Extensive Alternative Splicing Patterns in Systemic Lupus Erythematosus Highlight Sexual Differences
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Sequencing Study in SLE Patients and Healthy Individuals
2.2. Analysis of Differential Alternative Splicing (AS) Events
2.3. Differential Gene Expression and Pathway Analysis
3. Results
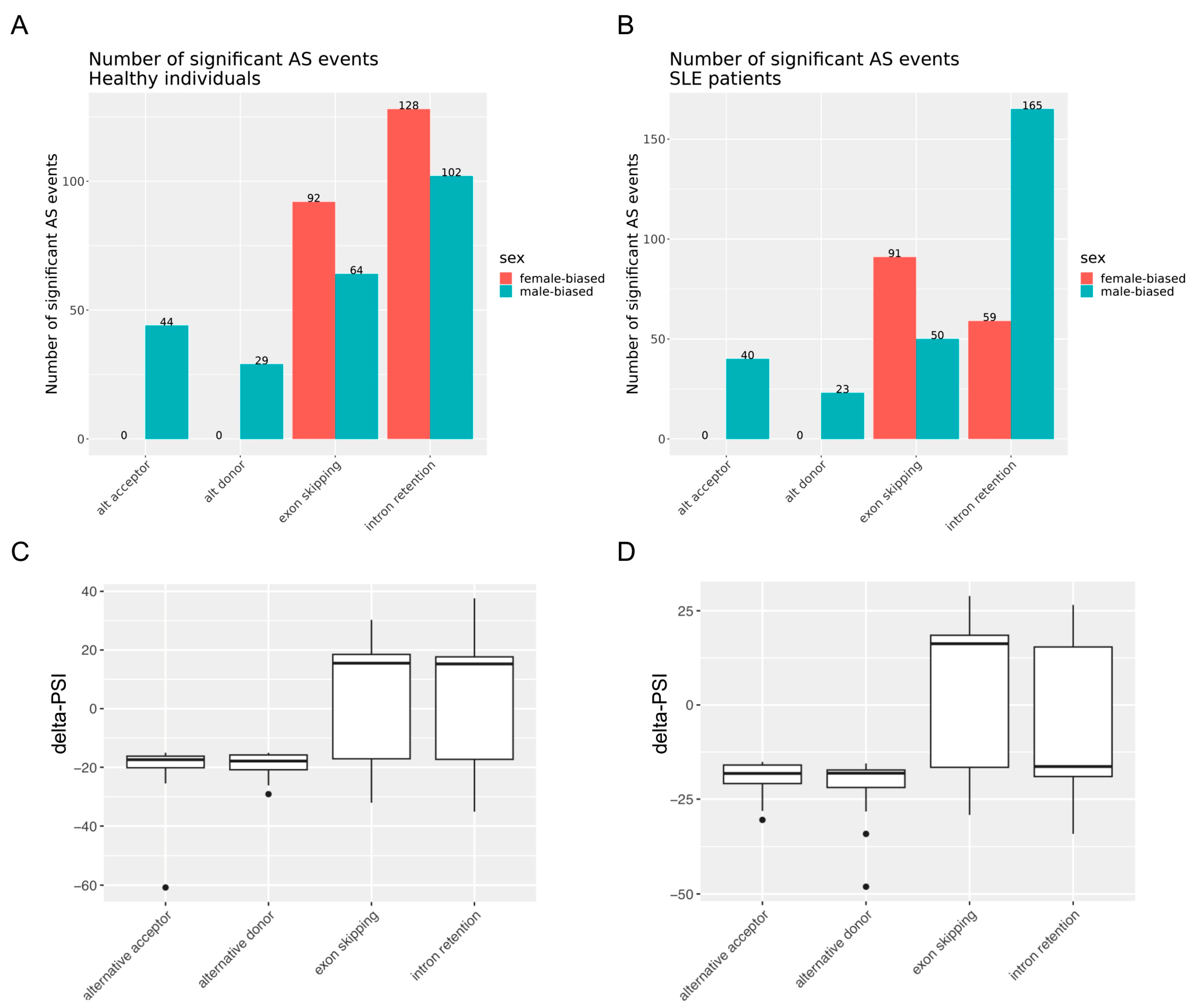
3.1. Sex Exerts Widespread Effects on RNA Splicing in SLE and the Healthy State with Enrichment for Intron Retention and Exon Skipping Events
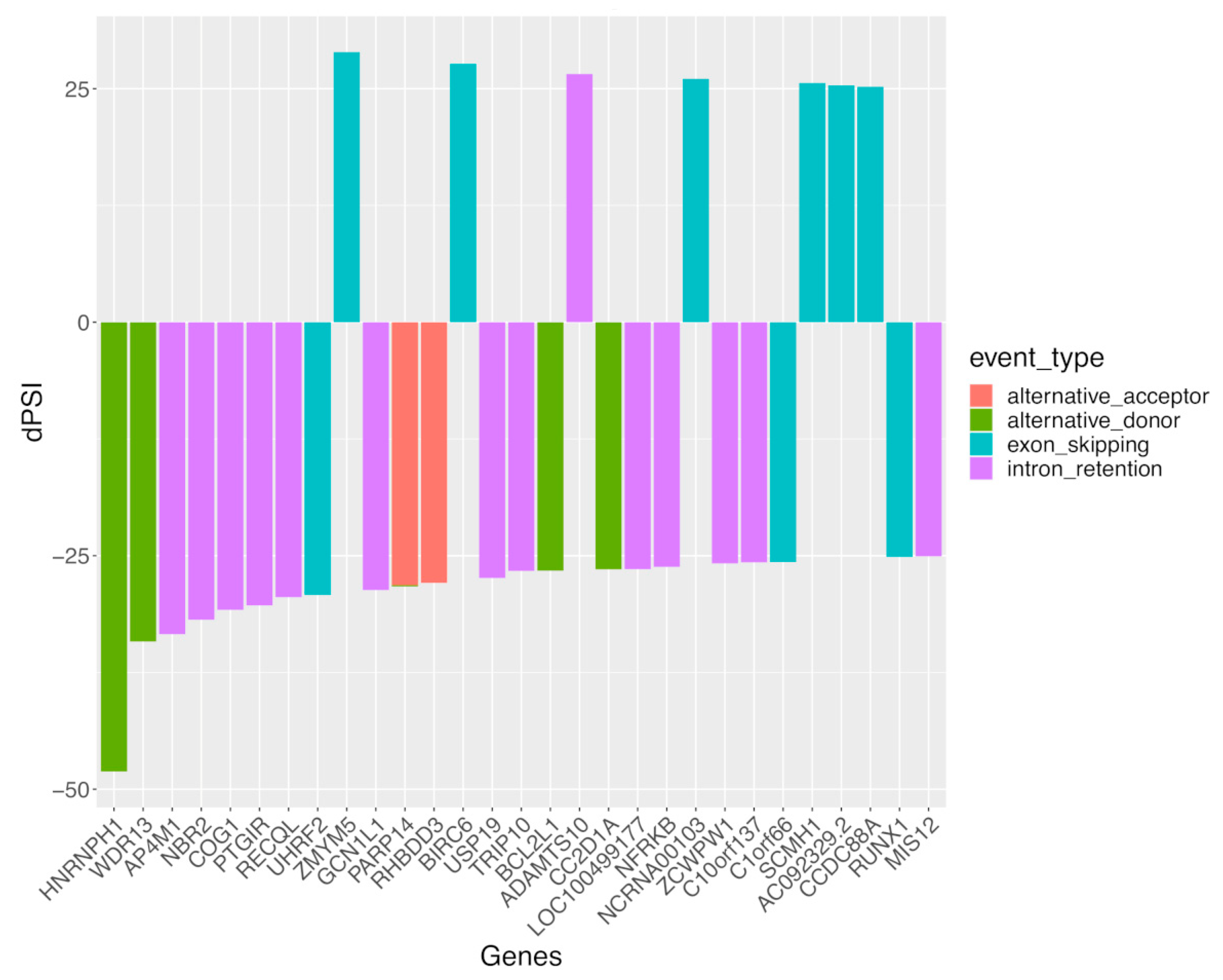
3.2. Alternative Splicing Events Implicate Distinct Molecular Pathways in SLE Disease and the Healthy State
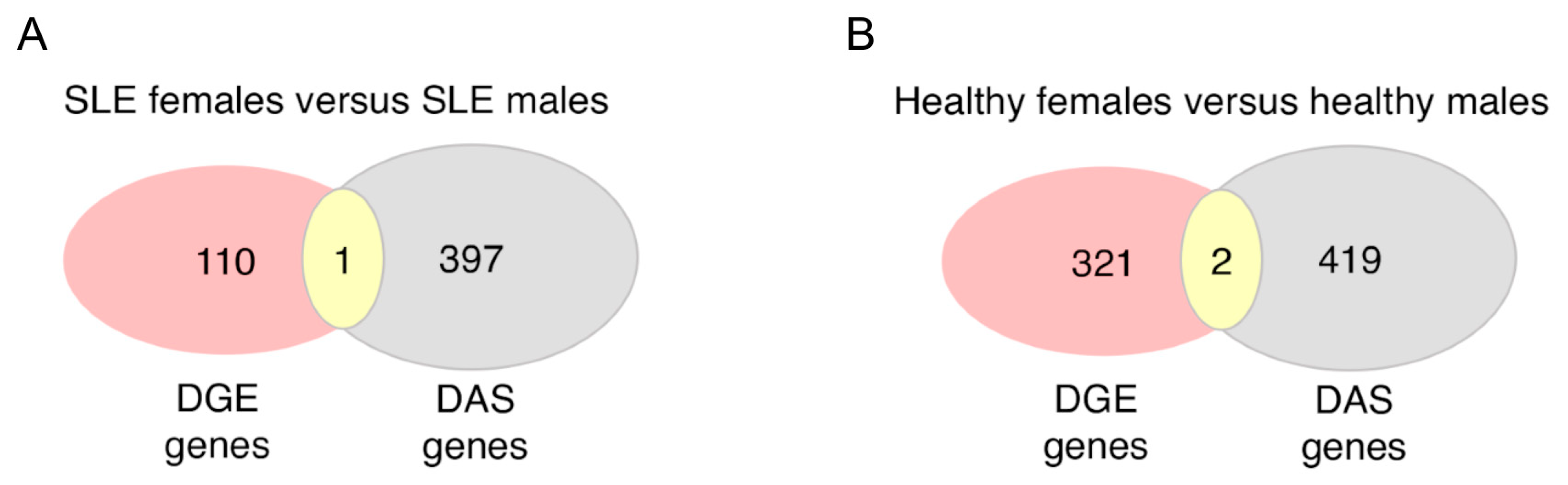
3.3. Sex Differences in Alternative Splicing Events Are More Extensive and May Underly Distinct Biological Processes Other Than Differential Gene Expression in SLE and Healthy Individuals
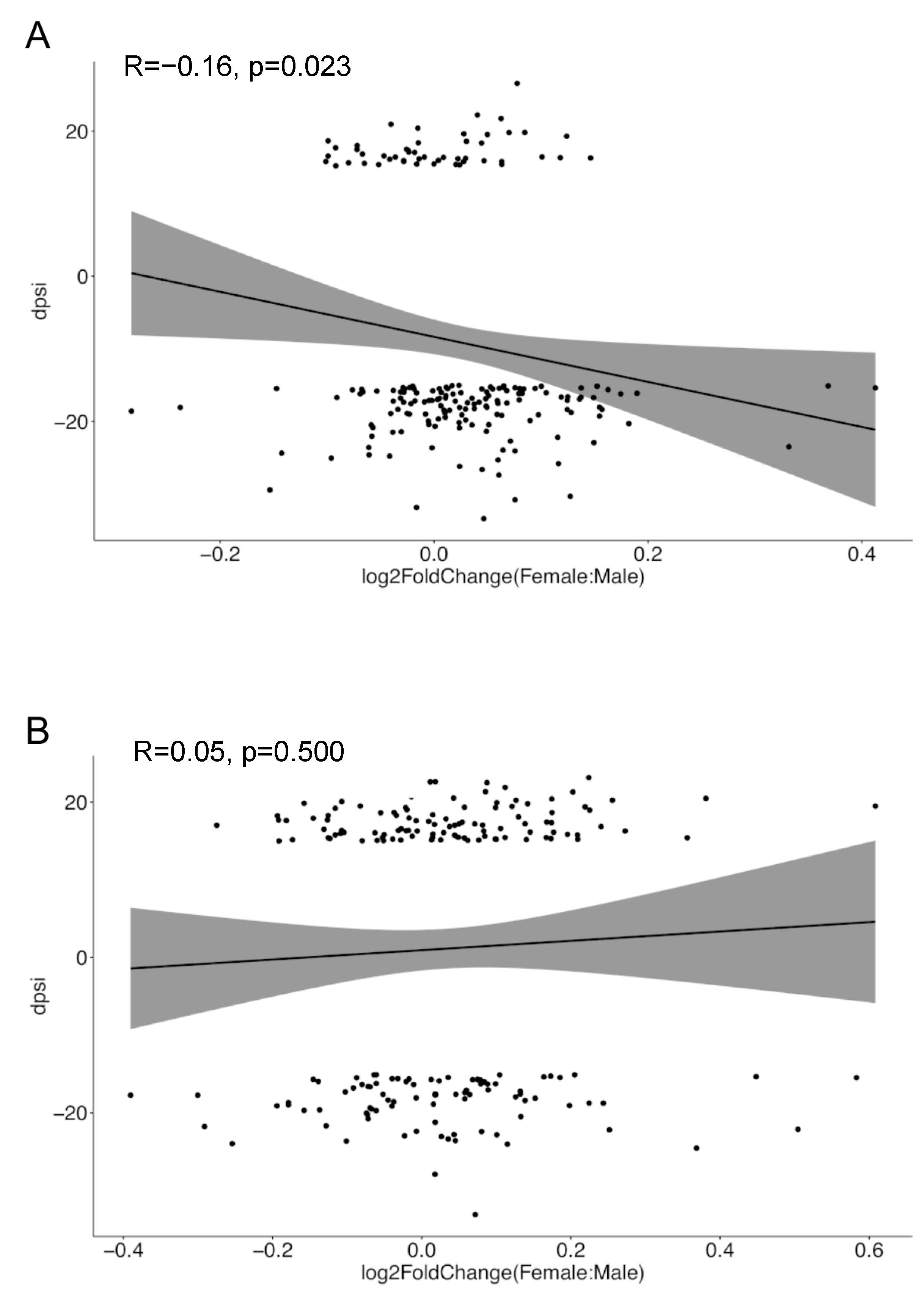
3.4. Sex-Biased Intron Retention Events Show Negative Correlation with Corresponding Gene Expression in SLE but Not in Healthy Individuals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wahren-Herlenius, M.; Dörner, T. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet 2013, 382, 819–831. [Google Scholar] [CrossRef]
- Whitacre, C.C. Sex differences in autoimmune disease. Nat. Immunol. 2001, 2, 777–780. [Google Scholar] [CrossRef]
- Fanouriakis, A.; Tziolos, N.; Bertsias, G.; Boumpas, D.T. Update omicronn the diagnosis and management of systemic lupus erythematosus. Ann. Rheum. Dis. 2021, 80, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Gergianaki, I.; Fanouriakis, A.; Repa, A.; Tzanakakis, M.; Adamichou, C.; Pompieri, A.; Spirou, G.; Bertsias, A.; Kabouraki, E.; Tzanakis, I.; et al. Epidemiology and burden of systemic lupus erythematosus in a Southern European population: Data from the community-based lupus registry of Crete, Greece. Ann. Rheum. Dis. 2017, 76, 1992–2000. [Google Scholar] [CrossRef] [PubMed]
- Margery-Muir, A.A.; Bundell, C.; Nelson, D.; Groth, D.M.; Wetherall, J.D. Gender balance in patients with systemic lupus erythematosus. Autoimmun. Rev. 2017, 16, 258–268. [Google Scholar] [CrossRef]
