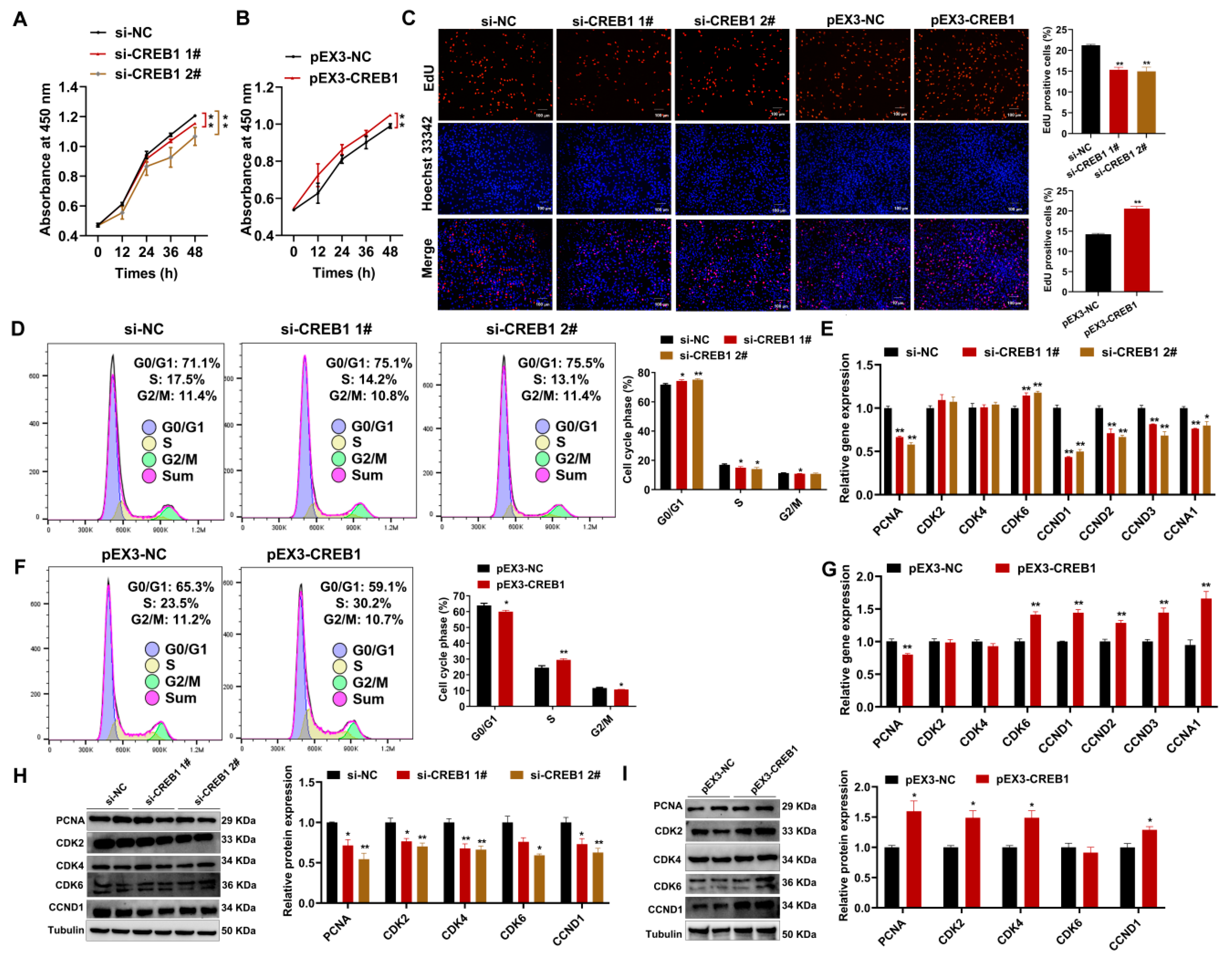
2.3. CREB1 Promotes Sheep Endometrial Stromal Cell Proliferation
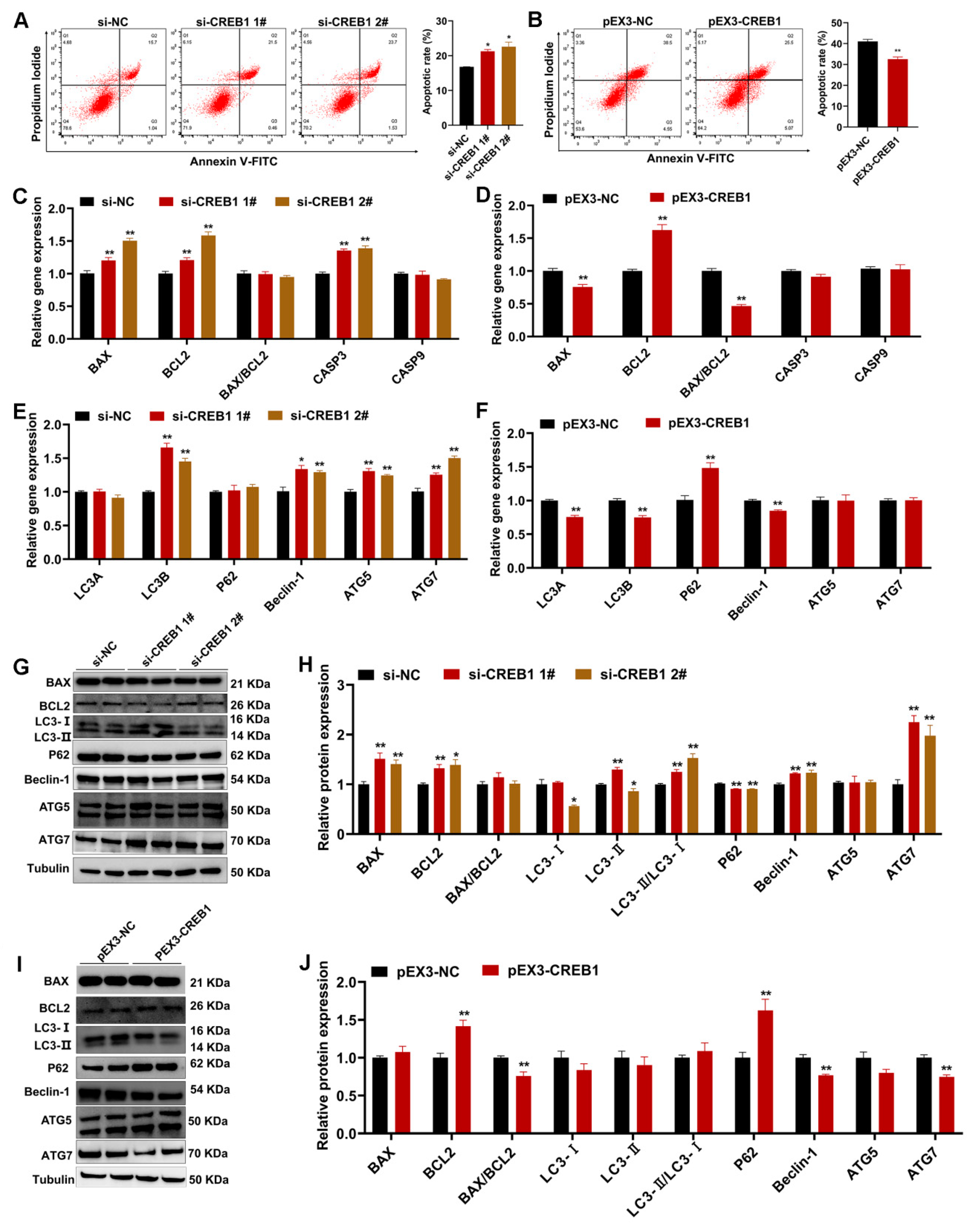
2.4. CREB1 Regulates Sheep Endometrial Stromal Cell Apoptosis and Autophagy
2.5. CREB1 Is a Direct Target of miR-134-5p
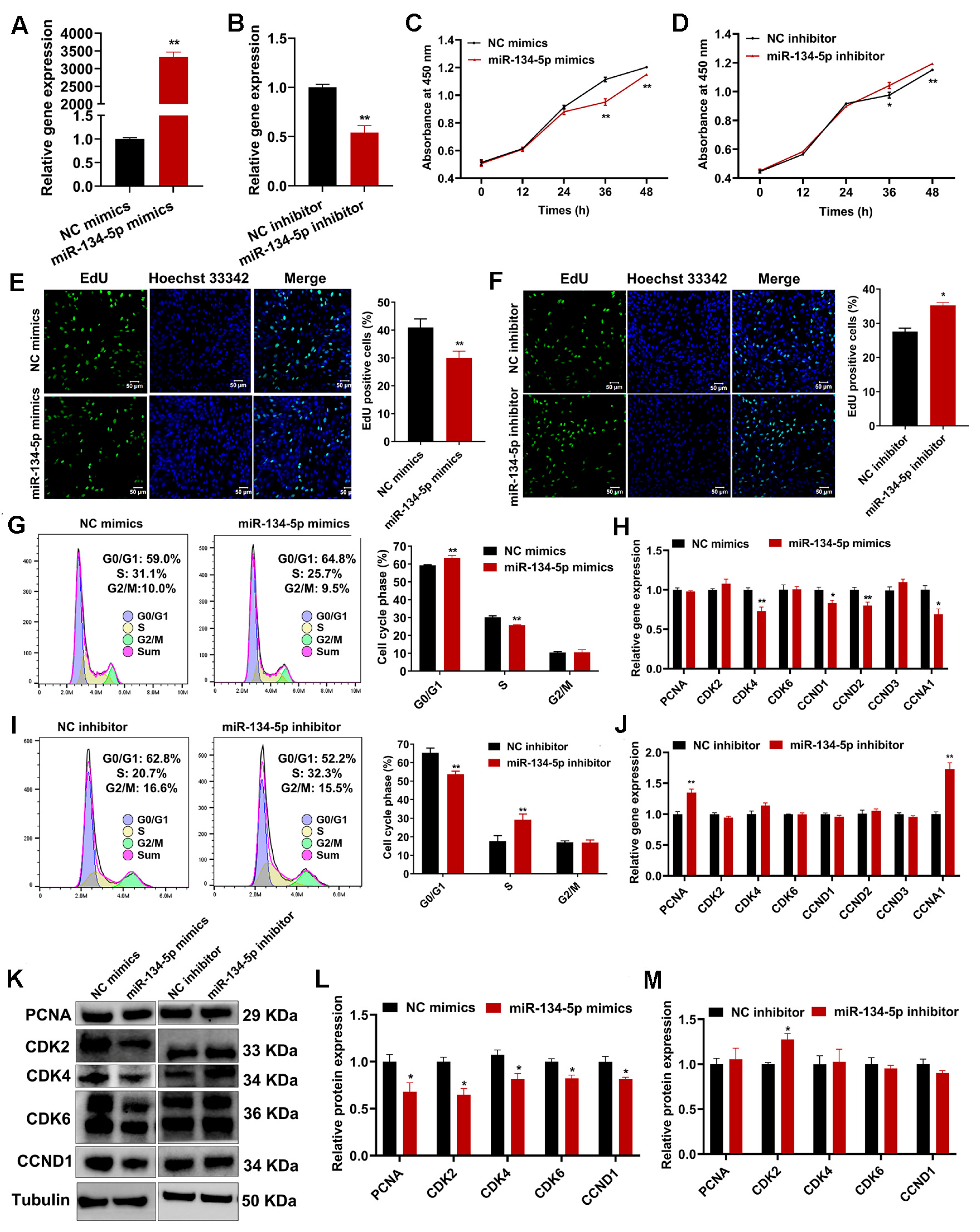
2.6. miR-134-5p Suppresses Sheep Endometrial Stromal Cell Proliferation
2.7. miR-134-5p Regulates Sheep Endometrial Stromal Cell Apoptosis and Autophagy
2.8. CREB1 Rescues miR-134-5p-Induced Cell Cycle Arrest, Apoptosis, and Autophagy in Sheep Endometrial Stromal Cells
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Animals, Tissue Collection
5.2. Morphological Analysis of Uterine Tissue
5.3. RNA-Seq and Bioinformatics Analysis
5.4. RNA Preparation and Real-Time Quantitative PCR (qRT-PCR)
5.5. Western Blotting
5.6. Immunohistochemistry (IHC)
5.7. Enzyme-Linked Immunosorbent Assay (ELISA)
5.8. Cell Culture and Treatment
5.9. Immunofluorescence (IF)
5.10. Cell Viability Assay
5.11. Cell Proliferation
5.12. Cell Cycle Analysis
5.13. Cell Apoptosis Analysis
5.14. Luciferase Activity Assay
5.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yue, G.H. Reproductive characteristics of Chinese Hu sheep. Anim. Reprod. Sci. 1996, 44, 223–230. [Google Scholar] [CrossRef]
- Feng, X.; Li, F.; Wang, F.; Zhang, G.; Pang, J.; Ren, C.; Zhang, T.; Yang, H.; Wang, Z.; Zhang, Y. Genome-wide differential expression profiling of mRNAs and lncRNAs associated with prolificacy in Hu sheep. Biosci. Rep. 2018, 38, BSR20171350. [Google Scholar] [CrossRef]
- Mahdavi, M.; Nanekarani, S.; Hosseini, S.D. Mutation in BMPR-IB gene is associated with litter size in Iranian Kalehkoohi sheep. Anim. Reprod. Sci. 2014, 147, 93–98. [Google Scholar] [CrossRef] [PubMed]
- El-Seedy, A.S.; Hashem, N.M.; El-Azrak, K.M.; Nour El-Din, A.; Ramadan, T.A.; Taha, T.A.; Salem, M.H. Genetic screening of FecB, FecX(G) and FecX(I) mutations and their linkage with litter size in Barki and Rahmani sheep breeds. Reprod. Domest. Anim. 2017, 52, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Di, R.; Wang, F.; Yu, P.; Wang, X.; He, X.; Mwacharo, J.M.; Pan, L.; Chu, M. Detection of Novel Variations Related to Litter Size in BMP15 Gene of Luzhong Mutton Sheep (Ovis aries). Animals 2021, 11, 3528. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chi, Z.; Jia, S.; Zhao, S.; Cao, G.; Purev, C.; Cang, M.; Yu, H.; Li, X.; Bao, S.; et al. Effects of novel variants in BMP15 gene on litter size in Mongolia and Ujimqin sheep breeds. Theriogenology 2023, 198, 1–11. [Google Scholar] [CrossRef]
- Tong, B.; Wang, J.; Cheng, Z.; Liu, J.; Wu, Y.; Li, Y.; Bai, C.; Zhao, S.; Yu, H.; Li, G. Novel Variants in GDF9 Gene Affect Promoter Activity and Litter Size in Mongolia Sheep. Genes 2020, 11, 375. [Google Scholar] [CrossRef]
- Vage, D.I.; Husdal, M.; Kent, M.P.; Klemetsdal, G.; Boman, I.A. A missense mutation in growth differentiation factor 9 (GDF9) is strongly associated with litter size in sheep. BMC Genet. 2013, 14, 1. [Google Scholar] [CrossRef]
