Liquid Biopsy in Head and Neck Cancer: Its Present State and Future Role in Africa
Abstract
1. Introduction
2. Circulating Tumor Cell in Head and Neck Cancer
3. Circulating Tumor DNA in Head and Neck Cancer
4. Circulating Tumor RNA in Head and Neck Cancer
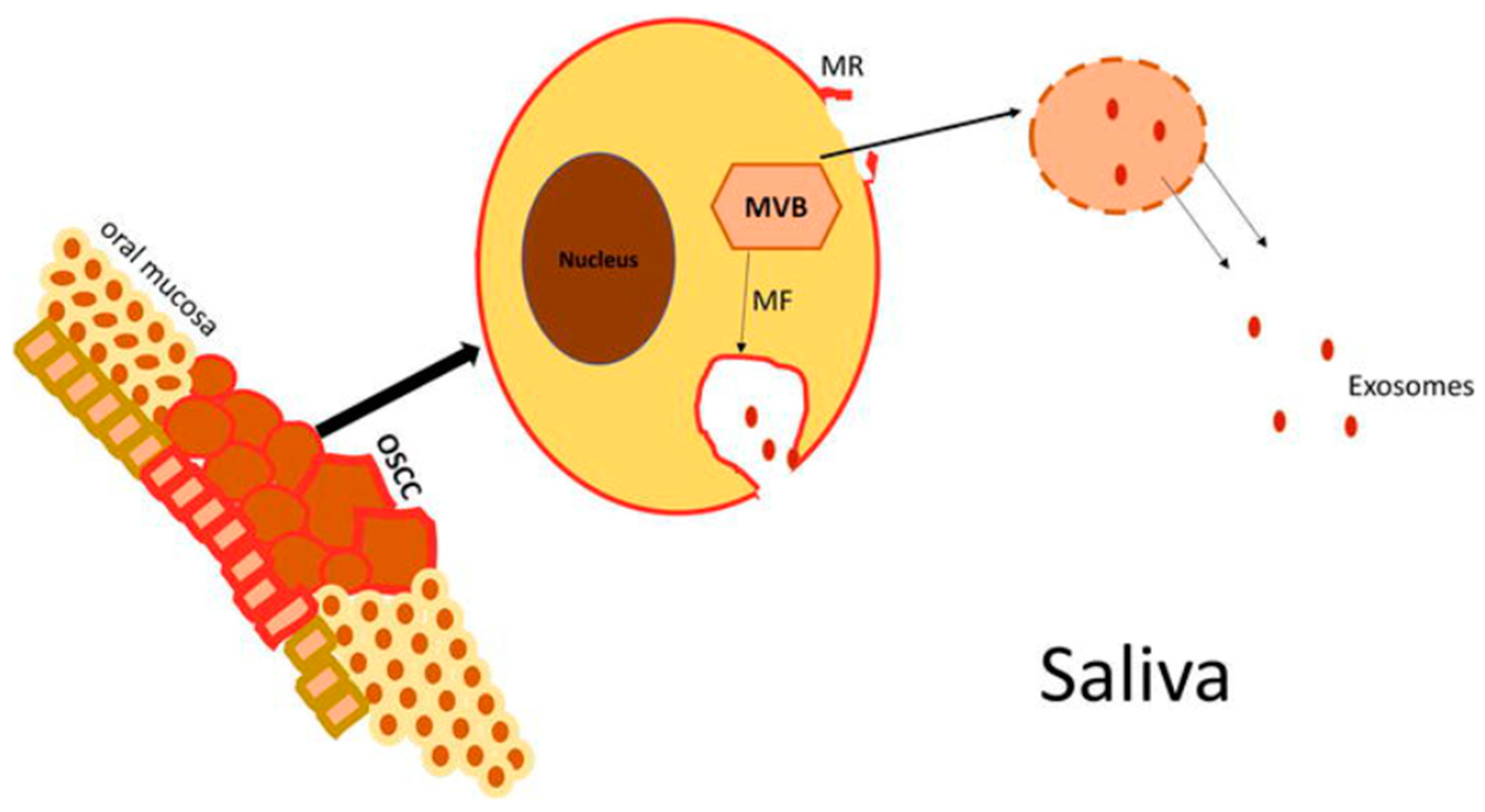
5. Exosomes in Head and Neck Cancer
6. Circulating Proteins and Peptides in Head and Neck Cancer
7. The Present State of Liquid Biopsy for Head and Neck Cancer in Africa
8. The Future of Liquid Biopsy for Head and Neck Cancer in Africa
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Barsouk, A.; Aluru, J.S.; Rawla, P.; Saginala, K.; Barsouk, A. Epidemiology, Risk Factors, and Prevention of Head and Neck Squamous Cell Carcinoma. Med. Sci. 2023, 11, 42. [Google Scholar] [CrossRef] [PubMed]
- Hoesseini, A.; Offerman, M.P.J.; van de Wall-Neecke, B.J.; Sewnaik, A.; Wieringa, M.H.; de Jong, R.J.B. Physicians’ clinical prediction of survival in head and neck cancer patients in the palliative phase. BMC Palliat. Care 2020, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- O’rorke, M.A.; Ellison, M.V.; Murray, L.J.; Moran, M.; James, J.; Anderson, L.A. Human papillomavirus related head and neck cancer survival: A systematic review and meta-analysis. Oral Oncol. 2012, 48, 1191–1201. [Google Scholar] [CrossRef]
- Benson, E.; Li, R.; Eisele, D.; Fakhry, C. The clinical impact of HPV tumor status upon head and neck squamous cell carcinomas. Oral Oncol. 2013, 50, 565–574. [Google Scholar] [CrossRef]
- Ibrahimovic, M.; Franzmann, E.; Mondul, A.M.; Weh, K.M.; Howard, C.; Hu, J.J.; Goodwin, W.J.; Kresty, L.A. Disparities in Head and Neck Cancer: A Case for Chemoprevention with Vitamin, D. Nutrients 2020, 12, 2638. [Google Scholar] [CrossRef]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef]
- Son, E.; Panwar, A.; Mosher, C.H.; Lydiatt, D. Cancers of the Major Salivary Gland. J. Oncol. Pract. 2018, 14, 99–108. [Google Scholar] [CrossRef]
- Gakunga, R.; Parkin, D.M.; Network, O.B.O.T.A.C.R. Cancer registries in Africa 2014: A survey of operational features and uses in cancer control planning. Int. J. Cancer 2015, 137, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, P.L.; Anchouche, S.; Gaffar, R.; Guadagno, E.; Ayad, T.; Poenaru, D. Barriers in Access to Care for Patients with Head and Neck Cancer in Resource-Limited Settings: A Systematic Review. JAMA Otolaryngol.-Head Neck Surg. 2020, 146, 291–297. [Google Scholar] [CrossRef]
- Korir, A.; Okerosi, N.; Ronoh, V.; Mutuma, G.; Parkin, M. Incidence of cancer in Nairobi, Kenya (2004–2008). Int. J. Cancer 2015, 137, 2053–2059. [Google Scholar] [CrossRef]
- Faggons, C.; Mabedi, C.; Shores, C.; Gopal, S. Review: Head and neck squamous cell carcinoma in sub-Saharan Africa. Malawi Med. J. 2015, 27, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Usman, S.; Jamal, A.; Teh, M.-T.; Waseem, A. Major Molecular Signaling Pathways in Oral Cancer Associated with Therapeutic Resistance. Front. Oral Health 2021, 1, 603160. [Google Scholar] [CrossRef]
- Rotimi, S.O.; Rotimi, O.A.; Salhia, B. A Review of Cancer Genetics and Genomics Studies in Africa. Front. Oncol. 2021, 10, 606400. [Google Scholar] [CrossRef] [PubMed]
- Coutts, K.A.; Seedat, J.; Vlok, E. The management of head and neck cancer in Africa. What lessons can be learned from African literature? S. Afr. J. Oncol. 2022, 6, 3. [Google Scholar] [CrossRef]
- Pickering, C.R.; Zhang, J.; Yoo, S.Y.; Bengtsson, L.; Moorthy, S.; Neskey, D.M.; Zhao, M.; Ortega Alves, M.V.; Chang, K.; Drummond, J.; et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013, 3, 770–781. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar]
- De Rubis, G.; Krishnan, S.R.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef]
- Alix-Panabières, C. The future of liquid biopsy. Nature 2020, 579, S9. [Google Scholar] [CrossRef]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: The future of cancer early detection. J. Transl. Med. 2023, 21, 118. [Google Scholar] [CrossRef]
- Heidrich, I.; Ačkar, L.; Mossahebi Mohammadi, P.; Pantel, K. Liquid biopsies: Potential and challenges. Int. J. Cancer 2021, 148, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Lianidou, E.; Pantel, K. Liquid biopsies. In Genes Chromosomes and Cancer; Blackwell Publishing Inc.: Hoboken, NJ, USA, 2019; Volume 58, pp. 219–232. [Google Scholar]