- Christou, E.A.A.; Banos, A.; Kosmara, D.; Bertsias, G.K.; Boumpas, D.T. Sexual dimorphism in SLE: Above and beyond sex hormones. Lupus 2019, 28, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Marasco, L.E.; Kornblihtt, A.R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 2023, 24, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Boidot, R.; Vegran, F. Alternative Splicing in Cancer and Immune Cells. Cancers 2022, 14, 1726. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, Y.; Zhang, X.; An, X.; Weng, M.; Shi, J.; Wang, S.; Liu, C.; Luo, S.; Zheng, T. An alternatively spliced STING isoform localizes in the cytoplasmic membrane and directly senses extracellular cGAMP. J. Clin. Investig. 2022, 132, e144339. [Google Scholar] [CrossRef]
- Sahoo, A.; Im, S.H. Interleukin and interleukin receptor diversity: Role of alternative splicing. Int. Rev. Immunol. 2010, 29, 77–109. [Google Scholar] [CrossRef] [PubMed]
- Tsuzaka, K.; Fukuhara, I.; Setoyama, Y.; Yoshimoto, K.; Suzuki, K.; Abe, T.; Takeuchi, T. TCR zeta mRNA with an alternatively spliced 3′-untranslated region detected in systemic lupus erythematosus patients leads to the down-regulation of TCR zeta and TCR/CD3 complex. J. Immunol. 2003, 171, 2496–2503. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.W. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 2004, 4, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, Q.; Zhao, Y.; Song, Y.; Leng, Y.; Chen, M.; Zhou, S.; Wang, Z. The regulatory role of alternative splicing in inflammatory bowel disease. Front. Immunol. 2023, 14, 1095267. [Google Scholar] [CrossRef]
- Manousou, P.; Kolios, G.; Drygiannakis, I.; Pyrovolaki, K.; Bourikas, L.; Papadaki, H.A.; Kouroumalis, E. Expression of a splice variant of CXCR3 in Crohn’s disease patients; indication for a lymphocyte—Epithelial cell interaction. J. Gastroenterol. Hepatol. 2008, 23, 1823–1833. [Google Scholar] [CrossRef]
- Ren, P.; Lu, L.; Cai, S.; Chen, J.; Lin, W.; Han, F. Alternative Splicing: A New Cause and Potential Therapeutic Target in Autoimmune Disease. Front. Immunol. 2021, 12, 713540. [Google Scholar] [CrossRef]
- Evsyukova, I.; Somarelli, J.A.; Gregory, S.G.; Garcia-Blanco, M.A. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biol. 2010, 7, 462–473. [Google Scholar] [CrossRef]