- Guan, F.; Liu, S.-R.; Shi, G.-Q.; Yang, L.-G. Polymorphism of FecB gene in nine sheep breeds or strains and its effects on litter size, lamb growth and development. Anim. Reprod. Sci. 2007, 99, 44–52. [Google Scholar] [CrossRef]
- Fogarty, N.M. A review of the effects of the Booroola gene (FecB) on sheep production. Small Rumin. Res. 2009, 85, 75–84. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, J.; Di, R.; Liu, Q.; Wang, X.; Gan, S.; Zhang, X.; Zhang, J.; Hu, W.; Chu, M. Comparative Transcriptomics Reveal Key Sheep (Ovis aries) Hypothalamus LncRNAs that Affect Reproduction. Animals 2019, 9, 152. [Google Scholar] [CrossRef] [PubMed]
- McBride, D.; Carré, W.; Sontakke, S.D.; Hogg, C.O.; Law, A.; Donadeu, F.X.; Clinton, M. Identification of miRNAs associated with the follicular-luteal transition in the ruminant ovary. Reproduction 2012, 144, 221–233. [Google Scholar] [CrossRef] [PubMed]
- La, Y.; Tang, J.; Guo, X.; Zhang, L.; Gan, S.; Zhang, X.; Zhang, J.; Hu, W.; Chu, M. Proteomic analysis of sheep uterus reveals its role in prolificacy. J. Proteom. 2020, 210, 103526. [Google Scholar] [CrossRef] [PubMed]
- Mihm, M.; Gangooly, S.; Muttukrishna, S. The normal menstrual cycle in women. Anim. Reprod. Sci. 2011, 124, 229–236. [Google Scholar] [CrossRef]
- Hernández-Montiel, W.; Collí-Dula, R.C.; Ramón-Ugalde, J.P.; Martínez-Núñez, M.A.; Zamora-Bustillos, R. RNA-seq Transcriptome Analysis in Ovarian Tissue of Pelibuey Breed to Explore the Regulation of Prolificacy. Genes 2019, 10, 358. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.; Zhao, H.; Qin, X. Co-expression analysis and identification of fecundity-related long non-coding RNAs in sheep ovaries. Sci. Rep. 2016, 6, 39398. [Google Scholar] [CrossRef]
- Yao, X.; Gao, X.; Bao, Y.; El-Samahy, M.A.; Yang, J.; Wang, Z.; Li, X.; Zhang, G.; Zhang, Y.; Liu, W.; et al. lncRNA FDNCR promotes apoptosis of granulosa cells by targeting the miR-543-3p/DCN/TGF-β signaling pathway in Hu sheep. Mol. Ther. Nucleic Acids 2021, 24, 223–240. [Google Scholar] [CrossRef]
- Stenhouse, C.; Newton, M.G.; Halloran, K.M.; Moses, R.M.; Sah, N.; Suva, L.J.; Bazer, F.W. Phosphate, calcium, and vitamin D signaling, transport, and metabolism in the endometria of cyclic ewes. J. Anim. Sci. Biotechnol. 2023, 14, 13. [Google Scholar] [CrossRef]
- Diedrich, K.; Fauser, B.C.; Devroey, P.; Griesinger, G. The role of the endometrium and embryo in human implantation. Hum. Reprod. Update 2007, 13, 365–377. [Google Scholar] [CrossRef]
- Spencer, T.E.; Forde, N.; Lonergan, P. Insights into conceptus elongation and establishment of pregnancy in ruminants. Reprod. Fertil. Dev. 2016, 29, 84–100. [Google Scholar] [CrossRef]
- Strassmann, B.I. The evolution of endometrial cycles and menstruation. Q. Rev. Biol. 1996, 71, 181–220. [Google Scholar] [CrossRef] [PubMed]
