The Sleep Quality- and Myopia-Linked PDE11A-Y727C Variant Impacts Neural Physiology by Reducing Catalytic Activity and Altering Subcellular Compartmentalization of the Enzyme
Abstract
:1. Introduction
2. Methods
2.1. Subjects
2.2. Tissue Isolation for Western Blot
2.3. Preparation of Samples for Western Blots
2.4. Western Blotting
2.5. Plasmid and Lentivirus Generation
2.6. Cell Culture
2.7. PDE Assay
2.8. Ocular Physiology Studies
2.9. Assessment of Cataracts
2.10. Sleep Studies
2.11. Subcellular Localization
2.12. Statistical Analysis
3. Results
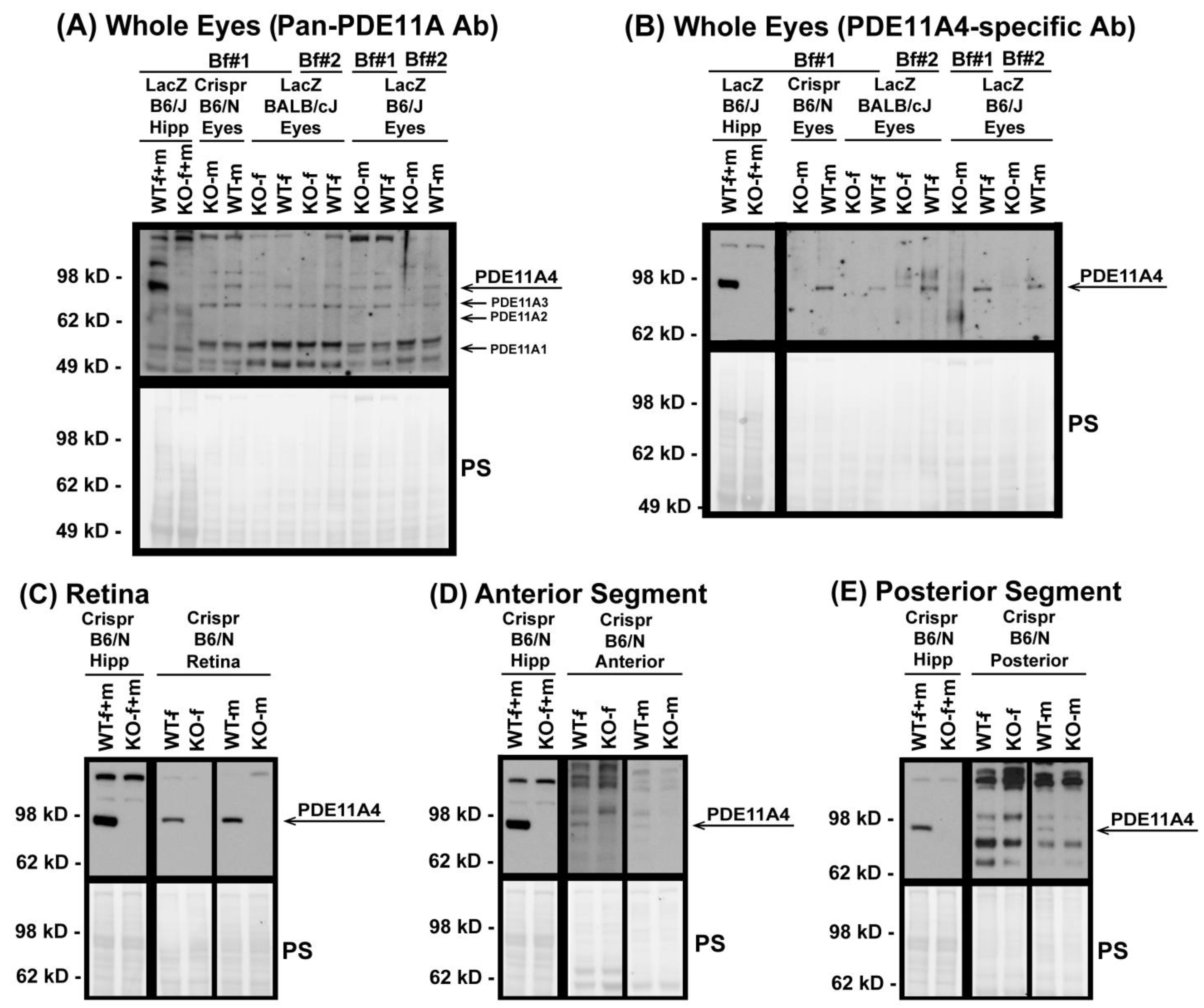
3.1. Western Blots Identify PDE11A4 Protein in Whole-Eye Tissue and Individual Eye Segments
3.2. PDE11A4 Expression in the Eye Changes with Age in A Segment-Specific Manner
3.3. PDE11A4-Y727C and -M878V Variants Exhibit Reduced cAMP-PDE and cGMP-PDE Activity in Neural Cells
3.4. Loss of PDE11A Does Not Alter Eye Growth
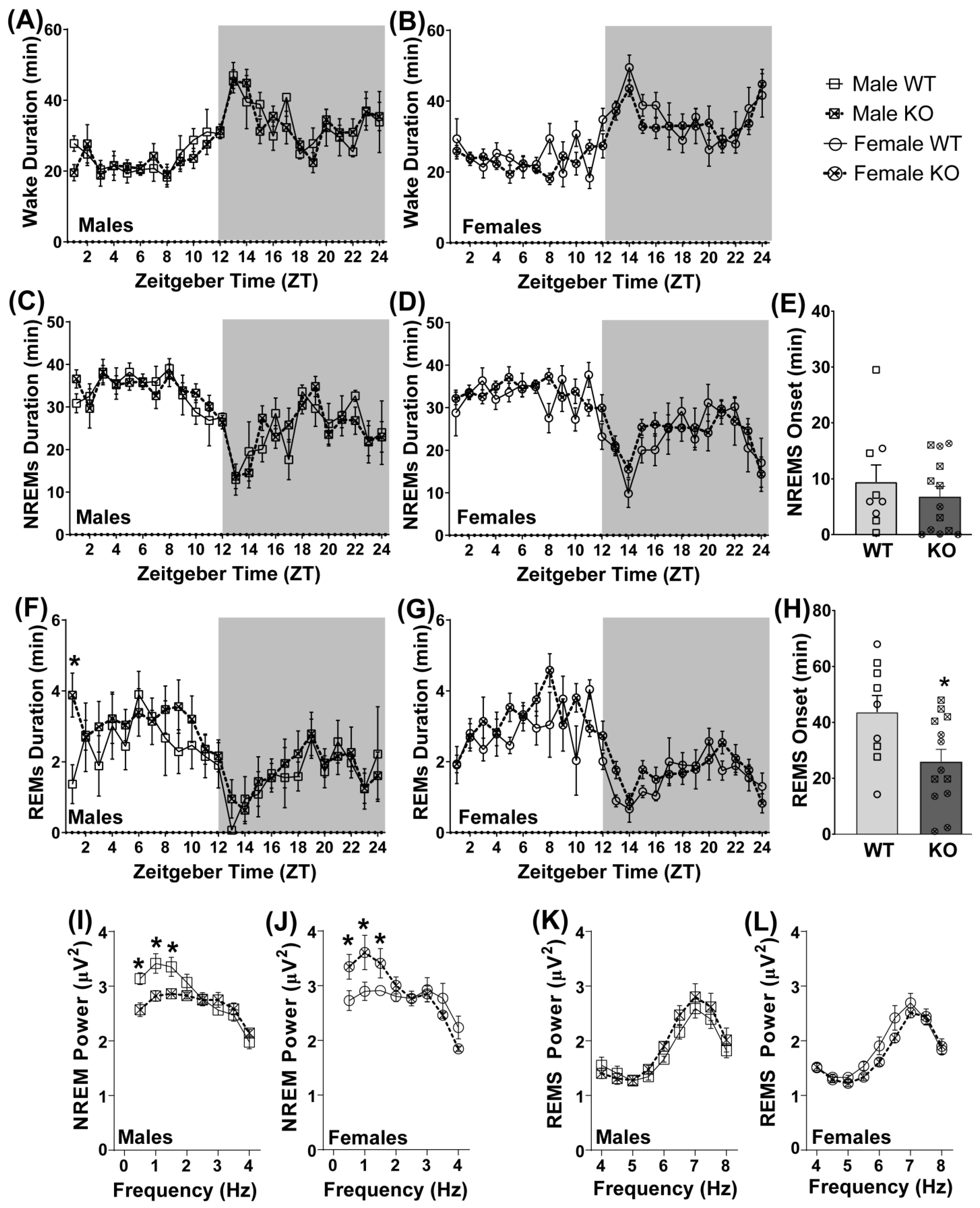
3.5. Pde11a Deletion Minimally Improves Sleep Quality
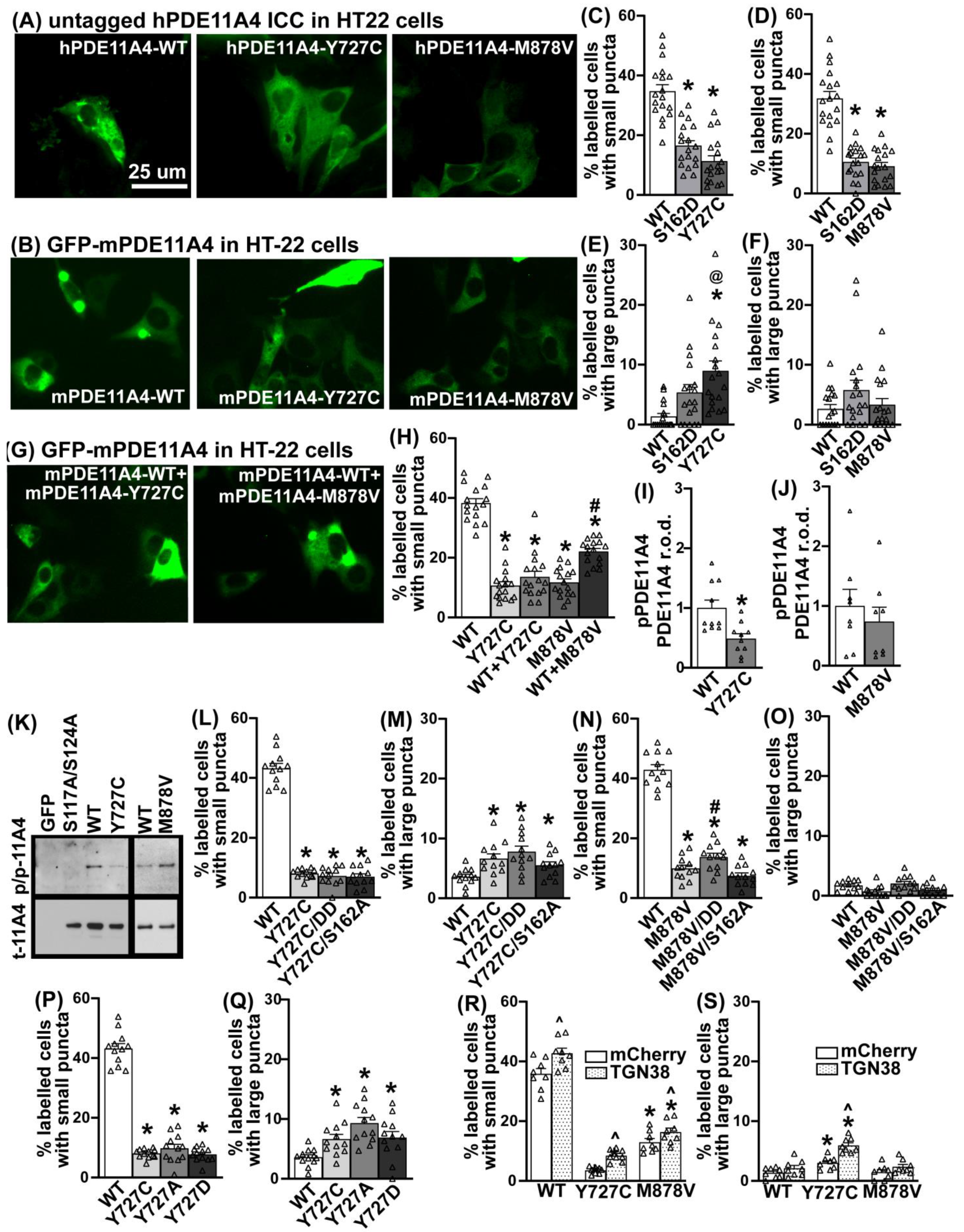
3.6. PDE11A4-Y727C and -M878V Variants Dramatically Alter the Subcellular Compartmentalization of the Enzyme
3.7. The Y727C Variant Dictates Localization of mPDE11A4-WT When Co-Expressed
3.8. The Effects of the Y727C and M878V Variants on PDE11A4 Subcellular Compartmentalization Do Not Require Phosphorylation of S162 nor Dephosphorylation of S117/S124
3.9. The Effects of Y727C on PDE11A4 Subcellular Compartmentalization Are Due to the Loss of the Tyrosine Impacting Processing via the Trans-Golgi Network
4. Discussion
4.1. PDE11A4 Expression Is Greatly Enriched in the Retina vs. Anterior or Posterior Segments of the Eye
4.2. PDE11A4 Expression in the Eye Changes with Age in a Segment-Specific Manner
4.3. The PDE11A4-Y727C and -M878V Variants Impair cAMP Hydrolysis More So than cGMP Hydrolysis
4.4. PDE11A4-Y727C May Reflect a Gain-of-Function Mutation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baillie, G.S.; Tejeda, G.S.; Kelly, M.P. Therapeutic targeting of 3′,5′-cyclic nucleotide phosphodiesterases: Inhibition and beyond. Nat. Rev. Drug Discov. 2019, 18, 770–796. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.P. Pde11a. In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 3804–3826. [Google Scholar] [CrossRef]
- Hegde, S.; Capell, W.R.; Ibrahim, B.A.; Klett, J.; Patel, N.S.; Sougiannis, A.T.; Kelly, M.P. Phosphodiesterase 11A (PDE11A), Enriched in Ventral Hippocampus Neurons, is Required for Consolidation of Social but not Nonsocial Memories in Mice. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2016, 41, 2920–2931. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.P.; Logue, S.F.; Brennan, J.; Day, J.P.; Lakkaraju, S.; Jiang, L.; Zhong, X.; Tam, M.; Sukoff Rizzo, S.J.; Platt, B.J.; et al. Phosphodiesterase 11A in brain is enriched in ventral hippocampus and deletion causes psychiatric disease-related phenotypes. Proc. Natl. Acad. Sci. USA 2010, 107, 8457–8462. [Google Scholar] [CrossRef] [PubMed]