- Martins, I.; Ribeiro, I.P.; Jorge, J.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Melo, J.B.; Carreira, I.M. Liquid Biopsies: Applications for Cancer Diagnosis and Monitoring. Genes 2021, 12, 349. [Google Scholar] [CrossRef] [PubMed]
- Karachaliou, N.; Mayo-De-Las-Casas, C.; Molina-Vila, M.A.; Rosell, R. Real-time liquid biopsies become a reality in cancer treatment. Ann. Transl. Med. 2015, 3, 36. [Google Scholar] [CrossRef]
- Moussa, S.A.; Guemira, F.; Buisine, M.P. Detection of circulating tumor cells in the peripheral blood of nasopharyngeal carcinoma patients by a nested reverse transcriptase polymerase assay for cytokeratin 19 mRNA. Afr. J. Biotechnol. 2014, 13, 378–384. [Google Scholar]
- Samb, M.D.; Mbaye, F.; Ndiaye, M.M.; Toure, S.; Sembene, M. Bat 26 Microsatellite Instability in Oral Cavity Cancers in Senegal. J. Cancer Ther. 2023, 14, 25–37. [Google Scholar] [CrossRef]
- Diaga, S.P.; Silly, T.; Elmostafa, E.F.; Abdoul, B.S.; Demba, D.J.P.; Yacouba, D.; Babacar, M.; Maguette, S.-N.; Alioune, D.; Rokhaya, N.-D.; et al. Targeting Mutational Landscape of TP53 in patients diagnosed with Oral Cancer living in Senegal. J. Cancer Genet. Biomarkers 2022, 1, 22–32. [Google Scholar] [CrossRef]
- Gihbid, A.; Benzeid, R.; Faouzi, A.; El Alami, I.; Tawfiq, N.; Benchakroun, N.; Bendahhou, K.; Benider, A.; Guensi, A.; Khaali, W.; et al. The Dynamic Change in Plasma Epstein–Barr Virus DNA Load over a Long-Term Follow-Up Period Predicts Prognosis in Nasopharyngeal Carcinoma. Viruses 2022, 15, 66. [Google Scholar] [CrossRef]
- Gihbid, A.; Benzeid, R.; Faouzi, A.; Nourlil, J.; Tawfiq, N.; Benchakroun, N.; Guensi, A.; Bendahhou, K.; Benider, A.; El Benna, N.; et al. Circulating cell-free epstein–barr virus DNA levels and clinical features in Moroccan patients with nasopharyngeal carcinoma. Infect. Agents Cancer 2021, 16, 15. [Google Scholar] [CrossRef]
- Azab, N.; Zahran, F.; Amin, A.; Rady, N. DNA integrity in diagnosis of premalignant lesions. Med. Oral Patol. Oral Y Cirugía Bucal 2021, 26, e445–e450. [Google Scholar] [CrossRef]
- Khlifi, R.; Kallel, I.; Hammami, B.; Hamza-Chaffai, A.; Rebai, A. DNA repair gene polymorphisms and risk of head and neck cancer in the Tunisian population. J. Oral Pathol. Med. 2013, 43, 217–224. [Google Scholar] [CrossRef]
- Hassen, E.; Farhat, K.; Gabbouj, S.; Bouaouina, N.; Abdelaziz, H.; Chouchane, L. Epstein–Barr virus DNA quantification and follow-up in Tunisian nasopharyngeal carcinoma patients. Biomarkers 2011, 16, 274–280. [Google Scholar] [CrossRef]
- Gara, S.; Abdennebi, M.; Chatti, S.; Touati, S.; Ladgham, A.; Guemira, F. Association of NAT2 gene substitution mutation T341C with increased risk for head and neck cancer in Tunisia. Acta Oncol. 2007, 46, 834–837. [Google Scholar] [CrossRef]
- Farag, A.; Sabry, D.; Hassabou, N.; Alaa EL-Din, Y. MicroRNA-134/MicroRNA-200a Derived Salivary Exosomes are Novel Diagnostic Biomarkers of Oral Squamous Cell Carcinoma. Egypt Dent. J. 2021, 67, 367–377. [Google Scholar] [CrossRef]
- Abdelwhab, A.; Shaker, O.; Aggour, R.L. Expression of Mucin1 in saliva in oral squamous cell carcinoma and oral potentially malignant disorders (case control study). Oral Dis. 2023, 29, 1487–1494. [Google Scholar] [CrossRef]
- Elwakeel, N.; Shaker, O. Salivary insulin like growth factor binding protein-3 and transferrin levels in patients with oral squamous cell carcinoma versus oral lichen planus. Egypt. Dent. J. 2017, 63, 579–589. [Google Scholar] [CrossRef]
- Zergoun, A.A.; Zebboudj, A.; Sellam, S.L.; Kariche, N.; Djennaoui, D.; Ouraghi, S.; Kerboua, E.; Amir-Tidadini, Z.C.; Chilla, D.; Asselah, F.; et al. IL-6/NOS2 inflammatory signals regulate MMP-9 and MMP-2 activity and disease outcome in nasopharyngeal carcinoma patients. Tumor Biol. 2016, 37, 3505–3514. [Google Scholar] [CrossRef] [PubMed]
- M’hamdi, H.; Baizig, N.M.; ELHadj, O.E.; M’hamdi, N.; Attia, Z.; Gritli, S.; Gamoudi, A.; EL May, M.V.; El May, A. Usefulness of IGF-1 serum levels as diagnostic marker of nasopharyngeal carcinoma. Immunobiology 2016, 221, 1304–1308. [Google Scholar] [CrossRef]
- El-Benhawy, S.A.; El-Sheredy, H.G. Adipocyte-fatty Acid Binding Protein is Associated with Clinical Stage and Inflammatory Markers in Head and Neck Cancer Patients. J. Cancer Res. Treat. 2016, 4, 41–48. Available online: http://pubs.sciepub.com/jcrt/4/3/2 (accessed on 10 September 2023).