- Groebner, A.E.; Rubio-Aliaga, I.; Schulke, K.; Reichenbach, H.D.; Daniel, H.; Wolf, E.; Meyer, H.H.; Ulbrich, S.E. Increase of essential amino acids in the bovine uterine lumen during preimplantation development. Reproduction 2011, 141, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.E.; Gopichandran, N.; Picton, H.M.; Leese, H.J.; Orsi, N.M. Nutrient concentrations in murine follicular fluid and the female reproductive tract. Theriogenology 2005, 64, 992–1006. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W. Uterine protein secretions: Relationship to development of the conceptus. J. Anim. Sci. 1975, 41, 1376–1382. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wu, G.; Spencer, T.E.; Johnson, G.A.; Li, X.; Bazer, F.W. Select nutrients in the ovine uterine lumen. I. Amino acids, glucose, and ions in uterine lumenal flushings of cyclic and pregnant ewes. Biol. Reprod. 2009, 80, 86–93. [Google Scholar] [CrossRef]
- Paria, B.C.; Reese, J.; Das, S.K.; Dey, S.K. Deciphering the cross-talk of implantation: Advances and challenges. Science 2002, 296, 2185–2188. [Google Scholar] [CrossRef]
- Lim, H.; Ma, L.; Ma, W.G.; Maas, R.L.; Dey, S.K. Hoxa-10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Mol. Endocrinol. 1999, 13, 1005–1017. [Google Scholar] [CrossRef]
- Su, L.; Liu, R.; Cheng, W.; Zhu, M.; Li, X.; Zhao, S.; Yu, M. Expression patterns of microRNAs in porcine endometrium and their potential roles in embryo implantation and placentation. PLoS ONE 2014, 9, e87867. [Google Scholar] [CrossRef]
- Xia, H.F.; Jin, X.H.; Cao, Z.F.; Hu, Y.; Ma, X. MicroRNA expression and regulation in the uterus during embryo implantation in rat. FEBS J. 2014, 281, 1872–1891. [Google Scholar] [CrossRef]
- Governini, L.; Luongo, F.P.; Haxhiu, A.; Piomboni, P.; Luddi, A. Main actors behind the endometrial receptivity and successful implantation. Tissue Cell 2021, 73, 101656. [Google Scholar] [CrossRef]
- Kinnear, S.; Salamonsen, L.A.; Francois, M.; Harley, V.; Evans, J. Uterine SOX17: A key player in human endometrial receptivity and embryo implantation. Sci. Rep. 2019, 9, 15495. [Google Scholar] [CrossRef] [PubMed]
- Plante, B.J.; Kannan, A.; Bagchi, M.K.; Yuan, L.; Young, S.L. Cyclic regulation of transcription factor C/EBP beta in human endometrium. Reprod. Biol. Endocrinol. 2009, 7, 15. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, J.; Wang, Y.; Zhao, W.; Wang, W.; Cui, J.; Yang, J.; Yue, Y.; Zhang, S.; Chu, M.; et al. A proteomic atlas of ligand-receptor interactions at the ovine maternal-fetal interface reveals the role of histone lactylation in uterine remodeling. J. Biol. Chem. 2022, 298, 101456. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liu, A.; Zhang, Y.; Nan, S.; Yin, R.; Lei, Q.; Zhu, H.; Chen, J.; Han, L.; Ding, M.; et al. Essential Role of CRIM1 on Endometrial Receptivity in Goat. Int. J. Mol. Sci. 2021, 22, 5323. [Google Scholar] [CrossRef]
- Feng, Y.; Tan, B.Z. LncRNA MALAT1 inhibits apoptosis of endometrial stromal cells through miR-126-5p-CREB1 axis by activating PI3K-AKT pathway. Mol. Cell Biochem. 2020, 475, 185–194. [Google Scholar] [CrossRef]