- Pilarzyk, K.; Porcher, L.; Capell, W.R.; Burbano, S.D.; Davis, J.; Fisher, J.L.; Gorny, N.; Petrolle, S.; Kelly, M.P. Conserved age-related increases in hippocampal PDE11A4 cause unexpected proteinopathies and cognitive decline of social associative memories. Aging Cell 2022, 21, e13687. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.P. Does phosphodiesterase 11A (PDE11A) hold promise as a future therapeutic target? Curr. Pharm. Des. 2015, 21, 389–416. [Google Scholar] [CrossRef] [PubMed]
- Hegde, S.; Ji, H.; Oliver, D.; Patel, N.S.; Poupore, N.; Shtutman, M.; Kelly, M.P. PDE11A regulates social behaviors and is a key mechanism by which social experience sculpts the brain. Neuroscience 2016, 335, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Pathak, G.; Agostino, M.J.; Bishara, K.; Capell, W.R.; Fisher, J.L.; Hegde, S.; Ibrahim, B.A.; Pilarzyk, K.; Sabin, C.; Tuczkewycz, T.; et al. PDE11A negatively regulates lithium responsivity. Mol. Psychiatry 2017, 22, 1714–1724. [Google Scholar] [CrossRef]
- Pilarzyk, K.; Capell, W.R.; Porcher, L.; Rips-Goodwin, A.; Kelly, M.P. Biologic that disrupts PDE11A4 homodimerization in hippocampus CA1 reverses age-related cognitive decline of social memories in mice. Neurobiol. Aging 2023, 131, 39–51. [Google Scholar] [CrossRef]
- Pilarzyk, K.; Farmer, R.; Porcher, L.; Kelly, M.P. The Role of PDE11A4 in Social Isolation-Induced Changes in Intracellular Signaling and Neuroinflammation. Front. Pharmacol. 2021, 12, 749628. [Google Scholar] [CrossRef]
- Pilarzyk, K.; Klett, J.; Pena, E.A.; Porcher, L.; Smith, A.J.; Kelly, M.P. Loss of Function of Phosphodiesterase 11A4 Shows that Recent and Remote Long-Term Memories Can Be Uncoupled. Curr. Biol. CB 2019, 29, 2307–2321.e5. [Google Scholar] [CrossRef]
- Smith, A.J.; Farmer, R.; Pilarzyk, K.; Porcher, L.; Kelly, M.P. A genetic basis for friendship? Homophily for membrane-associated PDE11A-cAMP-CREB signaling in CA1 of hippocampus dictates mutual social preference in male and female mice. Mol. Psychiatry 2021, 26, 7107–7117. [Google Scholar] [CrossRef]
- Tedja, M.S.; Wojciechowski, R.; Hysi, P.G.; Eriksson, N.; Furlotte, N.A.; Verhoeven, V.J.M.; Iglesias, A.I.; Meester-Smoor, M.A.; Tompson, S.W.; Fan, Q.; et al. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat. Genet. 2018, 50, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Hysi, P.G.; Choquet, H.; Khawaja, A.P.; Wojciechowski, R.; Tedja, M.S.; Yin, J.; Simcoe, M.J.; Patasova, K.; Mahroo, O.A.; Thai, K.K.; et al. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat. Genet. 2020, 52, 401–407. [Google Scholar] [CrossRef]
- Kiefer, A.K.; Tung, J.Y.; Do, C.B.; Hinds, D.A.; Mountain, J.L.; Francke, U.; Eriksson, N. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 2013, 9, e1003299. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; van Hees, V.T.; Mazzotti, D.R.; Marques-Vidal, P.; Sabia, S.; van der Spek, A.; Dashti, H.S.; Engmann, J.; Kocevska, D.; Tyrrell, J.; et al. Genetic studies of accelerometer-based sleep measures yield new insights into human sleep behaviour. Nat. Commun. 2019, 10, 1585. [Google Scholar] [CrossRef] [PubMed]
- Hartse, K.M.; Eisenhart, S.F.; Bergmann, B.M.; Rechtschaffen, A. Ventral hippocampus spikes during sleep, wakefulness, and arousal in the cat. Sleep 1979, 1, 231–246. [Google Scholar] [CrossRef]