- Ayadi, W.; Karray-Hakim, H.; Feki, L.; Khabir, A.; Boudawara, T.; Ghorbel, A.; Daoud, J.; Frikha, M.; Hammami, A. IgA antibodies against the Epstein-Barr nuclear antigen1 as a valuable biomarker for the diagnosis of nasopharyngeal carcinoma in Tunisian patients. J. Med. Virol. 2009, 81, 1412–1421. [Google Scholar] [CrossRef]
- Houali, K.; Wang, X.; Shimizu, Y.; Djennaoui, D.; Nicholls, J.; Fiorini, S.; Bouguermouh, A.; Ooka, T. A New Diagnostic Marker for Secreted Epstein-Barr Virus–Encoded LMP1 and BARF1 Oncoproteins in the Serum and Saliva of Patients with Nasopharyngeal Carcinoma. Clin. Cancer Res. 2007, 13, 4993–5000. [Google Scholar] [CrossRef] [PubMed]
- Karray, H.; Ayadi, W.; Fki, L.; Hammami, A.; Daoud, J.; Drira, M.; Frikha, M.; Jlidi, R.; Middeldorp, J. Comparison of three different serological techniques for primary diagnosis and monitoring of nasopharyngeal carcinoma in two age groups from Tunisia. J. Med. Virol. 2005, 75, 593–602. [Google Scholar] [CrossRef]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Peterse, J.L.; van’t Veer, L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 2005, 5, 591–602. [Google Scholar] [CrossRef]
- Lara, O.; Tong, X.; Zborowski, M.; Chalmers, J.J. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp. Hematol. 2004, 32, 891–904. [Google Scholar] [CrossRef]
- Liu, H.E.; Triboulet, M.; Zia, A.; Vuppalapaty, M.; Kidess-Sigal, E.; Coller, J.; Natu, V.S.; Shokoohi, V.; Che, J.; Renier, C.; et al. Workflow optimization of whole genome amplification and targeted panel sequencing for CTC mutation detection. Npj Genom. Med. 2017, 2, 34. [Google Scholar] [CrossRef]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating Tumor Cells in Patients with Breast Cancer Dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef]
- Micalizzi, D.S.; Maheswaran, S.; Haber, D.A. A conduit to metastasis: Circulating tumor cell biology. Genes Dev. 2017, 31, 1827–1840. [Google Scholar] [CrossRef]
- Yeung, K.T.; Yang, J. Epithelial–mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Gires, O.; Stoecklein, N.H. Dynamic EpCAM expression on circulating and disseminating tumor cells: Causes and consequences. Cell. Mol. Life Sci. 2014, 71, 4393–4402. [Google Scholar] [CrossRef]
- Dhar, M.; Lam, J.N.; Walser, T.; Dubinett, S.M.; Rettig, M.B.; Di Carlo, D. Functional profiling of circulating tumor cells with an integrated vortex capture and single-cell protease activity assay. Proc. Natl. Acad. Sci. USA 2018, 115, 9986–9991. [Google Scholar] [CrossRef] [PubMed]
- Rejniak, K.A. Investigating dynamical deformations of tumor cells in circulation: Predictions from a theoretical model. Front. Oncol. 2012, 2, 111. [Google Scholar] [CrossRef]
- McCarty, O.J.; Mousa, S.A.; Bray, P.F.; Konstantopoulos, K. Immobilized platelets support human colon carcinoma cell tethering, rolling, and firm adhesion under dynamic flow conditions. Blood J. Am. Soc. Hematol. 2000, 96, 1789–1797. [Google Scholar]
- Steinert, G.; Schölch, S.; Niemietz, T.; Iwata, N.; García, S.A.; Behrens, B.; Voigt, A.; Kloor, M.; Benner, A.; Bork, U.; et al. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res. 2014, 74, 1694–1704. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Martin-Killias, P.; Plückthun, A.; Zangemeister-Wittke, U. EpCAM-targeted delivery of nanocomplexed siRNA to tumor cells with designed ankyrin repeat proteins. Mol. Cancer Ther. 2009, 8, 2674–2683. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Kraan, J.; Bolt, J.; van der Spoel, P.; Elstrodt, F.; Schutte, M.; Martens, J.W.; Gratama, J.W.; Sleijfer, S.; Foekens, J.A. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J. Natl. Cancer Inst. 2009, 101, 61–66. [Google Scholar] [CrossRef]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Röse, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 1–13. [Google Scholar] [CrossRef]
- Clawson, G.A.; Kimchi, E.; Patrick, S.D.; Xin, P.; Harouaka, R.; Zheng, S.; Berg, A.; Schell, T.; Staveley-O’carroll, K.F.; Neves, R.I.; et al. Circulating Tumor Cells in Melanoma Patients. PLoS ONE 2012, 7, e41052. [Google Scholar] [CrossRef]
- Zhou, M.-D.; Hao, S.; Williams, A.J.; Harouaka, R.A.; Schrand, B.; Rawal, S.; Ao, Z.; Brenneman, R.; Gilboa, E.; Lu, B.; et al. Separable Bilayer Microfiltration Device for Viable Label-free Enrichment of Circulating Tumour Cells. Sci. Rep. 2014, 4, 7392. [Google Scholar] [CrossRef]
- Hofman, V.; Long, E.; Ilie, M.; Bonnetaud, C.; Vignaud, J.M.; Fléjou, J.F.; Lantuejoul, S.; Piaton, E.; Mourad, N.; Butori, C.; et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology 2011, 23, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Mao, X.; Imrali, A.; Syed, F.; Mutsvangwa, K.; Berney, D.; Cathcart, P.; Hines, J.; Shamash, J.; Lu, Y.-J. Optimization and Evaluation of a Novel Size Based Circulating Tumor Cell Isolation System. PLoS ONE 2015, 10, e0138032. [Google Scholar] [CrossRef] [PubMed]
- Gascoyne, P.R.C.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.I.; Musso, N.; Romano, A.; Caruso, G.; Petralia, S.; Lanzanò, L.; Broggi, G.; Camarda, M. The Role of Dielectrophoresis for Cancer Diagnosis and Prognosis. Cancers 2022, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.C.; Lin, H.; Huang, C.; Hsu, H.; Wu, T.M.; Lee, C.; Chen, M.; Wang, H.; Tseng, C. Prognostic value of circulating tumor cells with podoplanin expression in patients with locally advanced or metastatic head and neck squamous cell carcinoma. Head Neck 2015, 37, 1448–1455. [Google Scholar] [CrossRef]
- Grisanti, S.; Almici, C.; Consoli, F.; Verardi, R.; Buglione, M.; Ferrari, V.; Bolzoni-Villaret, A.; Nicolai, P.; Magrini, S.; Simoncini, E. Circulating Tumor Cells (CTCS) in Patients with Recurrent/Metastatic Head and Neck Squamous Cell Carcinoma (HNSCC): Frequency, Clinical Significance and EGFR Expression. Ann. Oncol. 2012, 23, ix338. [Google Scholar] [CrossRef]
- Hristozova, T.; Konschak, R.; Stromberger, C.; Fusi, A.; Liu, Z.; Weichert, W.; Stenzinger, A.; Budach, V.; Keilholz, U.; Tinhofer, I. The presence of circulating tumor cells (CTCs) correlates with lymph node metastasis in nonresectable squamous cell carcinoma of the head and neck region (SCCHN). Ann. Oncol. 2011, 22, 1878–1885. [Google Scholar] [CrossRef]
- Jatana, K.R.; Balasubramanian, P.; Lang, J.C.; Yang, L.; Jatana, C.A.; White, E.; Agrawal, A.; Ozer, E.; Schuller, D.E.; Teknos, T.N.; et al. Significance of circulating tumor cells in patients with squamous cell carcinoma of the head and neck: Initial results. Arch. Otolaryngol. -Head Neck Surg. 2010, 136, 1274–1279. [Google Scholar] [CrossRef]
- Buglione, M.; Grisanti, S.; Almici, C.; Mangoni, M.; Polli, C.; Consoli, F.; Verardi, R.; Costa, L.; Paiar, F.; Pasinetti, N.; et al. Circulating Tumour Cells in locally advanced head and neck cancer: Preliminary report about their possible role in predicting response to non-surgical treatment and survival. Eur. J. Cancer 2012, 48, 3019–3026. [Google Scholar] [CrossRef]
