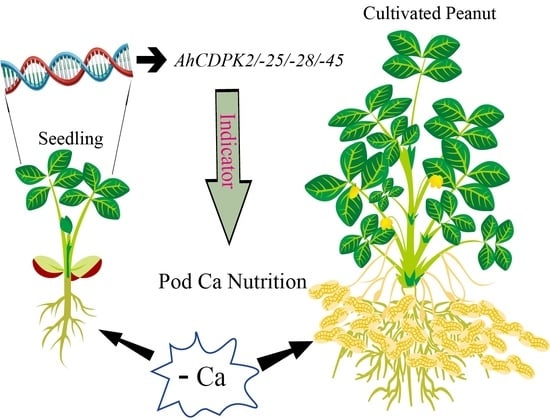
Genome-Wide Identification and Characterization of CDPK Gene Family in Cultivated Peanut (Arachis hypogaea L.) Reveal Their Potential Roles in Response to Ca Deficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Peanut CDPK Genes
2.2. Phylogenetic, Conserved Motifs, Gene Structure, and Protein Tertiary Structure Analysis of AhCDPKs
2.3. Gene Duplication Events and Synteny Analysis
2.4. Cis-Acting Regulatory Elements in Promoters and miRNA Target Predictions
2.5. Analysis of Protein Interaction Network and Functional Annotation Evaluation
2.6. RNA-Seq Data Analysis
2.7. Plant Growth and Treatments
2.8. RNA Extraction and qRT-PCR Analysis
3. Results
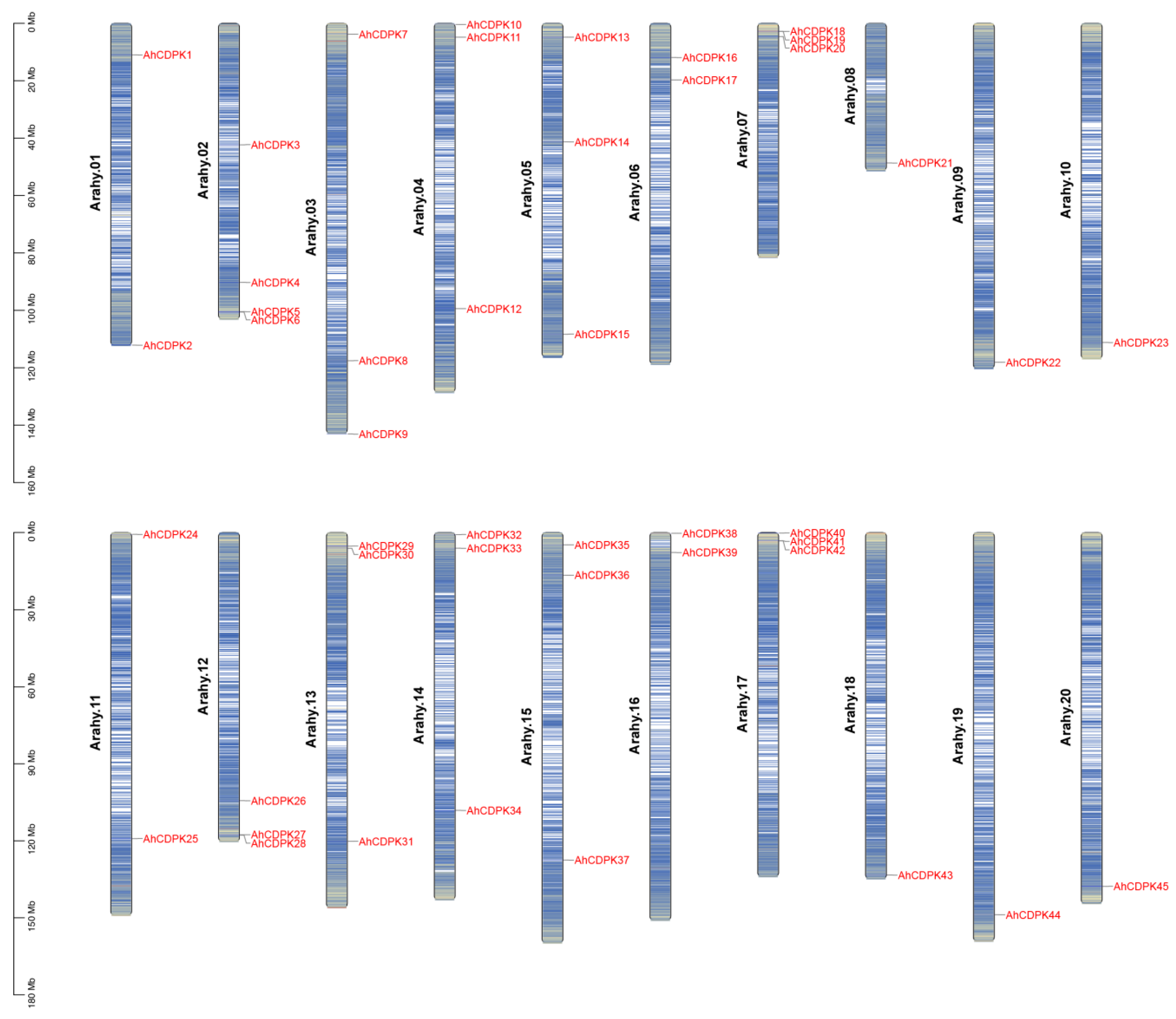
3.1. Genome-Wide Identification of 45 CDPK Genes in Peanut Genome
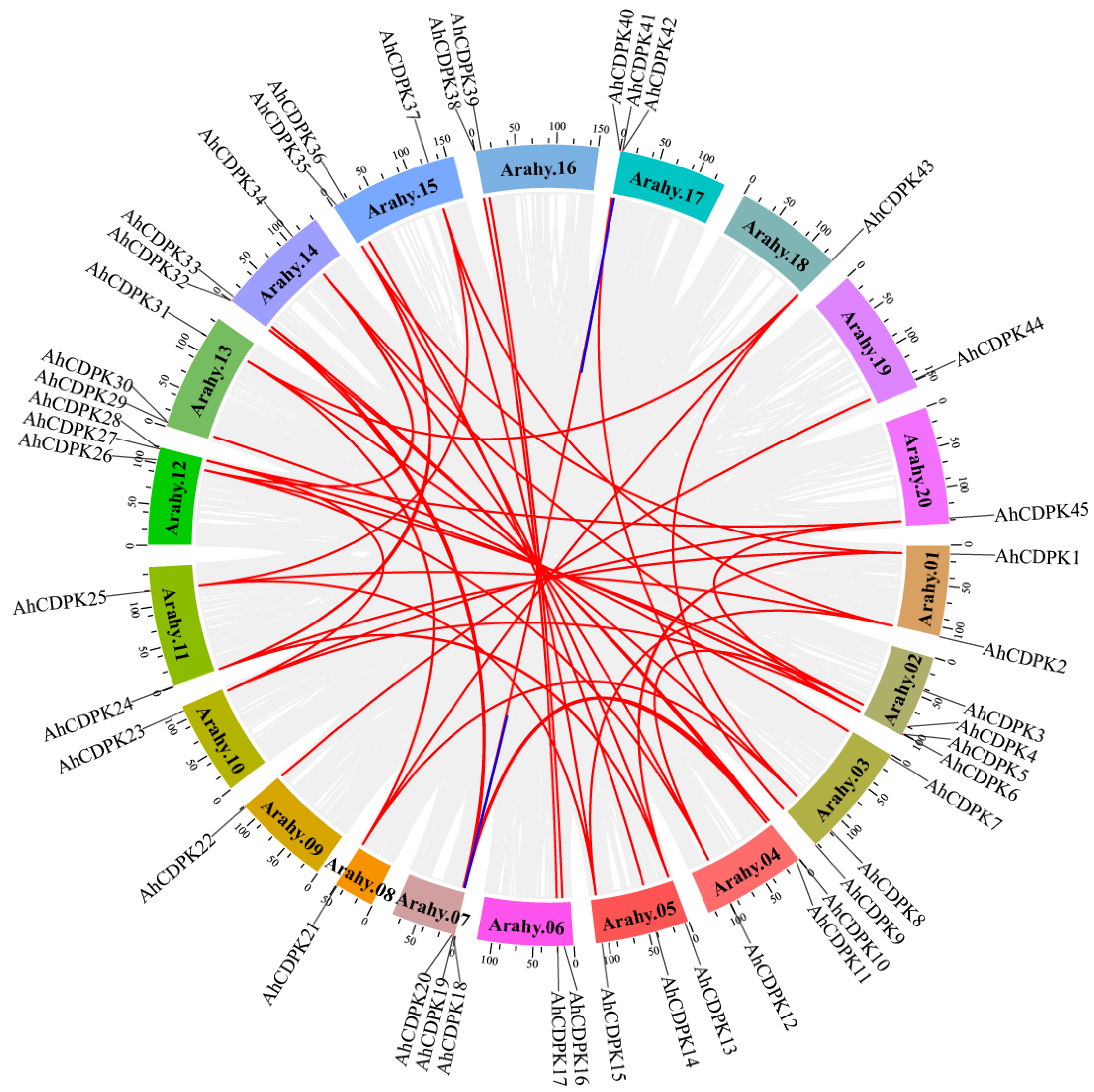
3.2. Synteny Analysis of CDPK Genes in Peanuts
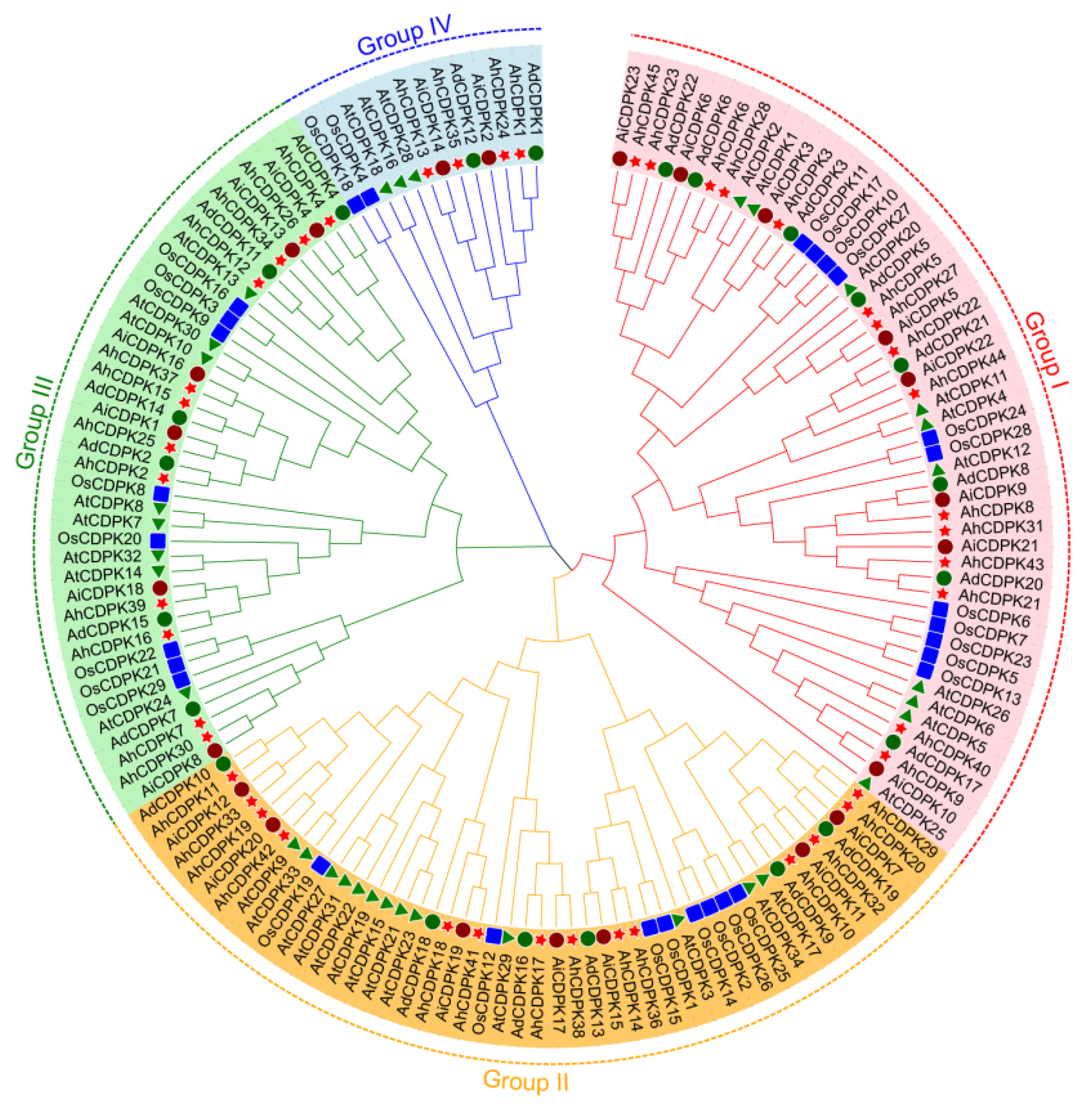
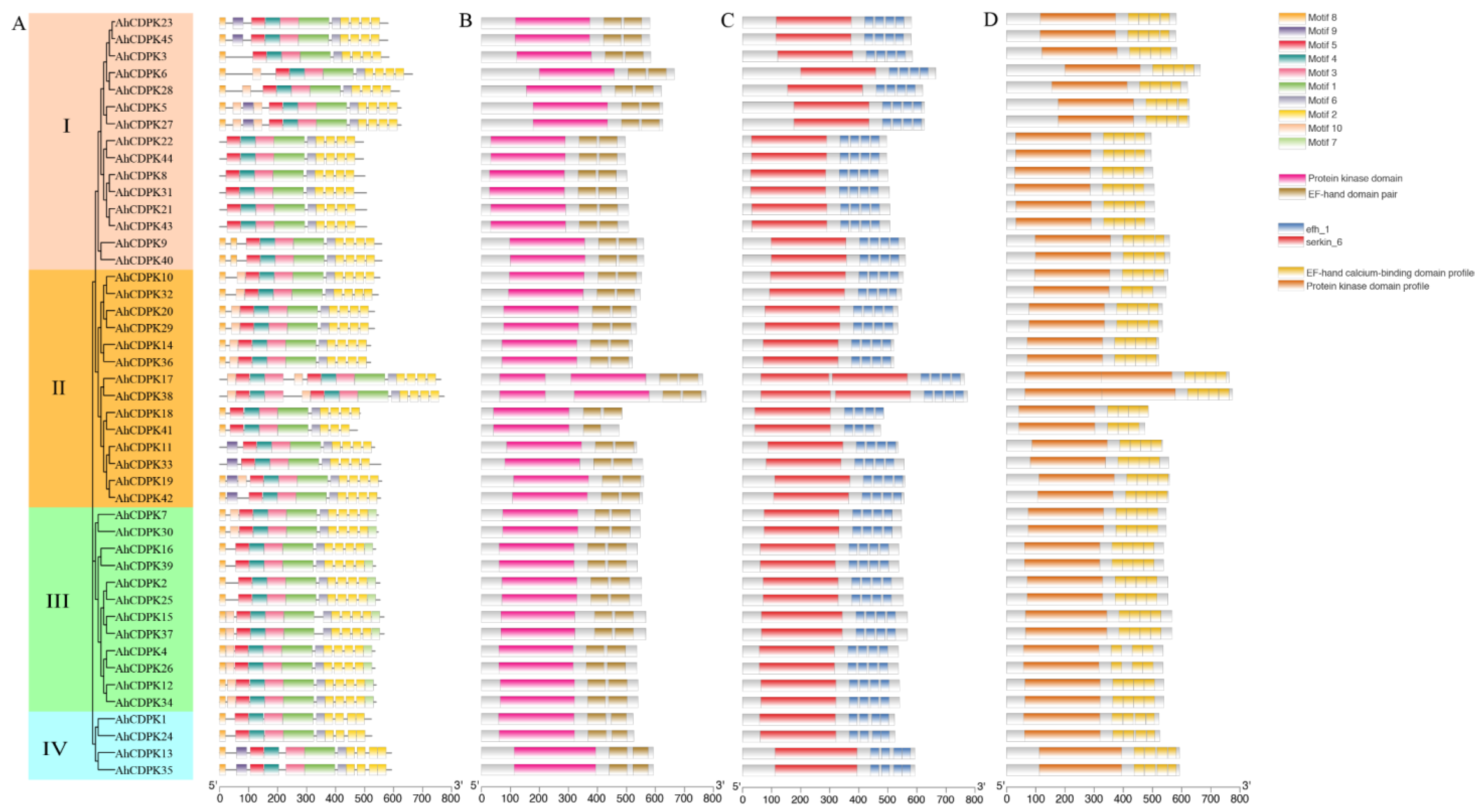
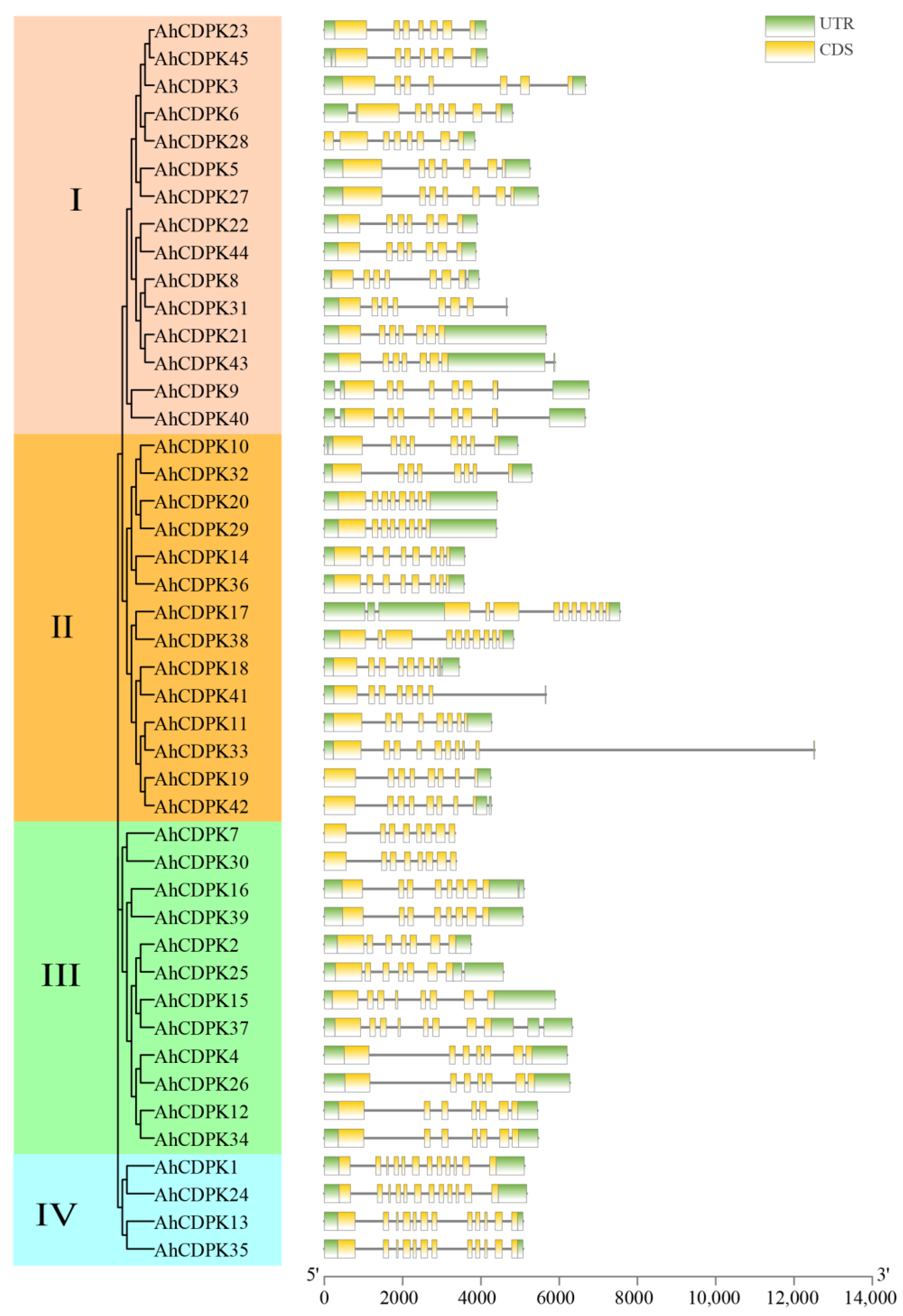
3.3. Phylogenetic Tree, Conserved Motifs, Gene Structure, and Tertiary Structures of AhCDPKs
3.4. Cis-Regulatory Elements Prediction in the Promotors of AhCDPKs
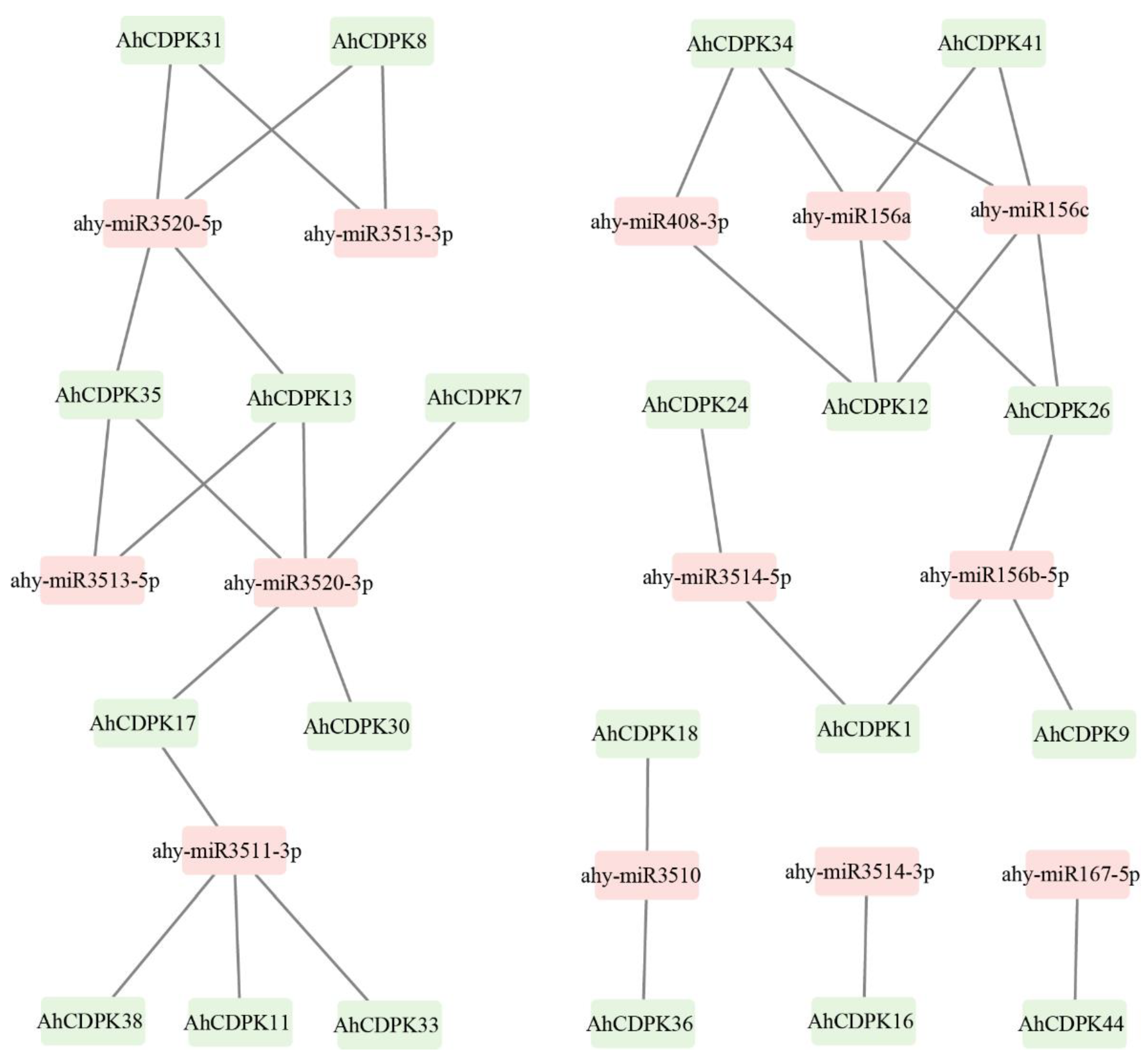
3.5. Prediction of miRNAs Targeting AhCDPK Genes
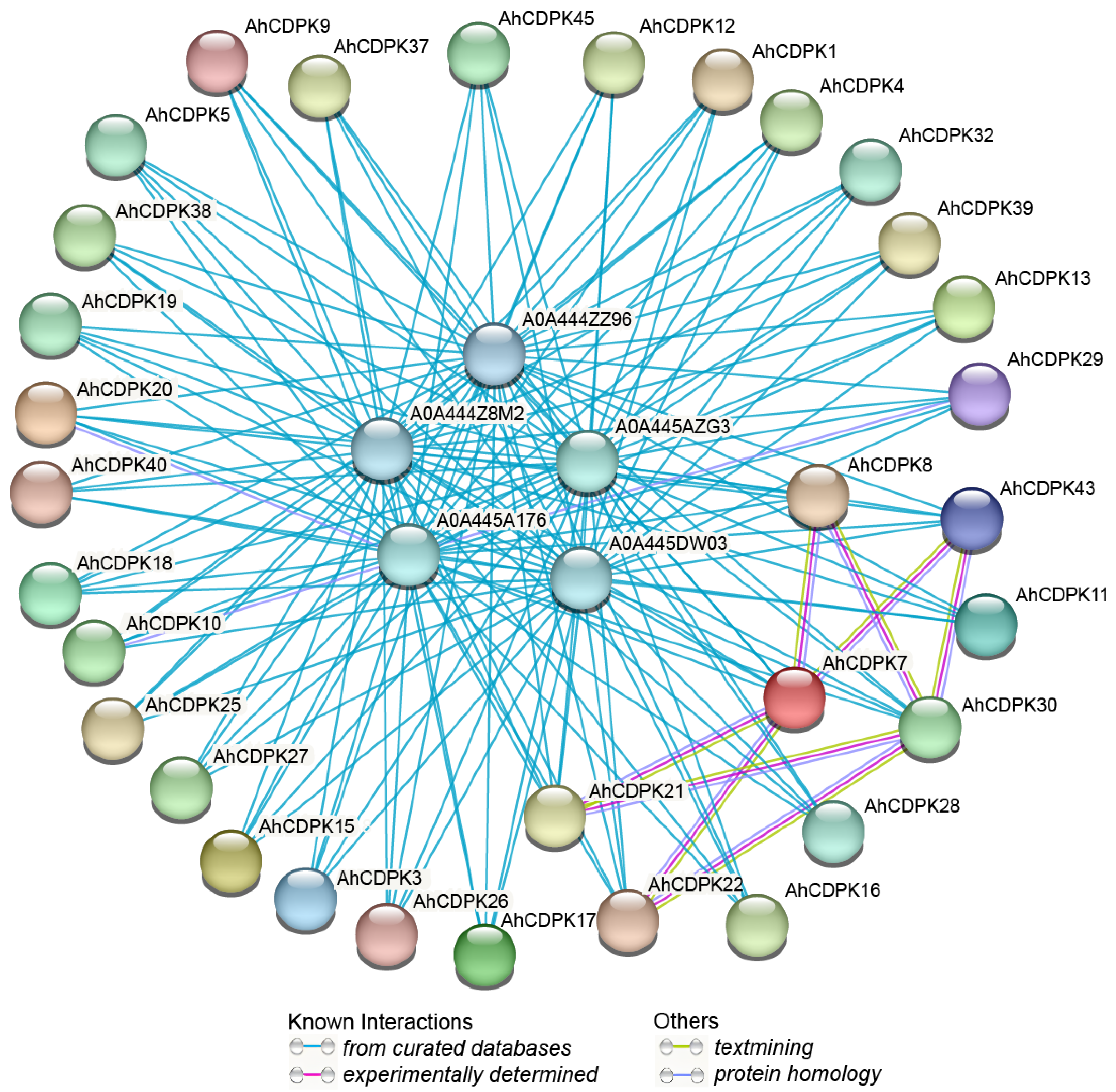
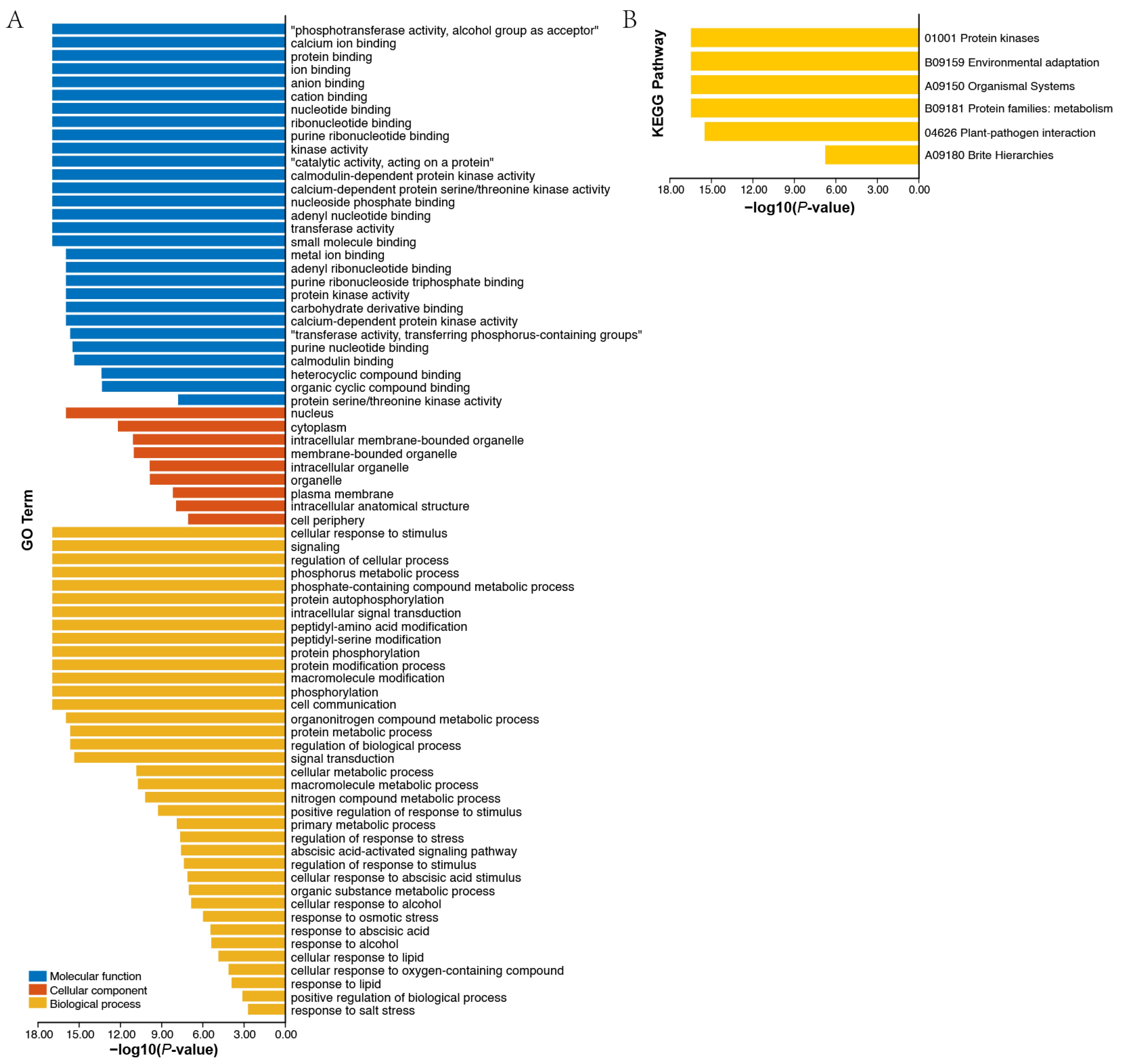
3.6. Protein Interaction Network and Functional Annotation Analysis of AhCDPKs
3.7. Spatial Expression Analysis of AhCDPKs
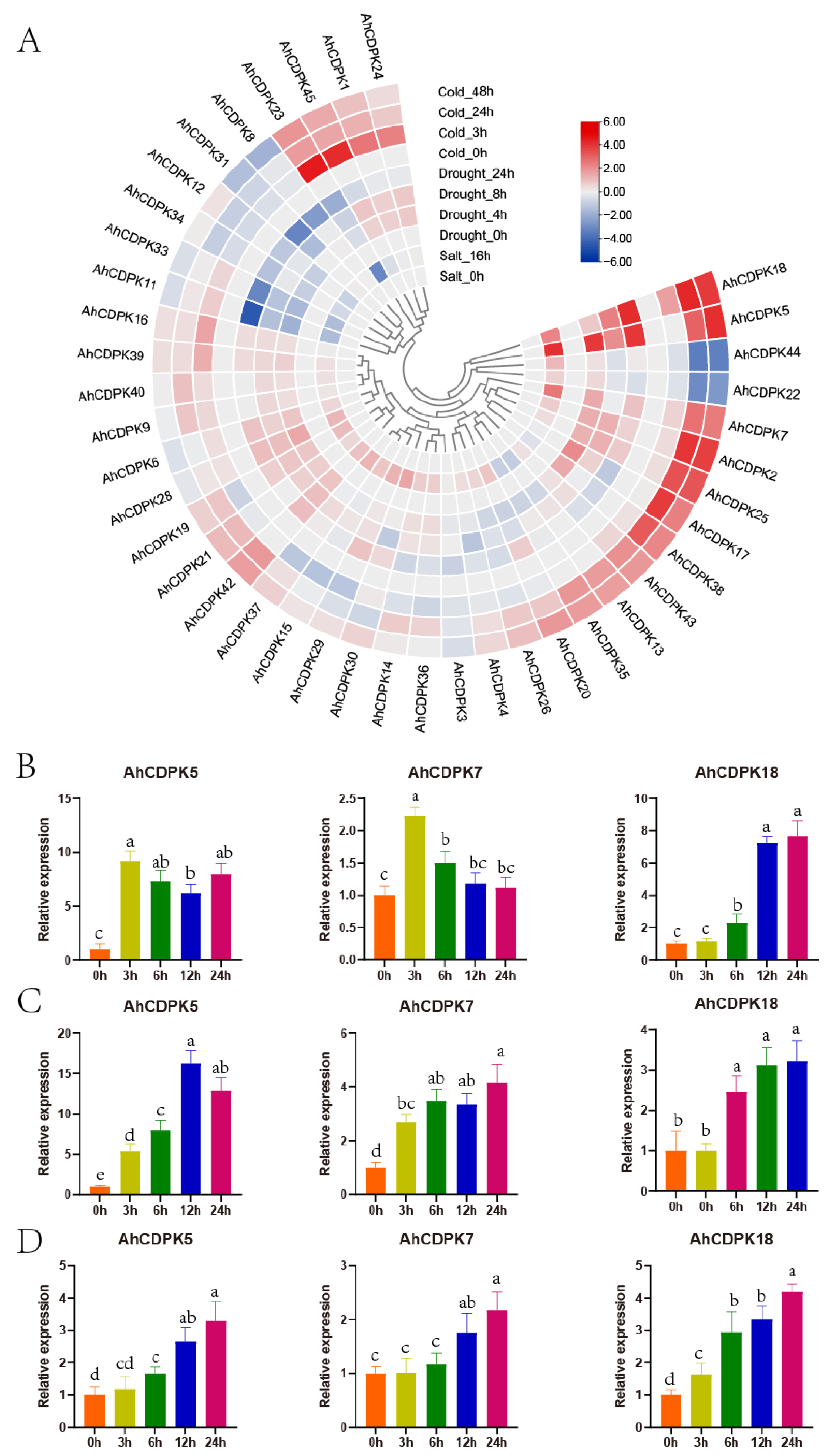
3.8. Expression Analysis of AhCDPKs under Abiotic Stresses
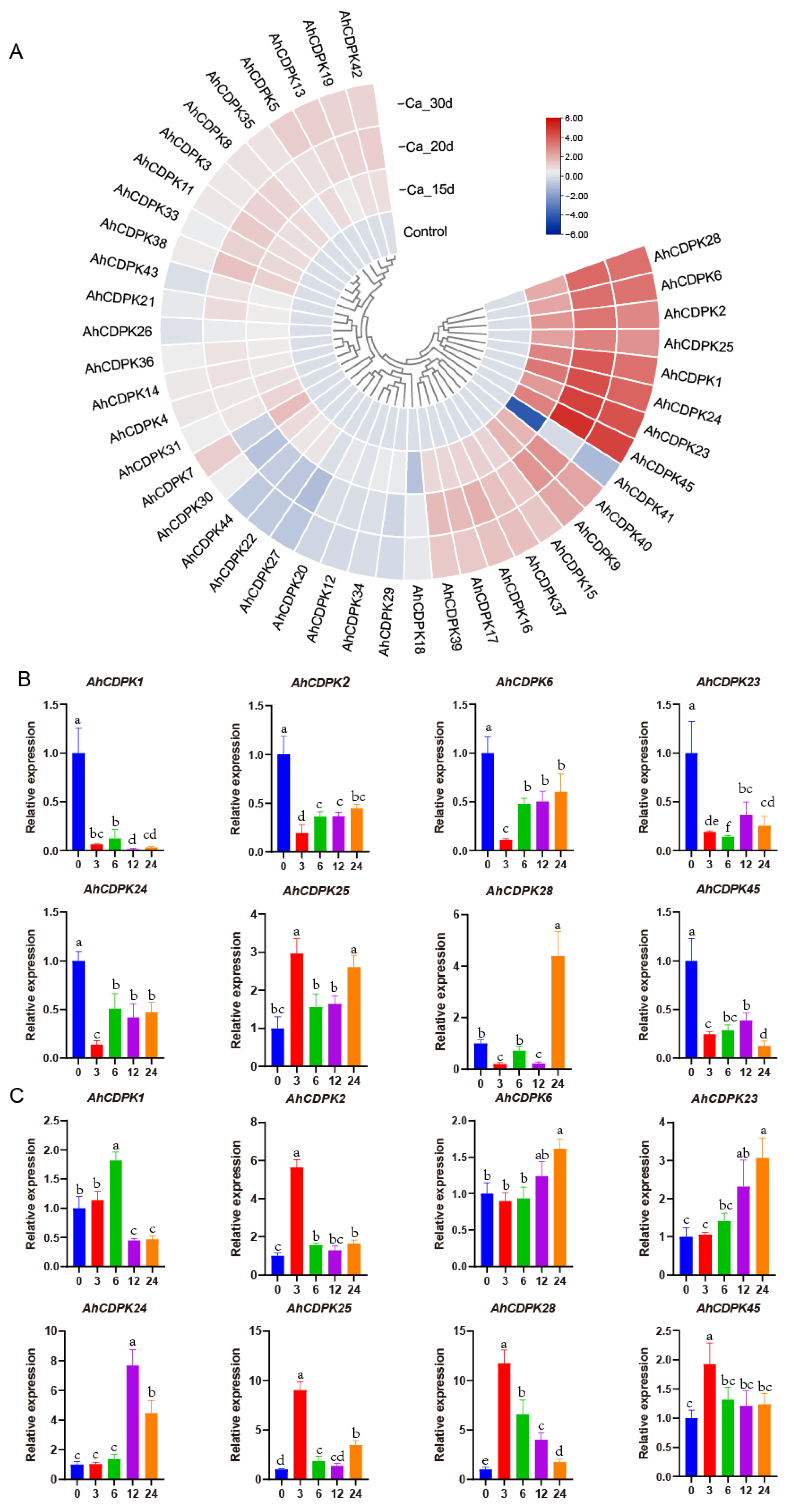
3.9. Expression Analysis of AhCDPKs in Response to Ca-Deficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef]