- Wagner, T.; Axmacher, N.; Lehnertz, K.; Elger, C.E.; Fell, J. Sleep-dependent directional coupling between human neocortex and hippocampus. Cortex A J. Devoted Study Nerv. Syst. Behav. 2010, 46, 256–263. [Google Scholar] [CrossRef]
- Vecsey, C.G.; Baillie, G.S.; Jaganath, D.; Havekes, R.; Daniels, A.; Wimmer, M.; Huang, T.; Brown, K.M.; Li, X.Y.; Descalzi, G.; et al. Sleep deprivation impairs cAMP signalling in the hippocampus. Nature 2009, 461, 1122–1125. [Google Scholar] [CrossRef]
- Navarro-Sanchis, C.; Brock, O.; Winsky-Sommerer, R.; Thuret, S. Modulation of Adult Hippocampal Neurogenesis by Sleep: Impact on Mental Health. Front. Neural Circuits 2017, 11, 74. [Google Scholar] [CrossRef]
- Kreckova, M.; Kemlink, D.; Sonka, K.; Krasensky, J.; Buskova, J.; Vaneckova, M.; Nemcova, V. Anterior hippocampus volume loss in narcolepsy with cataplexy. J. Sleep Res. 2019, 28, e12785. [Google Scholar] [CrossRef]
- Abel, T.; Havekes, R.; Saletin, J.M.; Walker, M.P. Sleep, plasticity and memory from molecules to whole-brain networks. Curr. Biol. CB 2013, 23, R774–R788. [Google Scholar] [CrossRef] [PubMed]
- Adamantidis, A.R.; de Lecea, L. Sleep and the hypothalamus. Science 2023, 382, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Lukowski, S.W.; Lo, C.Y.; Sharov, A.A.; Nguyen, Q.; Fang, L.; Hung, S.S.; Zhu, L.; Zhang, T.; Grünert, U.; Nguyen, T.; et al. A single-cell transcriptome atlas of the adult human retina. EMBO J. 2019, 38, e100811. [Google Scholar] [CrossRef] [PubMed]
- Cowan, C.S.; Renner, M.; De Gennaro, M.; Gross-Scherf, B.; Goldblum, D.; Hou, Y.; Munz, M.; Rodrigues, T.M.; Krol, J.; Szikra, T.; et al. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell 2020, 182, 1623–1640.e34. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.P.; Whitmore, S.S.; Flamme-Wiese, M.J.; Riker, M.J.; Wiley, L.A.; Tucker, B.A.; Stone, E.M.; Mullins, R.F.; Scheetz, T.E. Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp. Eye Res. 2019, 184, 234–242. [Google Scholar] [CrossRef]
- Voigt, A.P.; Mulfaul, K.; Mullin, N.K.; Flamme-Wiese, M.J.; Giacalone, J.C.; Stone, E.M.; Tucker, B.A.; Scheetz, T.E.; Mullins, R.F. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. USA 2019, 116, 24100–24107. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Zhang, P.; Zhu, H. Light Signaling and Myopia Development: A Review. Ophthalmol. Ther. 2022, 11, 939–957. [Google Scholar] [CrossRef]
- Landis, E.G.; Yang, V.; Brown, D.M.; Pardue, M.T.; Read, S.A. Dim Light Exposure and Myopia in Children. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4804–4811. [Google Scholar] [CrossRef]
- Klaus, C.; Caruso, G.; Gurevich, V.V.; Hamm, H.E.; Makino, C.L.; DiBenedetto, E. Phototransduction in retinal cones: Analysis of parameter importance. PLoS ONE 2021, 16, e0258721. [Google Scholar] [CrossRef]
- Chotikasemsri, P.; Tangtrakulwanich, B.; Sangkhathat, S. The Effect of Phototherapy on Cancer Predisposition Genes of Diabetic and Normal Human Skin Fibroblasts. BioMed. Res. Int. 2017, 2017, 7604861. [Google Scholar] [CrossRef] [PubMed]
- Ayaki, M.; Torii, H.; Tsubota, K.; Negishi, K. Decreased sleep quality in high myopia children. Sci. Rep. 2016, 6, 33902. [Google Scholar] [CrossRef] [PubMed]
- Jee, D.; Morgan, I.G.; Kim, E.C. Inverse relationship between sleep duration and myopia. Acta Ophthalmol. 2016, 94, e204–e210. [Google Scholar] [CrossRef]