- Mao, H.; Zhang, X.; Yin, L.; Ji, X.; Huang, C.; Wu, Q. Silencing of circ_0007299 suppresses proliferation, migration, and invasiveness and promotes apoptosis of ectopic endometrial stromal cells in endometriosis via miR-424-5p-dependent modulation of CREB1. Arch. Gynecol. Obstet. 2023, 307, 149–161. [Google Scholar] [CrossRef]
- Shan, J.; Li, D.J.; Wang, X.Q. Towards a Better Understanding of Endometriosis-Related Infertility: A Review on How Endometriosis Affects Endometrial Receptivity. Biomolecules 2023, 13, 430. [Google Scholar] [CrossRef]
- Chien, L.W.; Au, H.K.; Chen, P.L.; Xiao, J.; Tzeng, C.R. Assessment of uterine receptivity by the endometrial-subendometrial blood flow distribution pattern in women undergoing in vitro fertilization-embryo transfer. Fertil. Steril. 2002, 78, 245–251. [Google Scholar] [CrossRef]
- Filant, J.; Spencer, T.E. Uterine glands: Biological roles in conceptus implantation, uterine receptivity and decidualization. Int. J. Dev. Biol. 2014, 58, 107–116. [Google Scholar] [CrossRef]
- Gao, X.; Yao, X.; Wang, Z.; Li, X.; Li, X.; An, S.; Wei, Z.; Zhang, G.; Wang, F. Long non-coding RNA366.2 controls endometrial epithelial cell proliferation and migration by upregulating WNT6 as a ceRNA of miR-1576 in sheep uterus. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194606. [Google Scholar] [CrossRef]
- Nayak, R.; Chattopadhyay, T.; Mallick, B. Identification of potential repurposed drugs for treating endometriosis-associated infertility among women. Chem. Biol. Interact. 2022, 365, 110110. [Google Scholar] [CrossRef] [PubMed]
- Spencer, T.E. Biological roles of uterine glands in pregnancy. Semin. Reprod. Med. 2014, 32, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Demir, R.; Yaba, A.; Huppertz, B. Vasculogenesis and angiogenesis in the endometrium during menstrual cycle and implantation. Acta Histochem. 2010, 112, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, A.M.; DeMayo, F.J.; Spencer, T.E. Uterine Glands: Developmental Biology and Functional Roles in Pregnancy. Endocr. Rev. 2019, 40, 1424–1445. [Google Scholar] [CrossRef] [PubMed]
- Santi, A.; Felser, R.S.; Mueller, M.D.; Wunder, D.M.; McKinnon, B.; Bersinger, N.A. Increased endometrial placenta growth factor (PLGF) gene expression in women with successful implantation. Fertil. Steril. 2011, 96, 663–668. [Google Scholar] [CrossRef]
- Zhou, Y.; Bellingard, V.; Feng, K.T.; McMaster, M.; Fisher, S.J. Human cytotrophoblasts promote endothelial survival and vascular remodeling through secretion of Ang2, PlGF, and VEGF-C. Dev. Biol. 2003, 263, 114–125. [Google Scholar] [CrossRef]
- Passos, J.R.; D Guerreiro, D.; Otávio, K.; dos Santos-Neto, P.; Souza-Neves, M.; Cuadro, F.; Nuñez-Olivera, R.; Crispo, M.; Bezerra, M.J.; Silva, R.F.; et al. Global proteomic analysis of preimplantational ovine embryos produced in vitro. Reprod. Domest. Anim. 2022, 57, 784–797. [Google Scholar] [CrossRef]
- Tu, Z.; Wang, Q.; Cui, T.; Wang, J.; Ran, H.; Bao, H.; Lu, J.; Wang, B.; Lydon, J.P.; DeMayo, F.; et al. Uterine RAC1 via Pak1-ERM signaling directs normal luminal epithelial integrity conducive to on-time embryo implantation in mice. Cell Death Differ. 2016, 23, 169–181. [Google Scholar] [CrossRef]