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Lee, H.-J.; Seo, P.J. Ca2+ talyzing Initial Responses to Environmental Stresses. Trends Plant Sci. 2021, 26, 849–870. [Google Scholar] [CrossRef]
- Hamel, L.P.; Sheen, J.; Seguin, A. Ancient signals: Comparative genomics of green plant CDPKs. Trends Plant Sci. 2014, 19, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Harmon, A.C.; Gribskov, M.; Harper, J.F. CDPKs—A kinase for every Ca2+ signal? Trends Plant Sci. 2000, 5, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.-H.; Willmann, M.R.; Chen, H.-C.; Sheen, J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002, 129, 469–485. [Google Scholar] [CrossRef]
- Shi, S.; Li, S.; Asim, M.; Mao, J.; Xu, D.; Ullah, Z.; Liu, G.; Wang, Q.; Liu, H. The Arabidopsis Calcium-Dependent Protein Kinases (CDPKs) and Their Roles in Plant Growth Regulation and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 1900. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.L.; Busconi, L. Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 2000, 24, 429–435. [Google Scholar]
- Harper, J.F.; Sussman, M.R.; Schaller, G.E.; Putnam-Evans, C.; Charbonneau, H.; Harmon, A.C. A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science 1991, 252, 951–954. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef]
- Yip Delormel, T.; Boudsocq, M. Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 2019, 224, 585–604. [Google Scholar] [CrossRef]
- Wang, H.; Gong, J.; Su, X.; Li, L.; Pang, X.; Zhang, Z. MaCDPK7, a calcium-dependent protein kinase gene from banana is involved in fruit ripening and temperature stress responses. J. Hortic. Sci. Biotechnol. 2017, 92, 240–250. [Google Scholar] [CrossRef]