- Chakraborty, R.; Ostrin, L.A.; Nickla, D.L.; Iuvone, P.M.; Pardue, M.T.; Stone, R.A. Circadian rhythms, refractive development, and myopia. Ophthalmic Physiol. Opt. 2018, 38, 217–245. [Google Scholar] [CrossRef]
- Tkatchenko, T.V.; Shah, R.L.; Nagasaki, T.; Tkatchenko, A.V. Analysis of genetic networks regulating refractive eye development in collaborative cross progenitor strain mice reveals new genes and pathways underlying human myopia. BMC Med. Genom. 2019, 12, 113. [Google Scholar] [CrossRef]
- Liu, Z.; Xiu, Y.; Qiu, F.; Zhu, Z.; Zong, R.; Zhou, X.; An, J.; Wang, Q.; Reinach, P.S.; Li, W.; et al. Canonical Wnt Signaling Drives Myopia Development and Can Be Pharmacologically Modulated. Investig. Ophthalmol. Vis. Sci. 2021, 62, 21. [Google Scholar] [CrossRef]
- Xiao, H.; Lin, S.; Jiang, D.; Lin, Y.; Liu, L.; Zhang, Q.; He, J.; Chen, Y. Association of Extracellular Signal-Regulated Kinase Genes With Myopia: A Longitudinal Study of Chinese Children. Front. Genet. 2021, 12, 654869. [Google Scholar] [CrossRef]
- Yang, J.; Ouyang, X.; Fu, H.; Hou, X.; Liu, Y.; Xie, Y.; Yu, H.; Wang, G. Advances in biomedical study of the myopia-related signaling pathways and mechanisms. Biomed. Pharmacother. Biomed. Pharmacother. 2022, 145, 112472. [Google Scholar] [CrossRef]
- Du, J.; An, J.; Linton, J.D.; Wang, Y.; Hurley, J.B. How Excessive cGMP Impacts Metabolic Proteins in Retinas at the Onset of Degeneration. Adv. Exp. Med. Biol. 2018, 1074, 289–295. [Google Scholar] [CrossRef]
- Das, S.; Chen, Y.; Yan, J.; Christensen, G.; Belhadj, S.; Tolone, A.; Paquet-Durand, F. The role of cGMP-signalling and calcium-signalling in photoreceptor cell death: Perspectives for therapy development. Pflug. Arch. Eur. J. Physiol. 2021, 473, 1411–1421. [Google Scholar] [CrossRef]
- Traverso, V.; Bush, R.A.; Sieving, P.A.; Deretic, D. Retinal cAMP levels during the progression of retinal degeneration in rhodopsin P23H and S334ter transgenic rats. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1655–1661. [Google Scholar]
- Zhao, F.; Zhou, H.; Chen, W.; Zhao, C.; Zheng, Y.; Tao, Y.; Pan, M.; Reinach, P.S.; Zhu, J.; An, J.; et al. Declines in PDE4B activity promote myopia progression through downregulation of scleral collagen expression. Exp. Eye Res. 2021, 212, 108758. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, W.; Zhou, H.; Reinach, P.S.; Wang, Y.; Juo, S.H.; Yang, Z.; Xue, A.; Shi, Y.; Liang, C.L.; et al. PDE4B Proposed as a High Myopia Susceptibility Gene in Chinese Population. Front. Genet. 2021, 12, 775797. [Google Scholar] [CrossRef]
- Gopalakrishna, K.N.; Boyd, K.; Artemyev, N.O. Mechanisms of mutant PDE6 proteins underlying retinal diseases. Cell. Signal. 2017, 37, 74–80. [Google Scholar] [CrossRef]
- Zou, T.; Wang, T.; Zhen, F.; He, X.; Dong, S.; Zhang, H. Exogenous PDE5 Expression Rescues Photoreceptors in. Curr. Med. Chem. 2022, 29, 6115–6124. [Google Scholar] [CrossRef] [PubMed]
- Morshedian, A.; Sendek, G.; Ng, S.Y.; Boyd, K.; Radu, R.A.; Liu, M.; Artemyev, N.O.; Sampath, A.P.; Fain, G.L. Reproducibility of the Rod Photoreceptor Response Depends Critically on the Concentration of the Phosphodiesterase Effector Enzyme. J. Neurosci. Off. J. Soc. Neurosci. 2022, 42, 2180–2189. [Google Scholar] [CrossRef] [PubMed]