- Kawada, T.; Takahashi, H.; Sakakura, K.; Ida, S.; Mito, I.; Toyoda, M.; Chikamatsu, K. Circulating tumor cells in patients with head and neck squamous cell carcinoma: Feasibility of detection and quantitation. Head Neck 2017, 39, 2180–2186. [Google Scholar] [CrossRef]
- Garrel, R.; Mazel, M.; Perriard, F.; Vinches, M.; Cayrefourcq, L.; Guigay, J.; Digue, L.; Aubry, K.; Alfonsi, M.; Delord, J.-P.; et al. Circulating Tumor Cells as a Prognostic Factor in Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma: The Circutec Prospective Study. Clin. Chem. 2019, 65, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Tada, H.; Takahashi, H.; Ida, S.; Nagata, Y.; Chikamatsu, K. Epithelial–Mesenchymal Transition Status of Circulating Tumor Cells Is Associated With Tumor Relapse in Head and Neck Squamous Cell Carcinoma. Anticancer Res. 2020, 40, 3559–3564. [Google Scholar] [CrossRef] [PubMed]
- Grisanti, S.; Almici, C.; Consoli, F.; Buglione, M.; Verardi, R.; Bolzoni-Villaret, A.; Bianchetti, A.; Ciccarese, C.; Mangoni, M.; Ferrari, L.; et al. Circulating Tumor Cells in Patients with Recurrent or Metastatic Head and Neck Carcinoma: Prognostic and Predictive Significance. PLoS ONE 2014, 9, e103918. [Google Scholar] [CrossRef]
- Liu, K.; Chen, N.; Wei, J.; Ma, L.; Yang, S.; Zhang, X. Clinical significance of circulating tumor cells in patients with locally advanced head and neck squamous cell carcinoma. Oncol. Rep. 2020, 43, 1525–1535. [Google Scholar] [CrossRef]
- Strati, A.; Koutsodontis, G.; Papaxoinis, G.; Angelidis, I.; Zavridou, M.; Economopoulou, P.; Kotsantis, I.; Avgeris, M.; Mazel, M.; Perisanidis, C.; et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann. Oncol. 2017, 28, 1923–1933. [Google Scholar] [CrossRef]
- Chikamatsu, K.; Tada, H.; Takahashi, H.; Kuwabara-Yokobori, Y.; Ishii, H.; Ida, S.; Shino, M. Expression of immune-regulatory molecules in circulating tumor cells derived from patients with head and neck squamous cell carcinoma. Oral Oncol. 2018, 89, 34–39. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. CR Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Koffler, D.; Agnello, V.; Winchester, R.; Kunkel, H.G. The Occurrence of Single-Stranded DNA in the Serum of Patients with Systemic Lupus Erythematosus and Other Diseases. J. Clin. Investig. 1973, 52, 198–204. [Google Scholar] [CrossRef]
- Tan, E.M.; Schur, P.H.; Carr, R.I.; Kunkel, H.G. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J. Clin. Investig. 1966, 45, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef]
- Mouliere, F.; Mair, R.; Chandrananda, D.; Marass, F.; Smith, C.G.; Su, J.; Morris, J.; Watts, C.; Brindle, K.M.; Rosenfeld, N. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol. Med. 2018, 10, e9323. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2007, 1775, 181–232. [Google Scholar] [CrossRef]
- Sriram, K.B.; Relan, V.; Clarke, B.E.; Duhig, E.; Windsor, M.N.; Matar, K.S.; Naidoo, R.; Passmore, L.; McCaul, E.; Courtney, D.; et al. Pleural fluid cell-free DNA integrity index to identify cytologically negative malignant pleural effusions including mesotheliomas. BMC Cancer 2012, 12, 428. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomarkers Prev. 1994, 3, 67–71. [Google Scholar]
- Raja, R.; Kuziora, M.; Brohawn, P.; Higgs, B.; Gupta, A.; Dennis, P.; Ranade, K. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin. Cancer Res. 2018, 24, 6212–6222. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Wu, X.; Tong, X.; Wang, X.; Chang, Z.; Mao, Y.; Chen, X.; Sun, J.; Wang, Z.; Hong, Z.; et al. Circulating Tumor DNA Mutation Profiling by Targeted Next Generation Sequencing Provides Guidance for Personalized Treatments in Multiple Cancer Types. Sci. Rep. 2017, 7, 583. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef]
- Gao, Q.; Zeng, Q.; Wang, Z.; Li, C.; Xu, Y.; Cui, P.; Zhu, X.; Lu, H.; Wang, G.; Cai, S.; et al. Circulating cell-free DNA for cancer early detection. Innovation 2022, 3, 100259. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA Comprises an in Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Giacona, M.B.; Ruben, G.C.; Iczkowski, K.A.; Roos, T.B.; Porter, D.M.; Sorenson, G.D. Cell-free DNA in human blood plasma: Length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Umetani, N.; Giuliano, A.E.; Hiramatsu, S.H.; Amersi, F.; Nakagawa, T.; Martino, S.; Hoon, D.S. Prediction of Breast Tumor Progression by Integrity of Free Circulating DNA in Serum. J. Clin. Oncol. 2006, 24, 4270–4276. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Chan, K.C.A.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.F.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal Plasma DNA Sequencing Reveals the Genome-Wide Genetic and Mutational Profile of the Fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; Mouliere, F.; Gongora, C.; Ollier, J.; Robert, B.; Ychou, M.; Del Rio, M.; Molina, F. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010, 38, 6159–6175. [Google Scholar] [CrossRef]
- Trejo-Becerril, C.; Pérez-Cárdenas, E.; Taja-Chayeb, L.; Anker, P.; Herrera-Goepfert, R.; Medina-Velázquez, L.A.; Hidalgo-Miranda, A.; Pérez-Montiel, D.; Chávez-Blanco, A.; Cruz-Velázquez, J.; et al. Cancer Progression Mediated by Horizontal Gene Transfer in an In Vivo Model. PLoS ONE 2012, 7, e52754. [Google Scholar] [CrossRef]
- Raghuram, G.V.; Gupta, D.; Subramaniam, S.; Gaikwad, A.; Khare, N.K.; Nobre, M.; Nair, N.K.; Mittra, I. Physical shearing imparts biological activity to DNA and ability to transmit itself horizontally across species and kingdom boundaries. BMC Mol. Biol. 2017, 18, 21. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Hsieh, J.-S.; Chang, M.-Y.; Huang, T.-J.; Chen, F.-M.; Cheng, T.-L.; Alexandersen, K.; Huang, Y.-S.; Tzou, W.-S.; Lin, S.-R. Molecular Detection of APC, K-ras, and p53 Mutations in the Serum of Colorectal Cancer Patients as Circulating Biomarkers. Mol. Med. 2004, 28, 721–726. [Google Scholar] [CrossRef]
- Fujiwara, K.; Fujimoto, N.; Tabata, M.; Nishii, K.; Matsuo, K.; Hotta, K.; Kozuki, T.; Aoe, M.; Kiura, K.; Ueoka, H.; et al. Identification of Epigenetic Aberrant Promoter Methylation in Serum DNA Is Useful for Early Detection of Lung Cancer. Clin. Cancer Res. 2005, 11, 1219–1225. [Google Scholar] [CrossRef]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef]
- Tamkovich, S.N.; Cherepanova, A.V.; Kolesnikova, E.V.; Rykova, E.Y.; Pyshnyi, D.V.; Vlassov, V.V.; Laktionov, P.P. Circulating DNA and DNase Activity in Human Blood. Ann. New York Acad. Sci. 2006, 1075, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Dennis Lo, Y.M.; Zhang, J.; Leung, T.N.; Lau, T.K.; Chang, A.M.Z.; Magnus Hjelm, N. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 1999, 64, 218–224. [Google Scholar]
- Carvalho, A.L.; Jeronimo, C.; Kim, M.M.; Henrique, R.; Zhang, Z.; Hoque, M.O.; Chang, S.; Brait, M.; Nayak, C.S.; Jiang, W.-W.; et al. Evaluation of Promoter Hypermethylation Detection in Body Fluids as a Screening/Diagnosis Tool for Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2008, 14, 97–107. [Google Scholar] [CrossRef]