- Panousis, N.I.; Bertsias, G.K.; Ongen, H.; Gergianaki, I.; Tektonidou, M.G.; Trachana, M.; Romano-Palumbo, L.; Bielser, D.; Howald, C.; Pamfil, C.; et al. Combined genetic and transcriptome analysis of patients with SLE: Distinct, targetable signatures for susceptibility and severity. Ann. Rheum. Dis. 2019, 78, 1079–1089. [Google Scholar] [CrossRef]
- Odhams, C.A.; Cortini, A.; Chen, L.; Roberts, A.L.; Vinuela, A.; Buil, A.; Small, K.S.; Dermitzakis, E.T.; Morris, D.L.; Vyse, T.J.; et al. Mapping eQTLs with RNA-seq reveals novel susceptibility genes, non-coding RNAs and alternative-splicing events in systemic lupus erythematosus. Hum. Mol. Genet. 2017, 26, 1003–1017. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Z.; Li, Z.; Zeng, Z.; Peng, W.; Zhu, J.; Zhao, J.; Zhu, C.; Zeng, C.; Stearrett, N.; et al. Abnormalities in intron retention characterize patients with systemic lupus erythematosus. Sci. Rep. 2023, 13, 5141. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Bertsias, G.K.; Nikolaou, C. Extensive Changes in Transcription Dynamics Reflected on Alternative Splicing Events in Systemic Lupus Erythematosus Patients. Genes 2021, 12, 1260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hu, Q.; Liu, X.; Ji, Y.; Chao, H.P.; Liu, Y.; Tracz, A.; Kirk, J.; Buonamici, S.; Zhu, P.; et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat. Commun. 2020, 11, 2089. [Google Scholar] [CrossRef] [PubMed]
- Smart, A.C.; Margolis, C.A.; Pimentel, H.; He, M.X.; Miao, D.; Adeegbe, D.; Fugmann, T.; Wong, K.K.; Van Allen, E.M. Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol. 2018, 36, 1056–1058. [Google Scholar] [CrossRef]
- Inoue, D.; Polaski, J.T.; Taylor, J.; Castel, P.; Chen, S.; Kobayashi, S.; Hogg, S.J.; Hayashi, Y.; Pineda, J.M.B.; El Marabti, E.; et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat. Genet. 2021, 53, 707–718. [Google Scholar] [CrossRef]