- Coon, H.; Darlington, T.; Pimentel, R.; Smith, K.R.; Huff, C.D.; Hu, H.; Jerominski, L.; Hansen, J.; Klein, M.; Callor, W.B.; et al. Genetic risk factors in two Utah pedigrees at high risk for suicide. Transl. Psychiatry 2013, 3, e325. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.P.; Adamowicz, W.; Bove, S.; Hartman, A.J.; Mariga, A.; Pathak, G.; Reinhart, V.; Romegialli, A.; Kleiman, R.J. Select 3′,5′-cyclic nucleotide phosphodiesterases exhibit altered expression in the aged rodent brain. Cell. Signal. 2014, 26, 383–397. [Google Scholar] [CrossRef]
- Fosang, A.J.; Colbran, R.J. Transparency Is the Key to Quality. J. Biol. Chem. 2015, 290, 29692–29694. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Howell, M.T.; Kaminski, A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem. Sci. 1990, 15, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Rybalkin, S.D.; Hinds, T.R.; Beavo, J.A. Enzyme assays for cGMP hydrolyzing phosphodiesterases. Methods Mol. Biol. 2013, 1020, 51–62. [Google Scholar] [CrossRef]
- Roorda, A.; Bobier, W.R.; Campbell, M.C. An infrared eccentric photo-optometer. Vis. Res. 1998, 38, 1913–1924. [Google Scholar] [CrossRef]
- Schaeffel, F.; Burkhardt, E.; Howland, H.C.; Williams, R.W. Measurement of refractive state and deprivation myopia in two strains of mice. Optom. Vis. Sci. Off. Publ. Am. Acad. Optom. 2004, 81, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Janssen, C.F.; Maiello, P.; Wright, M.J.; Kracinovsky, K.B.; Newsome, J.T. Comparison of Atipamezole with Yohimbine for Antagonism of Xylazine in Mice Anesthetized with Ketamine and Xylazine. J. Am. Assoc. Lab Anim. Sci. 2017, 56, 142–147. [Google Scholar] [PubMed]
- Milosavljevic, S.; Smith, A.K.; Wright, C.J.; Valafar, H.; Pocivavsek, A. Kynurenine aminotransferase II inhibition promotes sleep and rescues impairments induced by neurodevelopmental insult. Transl. Psychiatry 2023, 13, 106. [Google Scholar] [CrossRef] [PubMed]
- Wellman, L.L.; Yang, L.; Sanford, L.D. Effects of corticotropin releasing factor (CRF) on sleep and temperature following predictable controllable and uncontrollable stress in mice. Front. Neurosci. 2015, 9, 258. [Google Scholar] [CrossRef]
- Missig, G.; Mokler, E.L.; Robbins, J.O.; Alexander, A.J.; McDougle, C.J.; Carlezon, W.A., Jr. Perinatal Immune Activation Produces Persistent Sleep Alterations and Epileptiform Activity in Male Mice. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2018, 43, 482–491. [Google Scholar] [CrossRef]
- Dittrich, L.; Morairty, S.R.; Warrier, D.R.; Kilduff, T.S. Homeostatic sleep pressure is the primary factor for activation of cortical nNOS/NK1 neurons. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2015, 40, 632–639. [Google Scholar] [CrossRef]
- Chen, F.; Duggal, P.; Klein, B.E.; Lee, K.E.; Truitt, B.; Klein, R.; Iyengar, S.K.; Klein, A.P. Variation in PTCHD2, CRISP3, NAP1L4, FSCB, and AP3B2 associated with spherical equivalent. Mol. Vis. 2016, 22, 783–796. [Google Scholar]
- Wojciechowski, R.; Cheng, C.Y. Involvement of multiple molecular pathways in the genetics of ocular refraction and myopia. Retina 2018, 38, 91–101. [Google Scholar] [CrossRef]