- La, Y.; Tang, J.; Di, R.; Wang, X.; Liu, Q.; Zhang, L.; Zhang, X.; Zhang, J.; Hu, W.; Chu, M. Differential Expression of Circular RNAs in Polytocous and Monotocous Uterus during the Reproductive Cycle of Sheep. Animals 2019, 9, 797. [Google Scholar] [CrossRef]
- Chen, S.; Guo, X.; He, X.; Di, R.; Zhang, X.; Zhang, J.; Wang, X.; Chu, M. Insight into Pituitary lncRNA and mRNA at Two Estrous Stages in Small Tail Han Sheep with Different FecB Genotypes. Front Endocrinol 2021, 12, 789564. [Google Scholar] [CrossRef]
- Chen, Q.; Ni, Y.; Han, M.; Zhou, W.J.; Zhu, X.B.; Zhang, A.J. Integrin-linked kinase improves uterine receptivity formation by activating Wnt/beta-catenin signaling and up-regulating MMP-3/9 expression. Am. J. Transl. Res. 2020, 12, 3011–3022. [Google Scholar] [PubMed]
- Zhang, L.; An, X.P.; Liu, X.R.; Fu, M.Z.; Han, P.; Peng, J.Y.; Hou, J.X.; Zhou, Z.Q.; Cao, B.Y.; Song, Y.X. Characterization of the Transcriptional Complexity of the Receptive and Pre-receptive Endometria of Dairy. Goats. Sci. Rep. 2015, 5, 14244. [Google Scholar] [CrossRef] [PubMed]
- Salker, M.S.; Singh, Y.; Durairaj, R.R.P.; Yan, J.; Alauddin, M.; Zeng, N.; Steel, J.H.; Zhang, S.; Nautiyal, J.; Webster, Z.; et al. LEFTY2 inhibits endometrial receptivity by downregulating Orai1 expression and store-operated Ca2+ entry. J. Mol. Med. 2018, 96, 173–182. [Google Scholar] [CrossRef]
- Luo, J.; Zhu, H.; Chang, H.M.; Lin, Y.M.; Yang, J.; Leung, P.C.K. The regulation of IGFBP3 by BMP2 has a role in human endometrial remodeling. FASEB J. 2020, 34, 15462–15479. [Google Scholar] [CrossRef] [PubMed]
- Kriseman, M.; Monsivais, D.; Agno, J.; Masand, R.P.; Creighton, C.J.; Matzuk, M.M. Uterine double-conditional inactivation of Smad2 and Smad3 in mice causes endometrial dysregulation, infertility, and uterine cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 3873–3882. [Google Scholar] [CrossRef]
- Tang, S.; Cope, D.I.; Vasquez, Y.M.; Monsivais, D. BMP/SMAD1/5 Signaling in the Endometrial Epithelium Is Essential for Receptivity and Early Pregnancy. Endocrinology 2022, 163, bqac043. [Google Scholar] [CrossRef]
- Lv, X.; Wang, D.; Ma, Y.; Long, Z. Analysis of the oncogene BRAF mutation and the correlation of the expression of wild-type BRAF and CREB1 in endometriosis. Int. J. Mol. Med. 2018, 41, 1349–1356. [Google Scholar] [CrossRef]
- Szaflik, T.; Smolarz, B.; Mroczkowska, B.; Kulig, B.; Soja, M.; Romanowicz, H.; Bryś, M.; Forma, E.; Szyłło, K. An Analysis of ESR2 and CYP19A1 Gene Expression Levels in Women with Endometriosis. In Vivo 2020, 34, 1765–1771. [Google Scholar] [CrossRef]
- Zhou, C.; Lv, M.; Wang, P.; Guo, C.; Ni, Z.; Bao, H.; Tang, Y.; Cai, H.; Lu, J.; Deng, W.; et al. Sequential activation of uterine epithelial IGF1R by stromal IGF1 and embryonic IGF2 directs normal uterine preparation for embryo implantation. J. Mol. Cell Biol. 2021, 13, 646–661. [Google Scholar] [CrossRef]