- Bongen, E.; Lucian, H.; Khatri, A.; Fragiadakis, G.K.; Bjornson, Z.B.; Nolan, G.P.; Utz, P.J.; Khatri, P. Sex Differences in the Blood Transcriptome Identify Robust Changes in Immune Cell Proportions with Aging and Influenza Infection. Cell Rep. 2019, 29, 1961–1973.e4. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Batista, S.; Brooks, A.I.; Tischfield, J.A.; Willemsen, G.; van Grootheest, G.; Hottenga, J.J.; Milaneschi, Y.; Mbarek, H.; Madar, V.; et al. Sex differences in the human peripheral blood transcriptome. BMC Genom. 2014, 15, 33. [Google Scholar] [CrossRef]
- Oliva, M.; Munoz-Aguirre, M.; Kim-Hellmuth, S.; Wucher, V.; Gewirtz, A.D.H.; Cotter, D.J.; Parsana, P.; Kasela, S.; Balliu, B.; Vinuela, A.; et al. The impact of sex on gene expression across human tissues. Science 2020, 369, eaba3066. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G.; et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018, 175, 1701–1715.e16. [Google Scholar] [CrossRef]
- Whitney, A.R.; Diehn, M.; Popper, S.J.; Alizadeh, A.A.; Boldrick, J.C.; Relman, D.A.; Brown, P.O. Individuality and variation in gene expression patterns in human blood. Proc. Natl. Acad. Sci. USA 2003, 100, 1896–1901. [Google Scholar] [CrossRef]
- Guy, K.; Diogo, F.T.V.; Anne Deslattes, M.; Christina, C.; Pablo Prieto, B.; Maria, C.; Anil, K.K.; Daniel, D.; Georgios, K.; Xingmin Aaron, Z.; et al. The impact of biological sex on alternative splicing. bioRxiv 2020, 490904. [Google Scholar] [CrossRef]
- Trabzuni, D.; Ramasamy, A.; Imran, S.; Walker, R.; Smith, C.; Weale, M.E.; Hardy, J.; Ryten, M.; North American Brain Expression, C. Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 2013, 4, 2771. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal transcriptome of the human brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef]
- Gladman, D.D.; Ibanez, D.; Urowitz, M.B. Systemic lupus erythematosus disease activity index 2000. J. Rheumatol. 2002, 29, 288–291. [Google Scholar] [PubMed]
- Mosca, M.; Bombardieri, S. Assessing remission in systemic lupus erythematosus. Clin. Exp. Rheumatol. 2006, 24, S-99-104. [Google Scholar] [PubMed]
- Irimia, M.; Weatheritt, R.J.; Ellis, J.D.; Parikshak, N.N.; Gonatopoulos-Pournatzis, T.; Babor, M.; Quesnel-Vallieres, M.; Tapial, J.; Raj, B.; O’Hanlon, D.; et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 2014, 159, 1511–1523. [Google Scholar] [CrossRef]