- Liu, M.; Wang, C.; Xu, Q.; Pan, Y.; Jiang, B.; Zhang, L.; Zhang, Y.; Tian, Z.; Lu, J.; Ma, C.; et al. Genome-wide identification of the CPK gene family in wheat (Triticum a estivum L.) and characterization of TaCPK40 associated with seed dormancy and germination. Plant Physiol. Biochem. 2023, 196, 608–623. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.-Y.; Yu, X.-C.; Wang, X.-J.; Zhao, R.; Li, Y.; Fan, R.-C.; Shang, Y.; Du, S.-Y.; Wang, X.-F.; Wu, F.-Q.; et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscis ic acid signal transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Bin, L.; Xu, Z.; Chu, Y.; Yan, Y.; Nie, X.; Song, W. Genome-wide analysis of calcium-dependent protein kinase (CDPK) family and functional characterization of TaCDPK25-U in response to drought stress in wheat. Environ. Exp. Bot. 2023, 209, 105277. [Google Scholar]
- Yu, T.-F.; Zhao, W.-Y.; Fu, J.-D.; Liu, Y.-W.; Chen, M.; Zhou, Y.-B.; Ma, Y.-Z.; Xu, Z.-S.; Xi, Y.-J. Genome-Wide Analysis of CDPK Family in Foxtail Millet and Determination of SiCDPK24 Functions in Drought Stress. Front. Plant Sci. 2018, 9, 651. [Google Scholar] [CrossRef]
- Wang, D.; Liu, Y.-X.; Yu, Q.; Zhao, S.-P.; Zhao, J.-Y.; Ru, J.-N.; Cao, X.-Y.; Fang, Z.-W.; Chen, J.; Zhou, Y.-B.; et al. Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses. Int. J. Mol. Sci. 2019, 20, 5909. [Google Scholar] [CrossRef]
- Ding, Y.; Yang, H.; Wu, S.; Fu, D.; Li, M.; Gong, Z.; Yang, S. CPK28-NLP7 module integrates cold-induced Ca2+ signal and transcriptional reprogramming in Arabidopsis. Sci. Adv. 2022, 8, eabn7901. [Google Scholar] [CrossRef]