- Lin, L.-H.; Chang, K.-W.; Kao, S.-Y.; Cheng, H.-W.; Liu, C.-J. Increased Plasma Circulating Cell-Free DNA Could Be a Potential Marker for Oral Cancer. Int. J. Mol. Sci. 2018, 19, 3303. [Google Scholar] [CrossRef] [PubMed]
- Rapado-González, Ó.; López-Cedrún, J.L.; Lago-Lestón, R.M.; Abalo, A.; Rubin-Roger, G.; Salgado-Barreira, Á.; López-López, R.; Muinelo-Romay, L.; Suárez-Cunqueiro, M.M. Integrity and quantity of salivary cell-free DNA as a potential molecular biomarker in oral cancer: A preliminary study. J. Oral Pathol. Med. 2022, 51, 429–435. [Google Scholar] [CrossRef]
- Flach, S.; Kumbrink, J.; Walz, C.; Hess, J.; Drexler, G.; Belka, C.; Canis, M.; Jung, A.; Baumeister, P. Analysis of genetic variants of frequently mutated genes in human papillomavirus-negative primary head and neck squamous cell carcinoma, resection margins, local recurrences and corresponding circulating cell-free DNA. J. Oral Pathol. Med. 2022, 51, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Coulet, F.; Blons, H.; Cabelguenne, A.; Lecomte, T.; Lacourreye, O.; Brasnu, D.; Beaune, P.; Zucman, J.; Laurent-Puig, P. Detection of plasma tumor DNA in head and neck squamous cell carcinoma by microsatellite typing and p53 mutation analysis. Cancer Res. 2000, 60, 707–711. [Google Scholar]
- Husain, N.; Verma, T.; Kumari, S.; Mishra, S.; Rastogi, M.; Tiwari, V.; Agarwal, G.R.; Anand, N. Circulating free DNA as a marker of response to chemoradiation in locally advanced head and neck squamous cell carcinoma. Indian J. Pathol. Microbiol. 2020, 63, 521–526. [Google Scholar] [CrossRef]
- Mazurek, A.M.; Rutkowski, T.; Fiszer-Kierzkowska, A.; Małusecka, E.; Składowski, K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. 2016, 54, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, R.; Imai, A.; Mima, M.; Yamada, S.; Takeuchi, K.; Mochizuki, D.; Shinmura, D.; Kita, J.-Y.; Nakagawa, T.; Kurokawa, T.; et al. Novel prognostic value and potential utility of opioid receptor gene methylation in liquid biopsy for oral cavity cancer. Curr. Probl. Cancer 2022, 46, 100834. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, A.; Hariharan, A.K.; Hasina, R.; Nair, J.R.; Katragadda, S.; Irusappan, S.; Ravichandran, A.; Veeramachaneni, V.; Bettadapura, R.; Bhati, M.; et al. Ultrasensitive detection of tumor-specific mutations in saliva of patients with oral cavity squamous cell carcinoma. Cancer 2021, 127, 1576–1589. [Google Scholar] [CrossRef] [PubMed]
- Burgener, J.M.; Zou, J.; Zhao, Z.; Zheng, Y.; Shen, S.Y.; Huang, S.H.; Keshavarzi, S.; Xu, W.; Liu, F.-F.; Liu, G.; et al. Tumor-Naïve multimodal profiling of circulating tumor DNA in head and neck squamous cell carcinoma. Clin. Cancer Res. 2021, 27, 4230–4244. [Google Scholar] [CrossRef] [PubMed]
- Galot, R.; van Marcke, C.; Helaers, R.; Mendola, A.; Goebbels, R.-M.; Caignet, X.; Ambroise, J.; Wittouck, K.; Vikkula, M.; Limaye, N.; et al. Liquid biopsy for mutational profiling of locoregional recurrent and/or metastatic head and neck squamous cell carcinoma. Oral Oncol. 2020, 104, 104631. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef]
- Schröck, A.; Leisse, A.; de Vos, L.; Gevensleben, H.; Dröge, F.; Franzen, A.; Wachendörfer, M.; Schröck, F.; Ellinger, J.; Teschke, M.; et al. Free-Circulating Methylated DNA in Blood for Diagnosis, Staging, Prognosis, and Monitoring of Head and Neck Squamous Cell Carcinoma Patients: An Observational Prospective Cohort Study. Clin. Chem. 2017, 63, 1288–1296. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, X.; Li, L. Biogenesis and function of extracellular miRNAs. ExRNA 2019, 1, 38. [Google Scholar] [CrossRef]
- Umu, S.U.; Langseth, H.; Bucher-Johannessen, C.; Fromm, B.; Keller, A.; Meese, E.; Lauritzen, M.; Leithaug, M.; Lyle, R.; Rounge, T.B. A comprehensive profile of circulating RNAs in human serum. RNA Biol. 2017, 15, 242–250. [Google Scholar] [CrossRef]
- Suraj, S.; Dhar, C.; Srivastava, S. Circulating nucleic acids: An analysis of their occurrence in malignancies (review). Biomed. Rep. 2017, 6, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Fok, E.T.; Scholefield, J.; Fanucchi, S.; Mhlanga, M.M.; Fadeel, B.; Achinger-Kawecka, J.; Clark, S.J.; Ziogas, D.E.; Katsios, C.; Roukos, D.H. The emerging molecular biology toolbox for the study of long noncoding RNA biology. Epigenomics 2017, 9, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Kao, S.Y.; Tu, H.F.; Tsai, M.M.; Chang, K.W.; Lin, S.C. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010, 16, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; John, M.A.R.S.; Wong, D.T.; Zhou, X.; Kim, Y.; Sinha, U.; Jordan, R.C.K.; Eisele, D.; Abemayor, E.; Elashoff, D.; et al. Salivary Transcriptome Diagnostics for Oral Cancer Detection. Clin. Cancer Res. 2004, 10, 8442–8450. [Google Scholar] [CrossRef]
- Fayda, M.; Isin, M.; Tambas, M.; Guveli, M.; Meral, R.; Altun, M.; Sahin, D.; Ozkan, G.; Sanli, Y.; Isin, H.; et al. Do circulating long non-coding RNAs (lncRNAs) (LincRNA-p21, GAS 5, HOTAIR) predict the treatment response in patients with head and neck cancer treated with chemoradiotherapy? Tumor Biol. 2016, 37, 3969–3978. [Google Scholar] [CrossRef]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef]
- Sheridan, C. Exosome cancer diagnostic reaches market. Nat. Biotechnol. 2016, 34, 359–360. [Google Scholar] [CrossRef]
- Vanni, I.; Alama, A.; Grossi, F.; Dal Bello, M.G.; Coco, S. Exosomes: A new horizon in lung cancer. Drug Discov. Today 2017, 22, 927–936. [Google Scholar] [CrossRef]
- Milane, L.; Singh, A.; Mattheolabakis, G.; Suresh, M.; Amiji, M.M. Exosome mediated communication within the tumor microenvironment. J. Control. Release 2015, 219, 278–294. [Google Scholar] [CrossRef]
- Adeola, H.A.; Holmes, H.; Temilola, D.O. Diagnostic Potential of Salivary Exosomes in Oral Cancer. In Oral Cancer—Current Concepts and Future Perspectives; Sridharan, G., Ed.; IntechOpen: London, UK, 2020. [Google Scholar]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Greening, D.W.; Zhu, H.-J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.; Simpson, R. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2019, 121, 109553. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.; Camussi, F.; Broccoletti, R.; Gambino, A.; Cabras, M.; Molinaro, L.; Carossa, S.; Camussi, G.; Arduino, P.G. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 2018, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Coon, J.; Kingsley, K.; Howard, K.M. miR-365 (microRNA): Potential Biomarker in Oral Squamous Cell Carcinoma Exosomes and Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 5317. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Guo, X.; Li, X.; Liao, C.; Wang, X.; He, K. Plasma-Derived Exosomal microRNA-130a Serves as a Noninvasive Biomarker for Diagnosis and Prognosis of Oral Squamous Cell Carcinoma. J. Oncol. 2021, 2021, 5547911. [Google Scholar] [CrossRef]