- Tapial, J.; Ha, K.C.H.; Sterne-Weiler, T.; Gohr, A.; Braunschweig, U.; Hermoso-Pulido, A.; Quesnel-Vallieres, M.; Permanyer, J.; Sodaei, R.; Marquez, Y.; et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017, 27, 1759–1768. [Google Scholar] [CrossRef]
- Han, H.; Braunschweig, U.; Gonatopoulos-Pournatzis, T.; Weatheritt, R.J.; Hirsch, C.L.; Ha, K.C.H.; Radovani, E.; Nabeel-Shah, S.; Sterne-Weiler, T.; Wang, J.; et al. Multilayered Control of Alternative Splicing Regulatory Networks by Transcription Factors. Mol. Cell 2017, 65, 539–553.e7. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Zhang, J.; Bouch, R.J.; Blekhman, M.G.; He, Z. USP19 Suppresses Th17-Driven Pathogenesis in Autoimmunity. J. Immunol. 2021, 207, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Alarcon-Riquelme, M.E. Role of RUNX in autoimmune diseases linking rheumatoid arthritis, psoriasis and lupus. Arthritis Res. Ther. 2004, 6, 169–173. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wong, W.F.; Kohu, K.; Nakamura, A.; Ebina, M.; Kikuchi, T.; Tazawa, R.; Tanaka, K.; Kon, S.; Funaki, T.; Sugahara-Tobinai, A.; et al. Runx1 deficiency in CD4+ T cells causes fatal autoimmune inflammatory lung disease due to spontaneous hyperactivation of cells. J. Immunol. 2012, 188, 5408–5420. [Google Scholar] [CrossRef]
- Monteuuis, G.; Wong, J.J.L.; Bailey, C.G.; Schmitz, U.; Rasko, J.E.J. The changing paradigm of intron retention: Regulation, ramifications and recipes. Nucleic Acids Res. 2019, 47, 11497–11513. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.J.; Schmitz, U. Intron retention: Importance, challenges, and opportunities. Trends Genet. 2022, 38, 789–792. [Google Scholar] [CrossRef]
- Le Billan, F.; Umogbai, G.; Cummins, C.L. Regulation of Alternative Splicing by Steroid Hormones. Endocrinology 2023, 164, bqad081. [Google Scholar] [CrossRef] [PubMed]
- Spruce, T.; Plass, M.; Gohr, A.; Ray, D.; Martinez de Lagran, M.; Rot, G.; Novoa, A.; Burguera, D.; Permanyer, J.; Miret, M.; et al. The X-linked splicing regulator MBNL3 has been co-opted to restrict placental growth in eutherians. PLoS Biol. 2022, 20, e3001615. [Google Scholar] [CrossRef]