- Zhao, L.; Xie, B.; Hou, Y.; Zhao, Y.; Zheng, Y.; Jin, P. Genome-wide identification of the CDPK gene family reveals the CDPK-RBOH pathway potential involved in improving chilling tolerance in peach fruit. Plant Physiol. Biochem. 2022, 191, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Tanaka, N.; Yang, G.; Hayashi, N.; Komatsu, S. Genome-wide Identification of the Rice Calcium-dependent Protein Kinase and its Closely Related Kinase Gene Families: Comprehensive Analysis of the CDPKs Gene Family in Rice. Plant Cell Physiol. 2005, 46, 356–366. [Google Scholar] [CrossRef]
- Zhao, P.; Liu, Y.; Kong, W.; Ji, J.; Cai, T.; Guo, Z. Genome-Wide Identification and Characterization of Calcium-Dependent Protein Kinase (CDPK) and CDPK-Related Kinase (CRK) Gene Families in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 1044. [Google Scholar] [CrossRef]
- Kong, X.; Lv, W.; Jiang, S.; Zhang, D.; Cai, G.; Pan, J.; Li, D. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genom. 2013, 14, 433. [Google Scholar] [CrossRef]
- Shi, G.; Zhu, X. Genome-wide identification and functional characterization of CDPK gene family reveal their involvement in response to drought stress in Gossypium barbadense. PeerJ 2022, 10, e12883. [Google Scholar] [CrossRef]
- Hettenhausen, C.; Sun, G.; He, Y.; Zhuang, H.; Sun, T.; Qi, J.; Wu, J. Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci. Rep. 2016, 6, 18973. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; He, Q.; Chai, M.; Huang, Y.; Chen, F.; Wang, X.; Liu, Y.; Cai, H.; Qin, Y. Genome-wide investigation of calcium-dependent protein kinase gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2020, 21, 72. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The Genome of Cultivated Peanut Provides Insight into Legume Karyotypes, Polyploid Evolution and Crop Domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The Genome Sequence of Segmental Allotetraploid Peanut Arachis Hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The Genome Sequences of Arachis Duranensis and Arachis Ipaensis, the Diploid Ancestors of Cultivated Peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Khandwal, D.; Choudhary, B.; Ardeshana, D.; Jha, R.K.; Tanna, B.; Yadav, S.; Mishra, A.; Varshney, R.K.; Siddique, K.H.M. Differential Physio-Biochemical and Metabolic Responses of Peanut (Arachis hypogaea L.) under Multiple Abiotic Stress Conditions. Int. J. Mol. Sci. 2022, 23, 660. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Li, L.; Zhang, B.; Zhang, W.; Zeng, N.; Zhang, H.; Liu, D.; Luo, Z. Gene co-expression network analysis identifies hub genes associated with different tolerance under calcium deficiency in two peanut cultivars. BMC Genom. 2023, 24, 421. [Google Scholar] [CrossRef]
- Li, Y.; Meng, J.; Yang, S.; Guo, F.; Zhang, J.; Geng, Y.; Cui, L.; Wan, S.; Li, X. Transcriptome Analysis of Calcium- and Hormone-Related Gene Expressions during Different Stages of Peanut Pod Development. Front. Plant Sci. 2017, 8, 1241. [Google Scholar] [CrossRef]
- Yang, S.; Li, L.; Zhang, J.; Geng, Y.; Guo, F.; Wang, J.; Meng, J.; Sui, N.; Wan, S.; Li, X. Transcriptome and Differential Expression Profiling Analysis of the Me chanism of Ca2+ Regulation in Peanut (Arachis hypogaea) Pod Development. Front. Plant Sci. 2017, 8, 1609. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Tang, Z.; Guo, F.; Zhang, Y.; Zhang, J.; Meng, J.; Zheng, L.; Wan, S.; Li, X. Transcriptome of peanut kernel and shell reveals the mechanism of calcium on peanut pod development. Sci. Rep. 2020, 10, 15723. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Q.; Fu, H.; Chen, K.; Zhao, S.; Zhang, C.; Cai, T.; Wang, L.; Lu, W.; Dang, H.; et al. Identification of Key Gene Networks and Deciphering Transcriptional Regulators Associated with Peanut Embryo Abortion Mediated by Calcium Deficiency. Front. Plant Sci. 2022, 13, 814015. [Google Scholar] [CrossRef]