- Guggenheim, J.A.; Clark, R.; Cui, J.; Terry, L.; Patasova, K.; Haarman, A.E.G.; Musolf, A.M.; Verhoeven, V.J.M.; Klaver, C.C.W.; Bailey-Wilson, J.E.; et al. Whole exome sequence analysis in 51 624 participants identifies novel genes and variants associated with refractive error and myopia. Hum. Mol. Genet. 2022, 31, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ju, M.; Cho, O.H.; Kim, Y.; Nam, K.T. Tyrosine-Rich Peptides as a Platform for Assembly and Material Synthesis. Adv. Sci. 2019, 6, 1801255. [Google Scholar] [CrossRef] [PubMed]
- Stanley, K.K.; Howell, K.E. TGN38/41: A molecule on the move. Trends Cell Biol. 1993, 3, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.K.; Civan, M.M.; To, C.H.; Do, C.W. cAMP Stimulates Transepithelial Short-Circuit Current and Fluid Transport Across Porcine Ciliary Epithelium. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6784–6794. [Google Scholar] [CrossRef] [PubMed]
- Meadows, M.A.; Balakrishnan, V.; Wang, X.; von Gersdorff, H. Glycine Release Is Potentiated by cAMP via EPAC2 and Ca. J. Neurosci. Off. J. Soc. Neurosci. 2021, 41, 9503–9520. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, L.A.; Samoiliuk, E.V.; Govardovskii, V.I.; Firsov, M.L. cAMP controls rod photoreceptor sensitivity via multiple targets in the phototransduction cascade. J. Gen. Physiol. 2012, 140, 421–433. [Google Scholar] [CrossRef]
- Ko, G.Y.; Ko, M.L.; Dryer, S.E. Circadian regulation of cGMP-gated channels of vertebrate cone photoreceptors: Role of cAMP and Ras. J. Neurosci. Off. J. Soc. Neurosci. 2004, 24, 1296–1304. [Google Scholar] [CrossRef]
- Michalakis, S.; Becirovic, E.; Biel, M. Retinal Cyclic Nucleotide-Gated Channels: From Pathophysiology to Therapy. Int. J. Mol. Sci. 2018, 19, 749. [Google Scholar] [CrossRef]
- Zhang, N.; Beuve, A.; Townes-Anderson, E. The nitric oxide-cGMP signaling pathway differentially regulates presynaptic structural plasticity in cone and rod cells. J. Neurosci. Off. J. Soc. Neurosci. 2005, 25, 2761–2770. [Google Scholar] [CrossRef]
- Caruso, G.; Gurevich, V.V.; Klaus, C.; Hamm, H.; Makino, C.L.; DiBenedetto, E. Local, nonlinear effects of cGMP and Ca2+ reduce single photon response variability in retinal rods. PLoS ONE 2019, 14, e0225948. [Google Scholar] [CrossRef]
- Rasmussen, M.; Welinder, C.; Schwede, F.; Ekström, P. The cGMP system in normal and degenerating mouse neuroretina: New proteins with cGMP interaction potential identified by a proteomics approach. J. Neurochem. 2021, 157, 2173–2186. [Google Scholar] [CrossRef]
- Fang, F.; Pan, M.; Yan, T.; Tao, Y.; Wu, H.; Liu, X.; Qu, J.; Zhou, X. The role of cGMP in ocular growth and the development of form-deprivation myopia in guinea pigs. Investig. Ophthalmol. Vis. Sci. 2013, 54, 7887–7902. [Google Scholar] [CrossRef]
- Lian, P.; Zhao, X.; Song, H.; Tanumiharjo, S.; Chen, J.; Wang, T.; Chen, S.; Lu, L. Metabolic characterization of human intraocular fluid in patients with pathological myopia. Exp. Eye Res. 2022, 222, 109184. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Pan, M.; Liu, S.; Fang, F.; Lu, R.; Lu, C.; Zheng, M.; An, J.; Xu, H.; Zhao, F.; et al. cAMP level modulates scleral collagen remodeling, a critical step in the development of myopia. PLoS ONE 2013, 8, e71441. [Google Scholar] [CrossRef] [PubMed]