- Dimas, A.S.; Nica, A.C.; Montgomery, S.B.; Stranger, B.E.; Raj, T.; Buil, A.; Giger, T.; Lappalainen, T.; Gutierrez-Arcelus, M.; Mu, T.C.; et al. Sex-biased genetic effects on gene regulation in humans. Genome Res. 2012, 22, 2368–2375. [Google Scholar] [CrossRef]
- Dery, K.J.; Kujawski, M.; Grunert, D.; Wu, X.; Ngyuen, T.; Cheung, C.; Yim, J.H.; Shively, J.E. IRF-1 regulates alternative mRNA splicing of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) in breast epithelial cells generating an immunoreceptor tyrosine-based inhibition motif (ITIM) containing isoform. Mol. Cancer 2014, 13, 64. [Google Scholar] [CrossRef]
- Louis, J.M.; Vaz, C.; Balaji, A.; Tanavde, V.; Talukdar, I. TNF-alpha regulates alternative splicing of genes participating in pathways of crucial metabolic syndromes; a transcriptome wide study. Cytokine 2020, 125, 154815. [Google Scholar] [CrossRef]
- Wu, W.; Syed, F.; Simpson, E.; Lee, C.C.; Liu, J.; Chang, G.; Dong, C.; Seitz, C.; Eizirik, D.L.; Mirmira, R.G.; et al. The Impact of Pro-Inflammatory Cytokines on Alternative Splicing Patterns in Human Islets. Diabetes, 2021; online ahead of print. [Google Scholar] [CrossRef]
- Butte, M.J.; Lee, S.J.; Jesneck, J.; Keir, M.E.; Haining, W.N.; Sharpe, A.H. CD28 costimulation regulates genome-wide effects on alternative splicing. PLoS ONE 2012, 7, e40032. [Google Scholar] [CrossRef]
- Thompson, M.G.; Dittmar, M.; Mallory, M.J.; Bhat, P.; Ferretti, M.B.; Fontoura, B.M.; Cherry, S.; Lynch, K.W. Viral-induced alternative splicing of host genes promotes influenza replication. Elife 2020, 9, e55500. [Google Scholar] [CrossRef]
- Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015, 84, 165–198. [Google Scholar] [CrossRef]
- Nieto Moreno, N.; Giono, L.E.; Cambindo Botto, A.E.; Munoz, M.J.; Kornblihtt, A.R. Chromatin, DNA structure and alternative splicing. FEBS Lett. 2015, 589, 3370–3378. [Google Scholar] [CrossRef]
- Ganez-Zapater, A.; Mackowiak, S.D.; Guo, Y.; Tarbier, M.; Jordan-Pla, A.; Friedlander, M.R.; Visa, N.; Ostlund Farrants, A.K. The SWI/SNF subunit BRG1 affects alternative splicing by changing RNA binding factor interactions with nascent RNA. Mol. Genet. Genom. 2022, 297, 463–484. [Google Scholar] [CrossRef]
- Batsche, E.; Yaniv, M.; Muchardt, C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006, 13, 22–29. [Google Scholar] [CrossRef]