- Tong, F.; Mao, X.; Zhang, S.; Xie, H.; Yan, B.; Wang, B.; Sun, J.; Wei, L. HPV + HNSCC-derived exosomal miR-9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer Lett. 2020, 478, 34–44. [Google Scholar] [CrossRef]
- Li, S.; Han, Y.; Lu, M.; Liu, Z.; Jin, J.; Guo, Q.; Wang, Y.; Liu, H. Mesenchymal stem cell-exosome-mediated matrix metalloproteinase 1 participates in oral leukoplakia and carcinogenesis by inducing angiogenesis. J. Oral Pathol. Med. 2022, 51, 638–648. [Google Scholar] [CrossRef]
- Ono, K.; Eguchi, T.; Sogawa, C.; Calderwood, S.K.; Futagawa, J.; Kasai, T.; Seno, M.; Okamoto, K.; Sasaki, A.; Kozaki, K. HSP-enriched properties of extracellular vesicles involve survival of metastatic oral cancer cells. J. Cell. Biochem. 2018, 119, 7350–7362. [Google Scholar] [CrossRef] [PubMed]
- Meckes, D.G., Jr.; Shair, K.H.Y.; Marquitz, A.R.; Kung, C.-P.; Edwards, R.H.; Raab-Traub, N. Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. USA 2010, 107, 20370–20375. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; McCarthy, M.I.; Schwenk, J.M. Genetics meets proteomics: Perspectives for large population-based studies. Nat. Rev. Genet. 2021, 22, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Basheeth, N.; Patil, N. Biomarkers in Head and Neck Cancer an Update. Indian J. Otolaryngol. Head Neck Surg. 2019, 71, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Amenábar, J.M.; Da Silva, B.M.; Punyadeera, C. Salivary protein biomarkers for head and neck cancer. Expert Rev. Mol. Diagn. 2020, 20, 305–313. [Google Scholar] [CrossRef]
- Yu, J.S.; Chen, Y.T.; Chiang, W.F.; Hsiao, Y.C.; Chu, L.J.; See, L.C.; Wu, C.S.; Tu, H.T.; Chen, H.W.; Chen, C.C.; et al. Saliva protein biomarkers to detect oral squamous cell carcinoma in a high-risk population in Taiwan. Proc. Natl. Acad. Sci. USA 2016, 113, 11549–11554. [Google Scholar] [CrossRef]
- Adeola, H.; Afrogheh, A.; Hille, J. The burden of head and neck cancer in Africa: The status quo and research prospects. S. Afr. Dent. J. 2018, 73, 477–488. [Google Scholar] [CrossRef]
- Fagan, J.J. Africa: A window on challenges and opportunities for head and neck cancer. Laryngoscope Investig. Otolaryngol. 2021, 6, 414–419. [Google Scholar] [CrossRef]
- Adeola, H.A.; Soyele, O.O.; Adefuye, A.O.; Jimoh, S.A.; Butali, A. Omics-based molecular techniques in oral pathology centred cancer: Prospect and challenges in Africa. Cancer Cell Int. 2017, 17, 61. [Google Scholar] [CrossRef]
- Ottersen, T.; Grépin, K.A.; Henderson, K.; Pinkstaff, C.B.; Norheim, O.F.; Røttingen, J.-A. New approaches to ranking countries for the allocation of development assistance for health: Choices, indicators and implications. Health Policy Plan. 2018, 33, i31–i46. [Google Scholar] [CrossRef]
- Kruk, M.E.; Gage, A.D.; Arsenault, C.; Jordan, K.; Leslie, H.H.; Roder-DeWan, S.; Adeyi, O.; Barker, P.; Daelmans, B.; Doubova, S.V.; et al. High-quality health systems in the Sustainable Development Goals era: Time for a revolution. Lancet Glob. Health 2018, 6, e1196–e1252. [Google Scholar] [CrossRef] [PubMed]
- Mourad, M.; Jetmore, T.; Jategaonkar, A.A.; Moubayed, S.; Moshier, E.; Urken, M.L. Epidemiological Trends of Head and Neck Cancer in the United States: A SEER Population Study. J. Oral Maxillofac. Surg. 2017, 75, 2562–2572. [Google Scholar] [CrossRef] [PubMed]
- Adeola, H.A.; Bello, I.O.; Aruleba, R.T.; Francisco, N.M.; Adekiya, T.A.; Adefuye, A.O.; Ikwegbue, P.C.; Musaigwa, F. The Practicality of the Use of Liquid Biopsy in Early Diagnosis and Treatment Monitoring of Oral Cancer in Resource-Limited Settings. Cancers 2022, 14, 1139. [Google Scholar] [CrossRef]
- Gillison, M.L.; Chaturvedi, A.K.; Anderson, W.F.; Fakhry, C. Epidemiology of Human Papillomavirus–Positive Head and Neck Squamous Cell Carcinoma. J. Clin. Oncol. 2015, 33, 3235–3242. [Google Scholar] [CrossRef]
- Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; Anderson, B.O.; et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016 a systematic analysis for the global burden of disease study global burden of disease cancer collaboration. JAMA Oncol. 2018, 4, 1553–1568. [Google Scholar]
- Kong, L.; Birkeland, A.C. Liquid Biopsies in Head and Neck Cancer: Current State and Future Challenges. Cancers 2021, 13, 1874. [Google Scholar] [CrossRef]
- Temilola, D.O.; Wium, M.; Coulidiati, T.H.; Adeola, H.A.; Carbone, G.M.; Catapano, C.V.; Zerbini, L.F. The Prospect and Challenges to the Flow of Liquid Biopsy in Africa. Cells 2019, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Bahnassy, A.A.; Zekri, A.-R.N.; El-Bastawisy, A.; Fawzy, A.; Shetta, M.; Hussein, N.; Omran, D.; Ahmed, A.A.S.; El-Labbody, S.S. Circulating tumor and cancer stem cells in hepatitis C virus-associated liver disease. World J. Gastroenterol. 2014, 20, 18240–18248. [Google Scholar] [CrossRef]
- Iyer, P.; Zekri, A.-R.; Hung, C.-W.; Schiefelbein, E.; Ismail, K.; Hablas, A.; Seifeldin, I.A.; Soliman, A.S. Concordance of DNA methylation pattern in plasma and tumor DNA of Egyptian hepatocellular carcinoma patients. Exp. Mol. Pathol. 2010, 88, 107–111. [Google Scholar] [CrossRef]
- Marchio, A.; Atsama, M.A.; Béré, A.; Komas, N.-P.; Noah, D.N.; Atangana, P.J.A.; Camengo-Police, S.-M.; Njouom, R.; Bekondi, C.; Pineau, P. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin. Exp. Med. 2018, 18, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.H.; Elkhoda, T.R.; Anwar, R.; Habeeb, M.R.; Mohammed, M.A. Regulation of Cancer Stem Cell Marker (CD133) by Transforming Growth Factor Beta in Hepatocellular Carcinoma. Int. J. Cancer Res. 2014, 10, 65–73. [Google Scholar] [CrossRef][Green Version]
- El-Yazeed, A.S.A.; Montaser, L.M.; Sonbol, A.; El-Gammal, A.S. Role of circulating endothelial progenitor cells in patients with breast cancer. Menoufia Med. J. 2019, 32, 770. [Google Scholar] [CrossRef]
- Elnagdy, M.H.; Farouk, O.; Seleem, A.K.; Nada, H.A. TFF1 and TFF3 mRNAs Are Higher in Blood from Breast Cancer Patients with Metastatic Disease than Those without. J. Oncol. 2018, 2018, 4793498. [Google Scholar] [CrossRef]
- Hussein, N.A.; Mohamed, S.N.; Ahmed, M.A. Plasma ALU-247, ALU-115, and cfDNA Integrity as Diagnostic and Prognostic Biomarkers for Breast Cancer. Appl. Biochem. Biotechnol. 2019, 187, 1028–1045. [Google Scholar] [CrossRef]
- Sabry, D.; El-Deek, S.E.; Maher, M.; El-Baz, M.A.; El-Bader, H.M.; Amer, E.; Hassan, E.A.; Fathy, W.; El-Deek, H.E. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: Impact of HIF-1α-VEGF signaling pathway. Mol. Cell Biochem. 2019, 454, 177–189. [Google Scholar] [CrossRef]
- Teama, S.H.; Agwa, S.H. Detection of circulating tumor cells by nested RT-PCR targeting EGFR/CEA/CK20mRNAs in colorectal carcinoma patients. Egypt. J. Med. Hum. Genet. 2010, 11, 173–180. [Google Scholar] [CrossRef][Green Version]