- Srinivasalu, N.; Lu, C.; Pan, M.; Reinach, P.S.; Wen, Y.; Hu, Y.; Qu, J.; Zhou, X. Role of Cyclic Adenosine Monophosphate in Myopic Scleral Remodeling in Guinea Pigs: A Microarray Analysis. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4318–4325. [Google Scholar] [CrossRef] [PubMed]
- Bolger, G.B. The PDE-Opathies: Diverse Phenotypes Produced by a Functionally Related Multigene Family. Trends Genet. 2021, 37, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Michy, K. Encyclopedia of Signaling Molecules, 2nd ed.; Choi, S., Ed.; Springer: Cham, Switzerland, 2017; pp. 3804–3822. [Google Scholar] [CrossRef]
- Chamessian, A.; Young, M.; Qadri, Y.; Berta, T.; Ji, R.R.; Van de Ven, T. Transcriptional Profiling of Somatostatin Interneurons in the Spinal Dorsal Horn. Sci. Rep. 2018, 8, 6809. [Google Scholar] [CrossRef] [PubMed]
- Horvath, A.; Boikos, S.; Giatzakis, C.; Robinson-White, A.; Groussin, L.; Griffin, K.J.; Stein, E.; Levine, E.; Delimpasi, G.; Hsiao, H.P.; et al. A genome-wide scan identifies mutations in the gene encoding phosphodiesterase 11A4 (PDE11A) in individuals with adrenocortical hyperplasia. Nat. Genet. 2006, 38, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Boikos, S.A.; Horvath, A.; Heyerdahl, S.; Stein, E.; Robinson-White, A.; Bossis, I.; Bertherat, J.; Carney, J.A.; Stratakis, C.A. Phosphodiesterase 11A expression in the adrenal cortex, primary pigmented nodular adrenocortical disease, and other corticotropin-independent lesions. Horm. Metab. Res. Horm. Stoffwechselforschung Horm. Metab. 2008, 40, 347–353. [Google Scholar] [CrossRef]
- Shivanna, M.; Srinivas, S.P. Elevated cAMP opposes (TNF-alpha)-induced loss in the barrier integrity of corneal endothelium. Mol. Vis. 2010, 16, 1781–1790. [Google Scholar]
- Gabelt, B.T.; Kaufman, P.L.; Rasmussen, C.A. Effect of nitric oxide compounds on monkey ciliary muscle in vitro. Exp. Eye Res. 2011, 93, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Vranka, J.A.; Kelley, M.J.; Acott, T.S.; Keller, K.E. Extracellular matrix in the trabecular meshwork: Intraocular pressure regulation and dysregulation in glaucoma. Exp. Eye Res. 2015, 133, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Di Pierdomenico, J.; Henderson, D.C.M.; Giammaria, S.; Smith, V.L.; Jamet, A.J.; Smith, C.A.; Hooper, M.L.; Chauhan, B.C. Age and intraocular pressure in murine experimental glaucoma. Prog. Retin. Eye Res. 2022, 88, 101021. [Google Scholar] [CrossRef]
- Jonas, J.B.; Nagaoka, N.; Fang, Y.X.; Weber, P.; Ohno-Matsui, K. Intraocular Pressure and Glaucomatous Optic Neuropathy in High Myopia. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5897–5906. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Grenell, A.; Zhong, F.; Yam, M.; Hauer, A.; Gregor, E.; Zhu, S.; Lohner, D.; Zhu, J.; Du, J. Metabolic signature of the aging eye in mice. Neurobiol. Aging 2018, 71, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Machamer, C.E. The Golgi complex in stress and death. Front. Neurosci. 2015, 9, 421. [Google Scholar] [CrossRef]