- Fotouhi, O.; Nizamuddin, S.; Falk, S.; Schilling, O.; Knuchel-Clarke, R.; Biniossek, M.L.; Timmers, H.T.M. Alternative mRNA Splicing Controls the Functions of the Histone H3K27 Demethylase UTX/KDM6A. Cancers 2023, 15, 3117. [Google Scholar] [CrossRef]
- Itoh, Y.; Golden, L.C.; Itoh, N.; Matsukawa, M.A.; Ren, E.; Tse, V.; Arnold, A.P.; Voskuhl, R.R. The X-linked histone demethylase Kdm6a in CD4+ T lymphocytes modulates autoimmunity. J. Clin. Investig. 2019, 129, 3852–3863. [Google Scholar] [CrossRef]
- Bradley, R.K.; Anczukow, O. RNA splicing dysregulation and the hallmarks of cancer. Nat. Rev. Cancer 2023, 23, 135–155. [Google Scholar] [CrossRef]
- Yang, X.; Coulombe-Huntington, J.; Kang, S.; Sheynkman, G.M.; Hao, T.; Richardson, A.; Sun, S.; Yang, F.; Shen, Y.A.; Murray, R.R.; et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 2016, 164, 805–817. [Google Scholar] [CrossRef]
- Iwata, H.; Goettsch, C.; Sharma, A.; Ricchiuto, P.; Goh, W.W.; Halu, A.; Yamada, I.; Yoshida, H.; Hara, T.; Wei, M.; et al. PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation. Nat. Commun. 2016, 7, 12849. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Shi, W.; Zhang, L.; Hu, Z.; Xu, C. USP19 suppresses cellular type I interferon signaling by targeting TRAF3 for deubiquitination. Future Microbiol. 2017, 12, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Gkirtzimanaki, K.; Kabrani, E.; Nikoleri, D.; Polyzos, A.; Blanas, A.; Sidiropoulos, P.; Makrigiannakis, A.; Bertsias, G.; Boumpas, D.T.; Verginis, P. IFNalpha Impairs Autophagic Degradation of mtDNA Promoting Autoreactivity of SLE Monocytes in a STING-Dependent Fashion. Cell Rep. 2018, 25, 921–933.e5. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Tian, S.; Chen, Y.; Zhang, C.; Xie, W.; Xia, X.; Cui, J.; Wang, R.F. USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. 2016, 35, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Rozen, E.J.; Ozeroff, C.D.; Allen, M.A. RUN(X) out of blood: Emerging RUNX1 functions beyond hematopoiesis and links to Down syndrome. Hum. Genom. 2023, 17, 83. [Google Scholar] [CrossRef]