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Buillard, V.; Cerutti, L.; Copley, R.; et al. New Developments in the InterPro Database. Nucleic Acids Res. 2007, 35, D224–D228. [Google Scholar] [CrossRef]
- Xie, Y.; Zheng, Y.; Li, H.; Luo, X.; He, Z.; Cao, S.; Shi, Y.; Zhao, Q.; Xue, Y.; Zuo, Z.; et al. GPS-Lipid: A robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 2016, 6, 28249. [Google Scholar] [CrossRef] [PubMed]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A Developmental Transcriptome Map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Pathak, B.P.; Harmon, A.C.; Tillman, B.L.; Gallo, M. Calcium dependent protein kinase (CDPK) expression during fruit develo pment in cultivated peanut (Arachis hypogaea) under Ca2+-sufficient and -deficient growth regimens. J. Plant Physiol. 2011, 168, 2272–2277. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Shi, P.; He, L.; Yang, W.; Fu, H.; Xu, Z. Bioinformatics Analysis of Calcium Dependent Protein Kinases Gene Family in Wild Peanut. Chin. J. Trop. Crops 2017, 38, 94–103. [Google Scholar]
- Deepika, D.; Poddar, N.; Kumar, S.; Singh, A. Molecular Characterization Reveals the Involvement of Calcium Dependent Protein Kinases in Abiotic Stress Signaling and Development in Chick pea (Cicer arietinum). Front. Plant Sci. 2022, 13, 831265. [Google Scholar] [CrossRef]