- El-Gayar, D.; El-Abd, N.; Hassan, N.; Ali, R. Increased Free Circulating DNA Integrity Index as a Serum Biomarker in Patients with Colorectal Carcinoma. Asian Pac. J. Cancer Prev. 2016, 17, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Soliman, S.E.-S.; Alhanafy, A.M.; Habib, M.S.E.; Hagag, M.; Ibrahem, R.A.L. Serum circulating cell free DNA as potential diagnostic and prognostic biomarker in non small cell lung cancer. Biochem. Biophys. Rep. 2018, 15, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Abd-El-Fattah, A.A.; Sadik, N.A.H.; Shaker, O.G.; Aboulftouh, M.L. Differential MicroRNAs Expression in Serum of Patients with Lung Cancer, Pulmonary Tuberculosis, and Pneumonia. Cell Biochem. Biophys. 2013, 67, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Hanekom, G.S.; Stubbings, H.M.; Johnson, C.A.; Kidson, S.H. The detection of circulating melanoma cells correlates with tumour thickness and ulceration but is not predictive of metastasis for patients with primary melanoma. Melanoma Res. 1999, 9, 465–473. [Google Scholar] [CrossRef]
- Hashad, D.; Elbanna, A.; Ibrahim, A.; Khedr, G. Evaluation of the Role of Circulating Long Non-Coding RNA H19 as a Promising Novel Biomarker in Plasma of Patients with Gastric Cancer. J. Clin. Lab. Anal. 2016, 30, 1100–1105. [Google Scholar] [CrossRef]
- Mahmoud, E.H.; Fawzy, A.; Elshimy, R.A. Serum MicroRNA-21 Negatively Relates to Expression of Programmed Cell Death-4 in Patients with Epithelial Ovarian Cancer. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 33–38. [Google Scholar] [CrossRef]
- Motawi, T.K.; Rizk, S.M.; Ibrahim, T.M.; Ibrahim, I.A.R. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochem Funct. 2016, 34, 142–148. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Hegazy, M.G.; Kotb, Y.M.; Essawy, N.O. Evaluation of urinary microRNA panel in bladder cancer diagnosis: Relation to bilharziasis. Transl. Res. 2015, 165, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, A.; Sweify, K.M.; El-Fayoumy, H.M.; Nofal, N. Quantitative analysis of plasma cell-free DNA and its DNA integrity in patients with metastatic prostate cancer using ALU sequence. J. Egypt. Natl. Cancer Inst. 2016, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- van der Vaart, M.; Semenov, D.V.; Kuligina, E.V.; Richter, V.A.; Pretorius, P.J. Characterisation of circulating DNA by parallel tagged sequencing on the 454 platform. Clin. Chim. Acta 2009, 409, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Hosny, G.; Farahat, N.; Hainaut, P. TP53 mutations in circulating free DNA from Egyptian patients with non-Hodgkin’s lymphoma. Cancer Lett. 2009, 275, 234–239. [Google Scholar] [CrossRef]
- Legason, I.D.; Ogwang, M.D.; Chamba, C.; Mkwizu, E.; El Mouden, C.; Mwinula, H.; Chirande, L.; Schuh, A.; Chiwanga, F. A protocol to clinically evaluate liquid biopsies as a tool to speed up diagnosis of children and young adults with aggressive infection-related lymphoma in East Africa “(AI-REAL)”. BMC Cancer 2022, 22, 484. [Google Scholar] [CrossRef]
- Elhamamsy, A.R.; El Sharkawy, M.S.; Zanaty, A.F.; Mahrous, M.A.; Mohamed, A.E.; Abushaaban, E.A. Circulating miR-92a, miR-143 and miR-342 in Plasma are Novel Potential Biomarkers for Acute Myeloid Leukemia. Int. J. Mol. Cell. Med. 2017, 6, 77–86. [Google Scholar] [CrossRef]
- Swellam, M.; El-Khazragy, N. Clinical impact of circulating microRNAs as blood-based marker in childhood acute lymphoblastic leukemia. Tumor Biol. 2016, 37, 10571–10576. [Google Scholar] [CrossRef]
- Marincowitz, G.J.O.; Marincowitz, C. Family physicians and the big question: A universal income grant for South Africa? S. Afr. Fam. Pract. 2023, 65, e1–e2. [Google Scholar] [CrossRef] [PubMed]
- Fusheini, A.; Eyles, J. Achieving universal health coverage in South Africa through a district health system approach: Conflicting ideologies of health care provision. BMC Health Serv. Res. 2016, 16, 558. [Google Scholar] [CrossRef] [PubMed]
- Okoturo, E.; Osasuyi, A.; Opaleye, T. Genetic Polymorphism of Head and Neck Cancers in African Populations: A Systematic Review. OTO Open 2020, 4, 2473974X20942202. [Google Scholar] [CrossRef] [PubMed]
- Koukourakis, I.M.; Xanthopoulou, E.; Koukourakis, M.I. Using Liquid Biopsy to Predict Relapse After Radiotherapy in Squamous Cell Head-Neck and Esophageal Cancer. Cancer Diagn. Progn. 2023, 3, 403–410. Available online: https://www.cancerdiagnosisprognosis.org/article/233/using-liquid-biopsy-to-predict-relapse-after-radiotherapy-in-squamous-cell-head-neck-and-esophageal-cancer (accessed on 10 September 2023). [CrossRef]
- Wu, C.-F.; Lin, L.; Mao, Y.-P.; Deng, B.; Lv, J.-W.; Zheng, W.-H.; Wen, D.-W.; Kou, J.; Chen, F.-P.; Yang, X.-L.; et al. Liquid biopsy posttreatment surveillance in endemic nasopharyngeal carcinoma: A cost-effective strategy to integrate circulating cell-free Epstein-Barr virus DNA. BMC Med. 2021, 19, 193. [Google Scholar] [CrossRef]
- Alshahrani, S.A.; Al-Qahtani, W.S.; Almufareh, N.A.; Domiaty, D.M.; Albasher, G.I.; Safhi, F.A.; AlQassim, F.A.; Alotaibi, M.A.; Al-Hazani, T.M.; Almutlaq, B.A. Oral cancer among Khat users: Finding evidence from DNA analysis of nine cancer-related gene mutations. BMC Oral Health 2021, 21, 626. [Google Scholar] [CrossRef]
- Perakis, S.; Speicher, M.R. Emerging concepts in liquid biopsies. BMC Med. 2017, 15, 75. [Google Scholar] [CrossRef]
- Shahid, N.; Iqbal, A.; Siddiqui, A.J.; Shoaib, M.; Musharraf, S.G. Plasma metabolite profiling and chemometric analyses of tobacco snuff dippers and patients with oral cancer: Relationship between metabolic signatures. Head Neck 2019, 41, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Wojakowska, A.; Zebrowska, A.; Skowronek, A.; Rutkowski, T.; Polanski, K.; Widlak, P.; Marczak, L.; Pietrowska, M. Metabolic Profiles of Whole Serum and Serum-Derived Exosomes Are Different in Head and Neck Cancer Patients Treated by Radiotherapy. J. Pers. Med. 2020, 10, 229. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Kooi, I.E.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef]
- Adeola, H.A.; Adefuye, A.; Soyele, O.; Butali, A. The dentist-scientist career pathway in Africa: Opportunities and obstacles. Korean J. Med. Educ. 2018, 30, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Umapathy, V.R.; Natarajan, P.M.; Swamikannu, B.; Moses, J.; Jones, S.; Chandran, M.P.; Anbumozhi, M.K. Emerging Biosensors for Oral Cancer Detection and Diagnosis—A Review Unravelling Their Role in Past and Present Advancements in the Field of Early Diagnosis. Biosensors 2022, 12, 498. [Google Scholar] [CrossRef] [PubMed]
- Uthoff, R.D.; Song, B.; Sunny, S.; Patrick, S.; Suresh, A.; Kolur, T.; Keerthi, G.; Spires, O.; Anbarani, A.; Wilder-Smith, P.; et al. Point-of-care, smartphone-based, dual-modality, dual-view, oral cancer screening device with neural network classification for low-resource communities. PLoS ONE 2018, 13, e0207493. [Google Scholar] [CrossRef]
- Mohamed, N.; van de Goor, R.; El-Sheikh, M.; Elrayah, O.; Osman, T.; Nginamau, E.S.; Johannessen, A.C.; Suleiman, A.; Costea, D.E.; Kross, K.W. Feasibility of a Portable Electronic Nose for Detection of Oral Squamous Cell Carcinoma in Sudan. Healthcare 2021, 9, 534. [Google Scholar] [CrossRef] [PubMed]