- Li, X.; Yao, X.; Xie, H.; Zhang, G.; Deng, M.; Deng, K.; Gao, X.; Bao, Y.; Li, K.; Wang, F. PPP2R2A affects embryonic implantation by regulating the proliferation and apoptosis of Hu sheep endometrial stromal cells. Theriogenology 2021, 176, 149–162. [Google Scholar] [CrossRef]
- Panicker, N.; Coutman, M.; Lawlor-O’Neill, C.; Kahl, R.G.S.; Roselli, S.; Verrills, N.M. Ppp2r2a Knockout Mice Reveal That Protein Phosphatase 2A Regulatory Subunit, PP2A-B55α, Is an Essential Regulator of Neuronal and Epidermal Embryonic Development. Front. Cell Dev. Biol. 2020, 8, 358. [Google Scholar] [CrossRef] [PubMed]
- Jaureguiberry, M.; Madoz, L.V.; Quintana, S.; Marín, M.; Burucúa, M.; Tizzano, M.; Rabaglino, B.; Rearte, R.; Pecoraro, M.; de la Sota, R.L. Endometrial expression of key genes related to fertility in repeat breeder and non-repeat breeder cows. Reprod. Domest. Anim. 2020, 55, 1660–1664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; Che, S.; Cui, J.; Liu, Y.; An, X.; Cao, B.; Song, Y. CircRNA-9119 regulates the expression of prostaglandin-endoperoxide synthase 2 (PTGS2) by sponging miR-26a in the endometrial epithelial cells of dairy goat. Reprod. Fertil. Dev. 2018, 30, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, T.; Hirota, Y.; Fukui, Y.; Gebril, M.; Kaku, T.; Aikawa, S.; Hirata, T.; Akaeda, S.; Matsuo, M.; Haraguchi, H.; et al. Differential roles of uterine epithelial and stromal STAT3 coordinate uterine receptivity and embryo attachment. Sci. Rep. 2020, 10, 15523. [Google Scholar] [CrossRef]
- Gao, X.; Yao, X.; Li, X.; Liang, Y.; Liu, Z.; Wang, Z.; Li, K.; Li, Y.; Zhang, G.; Wang, F. Roles of WNT6 in Sheep Endometrial Epithelial Cell Cycle Progression and Uterine Glands Organogenesis. Vet. Sci. 2021, 8, 316. [Google Scholar] [CrossRef]
- Torres, A.; Batista, M.; Diniz, P.; Mateus, L.; Lopes-da-Costa, L. Embryo-luteal cells co-culture: An in vitro model to evaluate steroidogenic and prostanoid bovine early embryo-maternal interactions. In Vitro Cell Dev. Biol. Anim. 2013, 49, 134–146. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, X.; Liu, J.; Ma, X.; Zhou, Z.; Song, Y.; Cao, B. miR-26a promoted endometrial epithelium cells (EECs) proliferation and induced stromal cells (ESCs) apoptosis via the PTEN-PI3K/AKT pathway in dairy goats. J. Cell Physiol. 2018, 233, 4688–4706. [Google Scholar] [CrossRef]
- Vogiagis, D.; Salamonsen, L.A.; Sandeman, R.M.; Squires, T.J.; Butt, A.R.; Fry, R.C. Effect of immunisation against leukaemia inhibitory factor on the establishment of pregnancy in sheep. Reprod. Nutr. Dev. 1997, 37, 459–468. [Google Scholar] [CrossRef]
- Zheng, P.; Huang, H.; Li, X.; Huang, F.; Adeniran, S.O.; Wang, Z.; Feng, R.; Zhang, G. LRH-A3 and HCG increase pregnancy rate during timed artificial insemination in dairy cows. Anim. Sci. J. 2021, 92, e13549. [Google Scholar] [CrossRef]
- Zhang, X.; Gong, S.; Li, H.; Jiang, J.; Jia, Y.; Zhang, R.; Liu, H.; Wang, A.; Jin, Y.; Lin, P. USP18 promotes endometrial receptivity via the JAK/STAT1 and the ISGylation pathway. Theriogenology 2023, 202, 110–118. [Google Scholar] [CrossRef]