- Qian, W.; Liao, B.-Y.; Chang, A.Y.-F.; Zhang, J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010, 26, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, X.; Li, J.; Guan, J.; Tan, Z.; Zhang, Z.; Shi, G. Genome-Wide Identification and Transcript Analysis Reveal Potential Roles of Oligopeptide Transporter Genes in Iron Deficiency Induced Cadmium Accumulation in Peanut. Front. Plant Sci. 2022, 13, 894848. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, N.; Zhang, Z.; Shi, G. Genome-Wide Identification and Expression Profile Reveal Potential Roles of Peanut ZIP Family Genes in Zinc/Iron-Deficiency Tolerance. Plants 2022, 11, 786. [Google Scholar] [CrossRef]
- Tan, Z.; Li, J.; Guan, J.; Wang, C.; Zhang, Z.; Shi, G. Genome-Wide Identification and Expression Analysis Reveals Roles of the NRAMP Gene Family in Iron/Cadmium Interactions in Peanut. Int. J. Mol. Sci. 2023, 24, 1713. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.A. Orthologs and paralogs—We need to get it right. Genome Biol. 2001, 2, interactions1002.1. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, L.; Zhang, H.; Liu, Q.; Zhai, H.; Zhao, N.; Gao, S.; He, S. Genome-Wide Identification and Characterization of CDPK Family Reveal Their Involvements in Growth and Development and Abiotic Stress in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2022, 23, 3088. [Google Scholar] [CrossRef]
- Ma, S.-Y.; Wu, W.-H. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 2007, 65, 511–518. [Google Scholar] [CrossRef]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef]
- Hu, C.-H.; Zeng, Q.-D.; Tai, L.; Li, B.-B.; Zhang, P.-P.; Nie, X.-M.; Wang, P.-Q.; Liu, W.-T.; Li, W.-Q.; Kang, Z.-S.; et al. Interaction between TaNOX7 and TaCDPK13 Contributes to Plant Fertility and Drought Tolerance by Regulating ROS Production. J. Agric. Food Chem. 2020, 68, 7333–7347. [Google Scholar] [CrossRef]