- Gattuso, G.; Crimi, S.; Lavoro, A.; Rizzo, R.; Musumarra, G.; Gallo, S.; Facciponte, F.; Paratore, S.; Russo, A.; Bordonaro, R.; et al. Liquid Biopsy and Circulating Biomarkers for the Diagnosis of Precancerous and Cancerous Oral Lesions. Non-Coding RNA 2022, 8, 60. [Google Scholar] [CrossRef] [PubMed]
| Sample Type | Study Design | Detection Method | Country | Study Findings | Authors (Year) |
|---|---|---|---|---|---|
| Circulating Tumor Cell | |||||
| Blood | 30 NPC patients and 20 healthy controls | RT-PCR of CK19 | Tunisia | The study found an improved CK19 RT-PCR assay that is sensitive and has a high clinical specificity of 73% to detect minimal metastatic disease in patients with NPC. | Moussa et al. (2014) [26] |
| Circulating Tumor DNA | |||||
| Blood | 40 OC patients and 52 healthy controls | NGS | Senegal | This study showed that the instability of the Bat 26 microsatellite could occur early in oral cavity cancers. | Samb et al. (2023) [27] |
| Blood | 88 OC patients and 94 healthy controls | RT-PCR quantification of TP53 gene | Senegal | The study found a strong linkage disequilibrium between the two most common cancer-related variants in Senegalese patients. | Diaga et al. (2022) [28] |
| Blood | 142 patients | RT-PCR for plasma EBV ctDNA | Morocco | The study reported a strong evidence that EBV DNA load testing is a promising dynamic and a minimally invasive biomarker for the prognosis and follow-up of NPC. | Gihbid et al. (2022) [29] |
| Blood | 121 NPC patients and 60 healthy controls | RT-PCR for plasma EBV ctDNA | Morocco | The study found that pre-treatment EBV DNA can be a useful prognostic biomarker in clinical decision-making and in improving the treatment of NPC in Morocco. | Gihbid et al. (2021) [30] |
| Saliva | 30 oral cancer patients, 21 OLP patients, 12 leukoplakia patients, and 30 healthy controls | DNA integrity index measured with RT-PCR | Egypt | The study showed that salivary DNA integrity index showed poor diagnostic abilities in differentiating between the oral cancer and premalignant lesions. | Azab et al. (2021) [31] |
| Blood | 169 HNC patients and 261 healthy controls | SNP PCR/RFLP analysis of XRCC1, ERCC2, and ERCC3 genes | Tunisia | The study showed that XRCC1 Arg399Gln polymorphism, which correlates with occupational exposure in Tunisian population, is associated with an increased risk of developing HNC. | Khlifi et al. (2013) [32] |
| Blood | 74 NPC patients | RT-PCR for plasma EBV ctDNA | Tunisia | The study found that the EBV DNA load quantification after treatment may be a useful predictor of disease progression and survival. | Hassen et al. (2011) [33] |
| Blood | 64 HNSSC patients and 160 healthy controls | PCR/RFLP analysis for NAT2 gene | Tunisia | The study found the T341C mutation of NAT2 gene to be associated with an elevated risk for head and neck cancer in Tunisian population. | Gara et al. (2007) [34] |
| Exosomes | |||||
| Saliva | 14 OSCC patients, 17 smokers, and 6 healthy controls | RT-PCR to assess the expression of microRNA-200a and microRNA-134 | Egypt | The study showed that isolated salivary exosomes can provide a stable and non-invasive route for the evaluation of different salivary biomarkers, which can be a useful tool in the early detection of oral cancer. | Farag et al. (2021) [35] |
| Circulating Proteins and Peptides | |||||
| Saliva | 40 OSCC patients and 20 healthy controls | Measurement of mucin1 expression using RT-PCR | Egypt | Th study found that the expression level of mucin1 in saliva might be a potential biomarker for diagnosing oral potentially malignant disorders and oral squamous cell carcinoma. | Abdelwhab et al. (2023) [36] |
| Saliva | 40 OSCC patients, 40 OLP patients, and 40 healthy controls | Measurement of TF and IGFbP-3 levels using enzyme-linked immunosorbent assay | Egypt | The study showed that IGFBP-3 and Tf seem to play a role in pathogenesis of both OSCC and OLP and could be considered as reliable markers in the diagnosis of OSCC and OLP. | Elwakeel et al. (2017) [37] |
| Blood | 17 NPC patients and 8 healthy controls | Measurement of IL-6 levels in plasma using enzyme-linked immunosorbent assay | Algeria | The study showed that the IL-6/NOS2 inflammatory signals are involved in the regulation of MMP-9- and MMP-2-dependent metastatic activity, and that high circulating nitrite levels in NPC patients may constitute a prognostic predictor for survival. | Zergoun et al. (2016) [38] |
| Blood | 82 NPC patients and 60 healthy controls | Measurement of IGF-1 levels using enzyme-linked immunosorbent assay | Tunisia | The study found that IGF-I could serve as a good NPC diagnostic marker. | M’hamdi et al. (2016) [39] |
| Blood | 50 HNSCC and 50 healthy controls | Measurement of plasma A-FABP levels using enzyme-linked immunosorbent assay | Egypt | The study found that a higher plasma level of A-FABP is associated with the clinical stage and risk of developing HNSCC. | El-Benhawy et al. (2016) [40] |
| Blood | 108 NPC patients, 18 lymphoma patients, 18 autoimmune diseases patients, and 55 healthy controls | Measurement of serum EBNA1 levels using enzyme-linked immunosorbent assay | Tunisia | The study found that IgA EBNA1 ELISA may be useful for the early diagnosis and mass screening of NPC in Tunisia, even in young patients. | Ayadi et al. (2009) [41] |
| Blood and saliva | 300 NPC patients and 50 healthy controls | Measurement of BARF1 protein and LMP1 levels using enzyme-linked immunosorbent assay | Algeria | The study showed LMP1 and BARF1 proteins to be good diagnostic markers of NPC, whereas BARF1 is a particularly promising marker for patients of all ages with NPC. | Houali et al. (2007) [42] |
| Blood | 117 NPC and 35 healthy controls | Measurement of EBV-VCA/EAlevels using enzyme-linked immunosorbent assay | Tunisia | The study found that VCA-p18 IgA ELISA seems suitable for the routine diagnosis and early detection of NPC complications. | Karray et al. (2005) [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Temilola, D.O.; Adeola, H.A.; Grobbelaar, J.; Chetty, M. Liquid Biopsy in Head and Neck Cancer: Its Present State and Future Role in Africa. Cells 2023, 12, 2663. https://doi.org/10.3390/cells12222663
Temilola DO, Adeola HA, Grobbelaar J, Chetty M. Liquid Biopsy in Head and Neck Cancer: Its Present State and Future Role in Africa. Cells. 2023; 12(22):2663. https://doi.org/10.3390/cells12222663
Chicago/Turabian StyleTemilola, Dada Oluwaseyi, Henry Ademola Adeola, Johan Grobbelaar, and Manogari Chetty. 2023. "Liquid Biopsy in Head and Neck Cancer: Its Present State and Future Role in Africa" Cells 12, no. 22: 2663. https://doi.org/10.3390/cells12222663
APA StyleTemilola, D. O., Adeola, H. A., Grobbelaar, J., & Chetty, M. (2023). Liquid Biopsy in Head and Neck Cancer: Its Present State and Future Role in Africa. Cells, 12(22), 2663. https://doi.org/10.3390/cells12222663