- Pyfrom, S.; Paneru, B.; Knox, J.J.; Cancro, M.P.; Posso, S.; Buckner, J.H.; Anguera, M.C. The dynamic epigenetic regulation of the inactive X chromosome in healthy human B cells is dysregulated in lupus patients. Proc. Natl. Acad. Sci. USA 2021, 118, e2024624118. [Google Scholar] [CrossRef]
- Shen, S.; Wang, Y.; Wang, C.; Wu, Y.N.; Xing, Y. SURVIV for survival analysis of mRNA isoform variation. Nat. Commun. 2016, 7, 11548. [Google Scholar] [CrossRef]
- Trincado, J.L.; Sebestyen, E.; Pages, A.; Eyras, E. The prognostic potential of alternative transcript isoforms across human tumors. Genome Med. 2016, 8, 85. [Google Scholar] [CrossRef]
- Ni, T.; Yang, W.; Han, M.; Zhang, Y.; Shen, T.; Nie, H.; Zhou, Z.; Dai, Y.; Yang, Y.; Liu, P.; et al. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 2016, 44, 6817–6829. [Google Scholar] [CrossRef]



| Main Group | Number of Patient Samples | Number of Subgroups |
|---|---|---|
| Healthy female | 48 | 5 |
| Healthy male | 10 | 1 |
| SLE female | 69 | 7 |
| SLE male | 10 | 1 |
| No. Genes | SLE Individuals | Healthy Individuals | Common |
|---|---|---|---|
| Sex-biased differential alternative splicing (DAS) | 398 | 421 | 56 |
| Sex-biased differential gene expression (DGE) | 111 | 323 | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosmara, D.; Papanikolaou, S.; Nikolaou, C.; Bertsias, G. Extensive Alternative Splicing Patterns in Systemic Lupus Erythematosus Highlight Sexual Differences. Cells 2023, 12, 2678. https://doi.org/10.3390/cells12232678
Kosmara D, Papanikolaou S, Nikolaou C, Bertsias G. Extensive Alternative Splicing Patterns in Systemic Lupus Erythematosus Highlight Sexual Differences. Cells. 2023; 12(23):2678. https://doi.org/10.3390/cells12232678
Chicago/Turabian StyleKosmara, Despoina, Sofia Papanikolaou, Christoforos Nikolaou, and George Bertsias. 2023. "Extensive Alternative Splicing Patterns in Systemic Lupus Erythematosus Highlight Sexual Differences" Cells 12, no. 23: 2678. https://doi.org/10.3390/cells12232678
APA StyleKosmara, D., Papanikolaou, S., Nikolaou, C., & Bertsias, G. (2023). Extensive Alternative Splicing Patterns in Systemic Lupus Erythematosus Highlight Sexual Differences. Cells, 12(23), 2678. https://doi.org/10.3390/cells12232678