- Zhao, C.-Z.; Xia, H.; Frazier, T.P.; Yao, Y.-Y.; Bi, Y.-P.; Li, A.-Q.; Li, M.-J.; Li, C.-S.; Zhang, B.-H.; Wang, X.-J. Deep Sequencing Identifies Novel and Conserved MicroRNAs in Peanuts (Arachis hypogaea L.). BMC Plant Biol. 2010, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wang, P.; Zhao, S.; Zhao, C.; Xia, H.; Hou, L.; Ju, Z.; Zhang, Y.; Li, C.; Wang, X. Small RNA Profiling and Degradome Analysis Reveal Regulation of MicroRNA in Peanut Embryogenesis and Early Pod Development. BMC Genom. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, Q.; Chen, K.; Zhao, S.; Zhang, C.; Pan, R.; Cai, T.; Deng, Y.; Wang, X.; Chen, Y.; et al. Integrated MicroRNA and Transcriptome Profiling Reveals a MiRNA-Mediated Regulatory Network of Embryo Abortion under Calcium Deficiency in Peanut (Arachis hypogaea L.). BMC Genom. 2019, 20, 392. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Yang, S.; Li, G.; Wan, S. Genome-Wide Identification and Characterization of CDPK Gene Family in Cultivated Peanut (Arachis hypogaea L.) Reveal Their Potential Roles in Response to Ca Deficiency. Cells 2023, 12, 2676. https://doi.org/10.3390/cells12232676
Fan S, Yang S, Li G, Wan S. Genome-Wide Identification and Characterization of CDPK Gene Family in Cultivated Peanut (Arachis hypogaea L.) Reveal Their Potential Roles in Response to Ca Deficiency. Cells. 2023; 12(23):2676. https://doi.org/10.3390/cells12232676
Chicago/Turabian StyleFan, Shikai, Sha Yang, Guowei Li, and Shubo Wan. 2023. "Genome-Wide Identification and Characterization of CDPK Gene Family in Cultivated Peanut (Arachis hypogaea L.) Reveal Their Potential Roles in Response to Ca Deficiency" Cells 12, no. 23: 2676. https://doi.org/10.3390/cells12232676