- Seo, H.; Frank, J.W.; Burghardt, R.C.; Bazer, F.W.; Johnson, G.A. Integrins and OPN localize to adhesion complexes during placentation in sheep. Reproduction 2020, 160, 521–532. [Google Scholar] [CrossRef]
- Johnson, G.A.; Burghardt, R.C.; Bazer, F.W. Osteopontin: A leading candidate adhesion molecule for implantation in pigs and sheep. J. Anim. Sci. Biotechnol. 2014, 5, 56. [Google Scholar] [CrossRef]
- Cheng, J.; Li, C.; Ying, Y.; Lv, J.; Qu, X.; McGowan, E.; Lin, Y.; Zhu, X. Metformin Alleviates Endometriosis and Potentiates Endometrial Receptivity via Decreasing VEGF and MMP9 and Increasing Leukemia Inhibitor Factor and HOXA10. Front. Pharmacol. 2022, 13, 750208. [Google Scholar] [CrossRef] [PubMed]
- Pabona, J.M.; Zeng, Z.; Simmen, F.A.; Simmen, R.C. Functional differentiation of uterine stromal cells involves cross-regulation between bone morphogenetic protein 2 and Kruppel-like factor (KLF) family members KLF9 and KLF13. Endocrinology 2010, 151, 3396–3406. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.S.; Han, B.; Zhao, J.X.; Tao, N.; Dong, C.Y. MiR-155-5p affects Wilms’ tumor cell proliferation and apoptosis via targeting CREB1. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 6826. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.R.; Raza, S.H.A.; Liang, C.C.; Wang, X.Y.; Wang, J.F.; Zhang, W.Z.; Zan, L. CREB1 promotes proliferation and differentiation by mediating the transcription of CCNA2 and MYOG in bovine myoblasts. Int. J. Biol. Macromol. 2022, 216, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, J.; Lang, H.; Wang, W.; Liu, X.; Liu, H.; Tan, C.; Li, X.; Zhao, Y.; Wu, X. Knockdown of CREB1 promotes apoptosis and decreases estradiol synthesis in mouse granulosa cells. Biomed. Pharmacother. 2018, 105, 1141–1146. [Google Scholar] [CrossRef]
- Kuwana, T.; King, L.E.; Cosentino, K.; Suess, J.; Garcia-Saez, A.J.; Gilmore, A.P.; Newmeyer, D.D. Mitochondrial residence of the apoptosis inducer BAX is more important than BAX oligomerization in promoting membrane permeabilization. J. Biol. Chem. 2020, 295, 1623–1636. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Z.; Liu, Z.; Chen, R.; Peng, H.; Guo, J.; Chen, X.; Zhang, H. MTOR inhibition attenuates DNA damage and apoptosis through autophagy-mediated suppression of CREB1. Autophagy 2013, 9, 2069–2086. [Google Scholar] [CrossRef]
- Yu, S.L.; Kang, Y.; Jeong, D.U.; Lee, D.C.; Jeon, H.J.; Kim, T.H.; Lee, S.K.; Han, A.R.; Kang, J.; Park, S.R. The miR-182-5p/NDRG1 Axis Controls Endometrial Receptivity through the NF-κB/ZEB1/E-Cadherin Pathway. Int. J. Mol. Sci. 2022, 23, 12303. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, X.; Liu, J.; Zhou, Z.; Song, Y.; Cao, B.; An, X. miR-182 aids in receptive endometrium development in dairy goats by down-regulating PTN expression. PLoS ONE 2017, 12, e0179783. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, H.; Yuan, M.; Li, D.; Sun, C.; Wang, G. Serum Exosomal MicroRNAs as Potential Circulating Biomarkers for Endometriosis. Dis. Markers 2020, 2020, 2456340. [Google Scholar] [CrossRef]