- Santone, R.; Giorgi, M.; Maccarone, R.; Basso, M.; Deplano, S.; Bisti, S. Gene expression and protein localization of calmodulin-dependent phosphodiesterase in adult rat retina. J. Neurosci. Res. 2006, 84, 1020–1026. [Google Scholar] [CrossRef]
- Diederen, R.M.; La Heij, E.C.; Markerink-van Ittersum, M.; Kijlstra, A.; Hendrikse, F.; de Vente, J. Selective blockade of phosphodiesterase types 2, 5 and 9 results in cyclic 3′5′ guanosine monophosphate accumulation in retinal pigment epithelium cells. Br. J. Ophthalmol. 2007, 91, 379–384. [Google Scholar] [CrossRef]
- Foresta, C.; Caretta, N.; Zuccarello, D.; Poletti, A.; Biagioli, A.; Caretti, L.; Galan, A. Expression of the PDE5 enzyme on human retinal tissue: New aspects of PDE5 inhibitors ocular side effects. Eye 2008, 22, 144–149. [Google Scholar] [CrossRef]
- Whitaker, C.M.; Cooper, N.G. The novel distribution of phosphodiesterase-4 subtypes within the rat retina. Neuroscience 2009, 163, 1277–1291. [Google Scholar] [CrossRef]
- Hetman, J.M.; Soderling, S.H.; Glavas, N.A.; Beavo, J.A. Cloning and characterization of PDE7B, a cAMP-specific phosphodiesterase. Proc. Natl. Acad. Sci. USA 2000, 97, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Soderling, S.H.; Bayuga, S.J.; Beavo, J.A. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc. Natl. Acad. Sci. USA 1998, 95, 8991–8996. [Google Scholar] [CrossRef]
- Fu, Y.; Yau, K.W. Phototransduction in mouse rods and cones. Pflug. Arch. Eur. J. Physiol. 2007, 454, 805–819. [Google Scholar] [CrossRef]



| Batch | Segment | Sex | Acetone | Sample Concentration for Western Blot (µg/µL) |
|---|---|---|---|---|
| 1 | Anterior | male | yes | 2.6 |
| 2 | Anterior | female | no | 2.32 |
| 3 | Anterior | male | no | 2.6 |
| 4 | Anterior | female | no | 2.44 |
| 1 | Posterior | male | yes | 3 |
| 2 | Posterior | female | no | 2.72 |
| 3 | Posterior | male | no | 3 |
| 4 | Posterior | female | no | 3 |
| 1 | Retina | male | yes | 2.44 |
| 2 | Retina | female | no | 2.44 |
| 3 | Retina | male | no | 2.6 |
| 4 | Retina | female | no | 2.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sbornova, I.; van der Sande, E.; Milosavljevic, S.; Amurrio, E.; Burbano, S.D.; Das, P.K.; Do, H.H.; Fisher, J.L.; Kargbo, P.; Patel, J.; et al. The Sleep Quality- and Myopia-Linked PDE11A-Y727C Variant Impacts Neural Physiology by Reducing Catalytic Activity and Altering Subcellular Compartmentalization of the Enzyme. Cells 2023, 12, 2839. https://doi.org/10.3390/cells12242839
Sbornova I, van der Sande E, Milosavljevic S, Amurrio E, Burbano SD, Das PK, Do HH, Fisher JL, Kargbo P, Patel J, et al. The Sleep Quality- and Myopia-Linked PDE11A-Y727C Variant Impacts Neural Physiology by Reducing Catalytic Activity and Altering Subcellular Compartmentalization of the Enzyme. Cells. 2023; 12(24):2839. https://doi.org/10.3390/cells12242839
Chicago/Turabian StyleSbornova, Irina, Emilie van der Sande, Snezana Milosavljevic, Elvis Amurrio, Steven D. Burbano, Prosun K. Das, Helen H. Do, Janet L. Fisher, Porschderek Kargbo, Janvi Patel, and et al. 2023. "The Sleep Quality- and Myopia-Linked PDE11A-Y727C Variant Impacts Neural Physiology by Reducing Catalytic Activity and Altering Subcellular Compartmentalization of the Enzyme" Cells 12, no. 24: 2839. https://doi.org/10.3390/cells12242839