- Yang, J.; Liu, S.; Wang, H.; Liu, Y.; Liu, Y. miR-134-5p inhibition reduces infarct-induced cardiomyocyte apoptosis via Creb1 upregulation. J. Stroke Cerebrovasc. Dis. 2020, 29, 104850. [Google Scholar] [CrossRef]
- Okada, N.; Lin, C.P.; Ribeiro, M.C.; Biton, A.; Lai, G.; He, X.; Bu, P.; Vogel, H.; Jablons, D.M.; Keller, A.C.; et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes. Dev. 2014, 28, 438–450. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, Z.; Yang, H.; Yao, X.; Yang, P.; Ren, C.; Wang, F.; Zhang, Y. Pituitary Transcriptomic Study Reveals the Differential Regulation of lncRNAs and mRNAs Related to Prolificacy in Different FecB Genotyping Sheep. Genes 2019, 10, 157. [Google Scholar] [CrossRef]
- Hewitt, S.C.; Korach, K.S. Oestrogen receptor knockout mice: Roles for oestrogen receptors alpha and beta in reproductive tissues. Reproduction 2003, 125, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Liu, D.; Zhou, H.; Dai, C.; Wang, T.; Fang, Y.; Jia, Y.; Li, K. Melatonin improves pregnancy outcomes in adenomyosis mice by restoring endometrial receptivity via NF-kappaB/apoptosis signaling. Ann. Transl. Med. 2022, 10, 1317. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.G.; Soares, J.M., Jr.; Motta, E.L.; Simões, M.J.; Oliveira-Filho, R.M.; Haidar, M.A.; Rodrigues de Lima, G.; Baracat, E.C. Metoclopramide-induced hyperprolactinemia affects mouse endometrial morphology. Gynecol. Obstet. Investig. 2002, 54, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Nitzsche, B.; Frey, S.; Collins, L.D.; Seeger, J.; Lobsien, D.; Dreyer, A.; Kirsten, H.; Stoffel, M.H.; Fonov, V.S.; Boltze, J. A stereotaxic, population-averaged T1w ovine brain atlas including cerebral morphology and tissue volumes. Front. Neuroanat. 2015, 9, 69. [Google Scholar] [CrossRef]
- Li, X.; Yao, X.; Xie, H.; Deng, M.; Gao, X.; Deng, K.; Bao, Y.; Wang, Q.; Wang, F. Effects of SPATA6 on proliferation, apoptosis and steroidogenesis of Hu sheep Leydig cells in vitro. Theriogenology 2021, 166, 9–20. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yao, X.; Li, K.; Guo, J.; Deng, K.; Liu, Z.; Yang, F.; Fan, Y.; Yang, Y.; Zhu, H.; et al. CREB1 Is Involved in miR-134-5p-Mediated Endometrial Stromal Cell Proliferation, Apoptosis, and Autophagy. Cells 2023, 12, 2554. https://doi.org/10.3390/cells12212554
Li X, Yao X, Li K, Guo J, Deng K, Liu Z, Yang F, Fan Y, Yang Y, Zhu H, et al. CREB1 Is Involved in miR-134-5p-Mediated Endometrial Stromal Cell Proliferation, Apoptosis, and Autophagy. Cells. 2023; 12(21):2554. https://doi.org/10.3390/cells12212554
Chicago/Turabian StyleLi, Xiaodan, Xiaolei Yao, Kang Li, Jiahe Guo, Kaiping Deng, Zhipeng Liu, Fan Yang, Yixuan Fan, Yingnan Yang, Huabin Zhu, and et al. 2023. "CREB1 Is Involved in miR-134-5p-Mediated Endometrial Stromal Cell Proliferation, Apoptosis, and Autophagy" Cells 12, no. 21: 2554. https://doi.org/10.3390/cells12212554





