Prevalence of Olfactory Dysfunction with the Omicron Variant of SARS-CoV-2: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
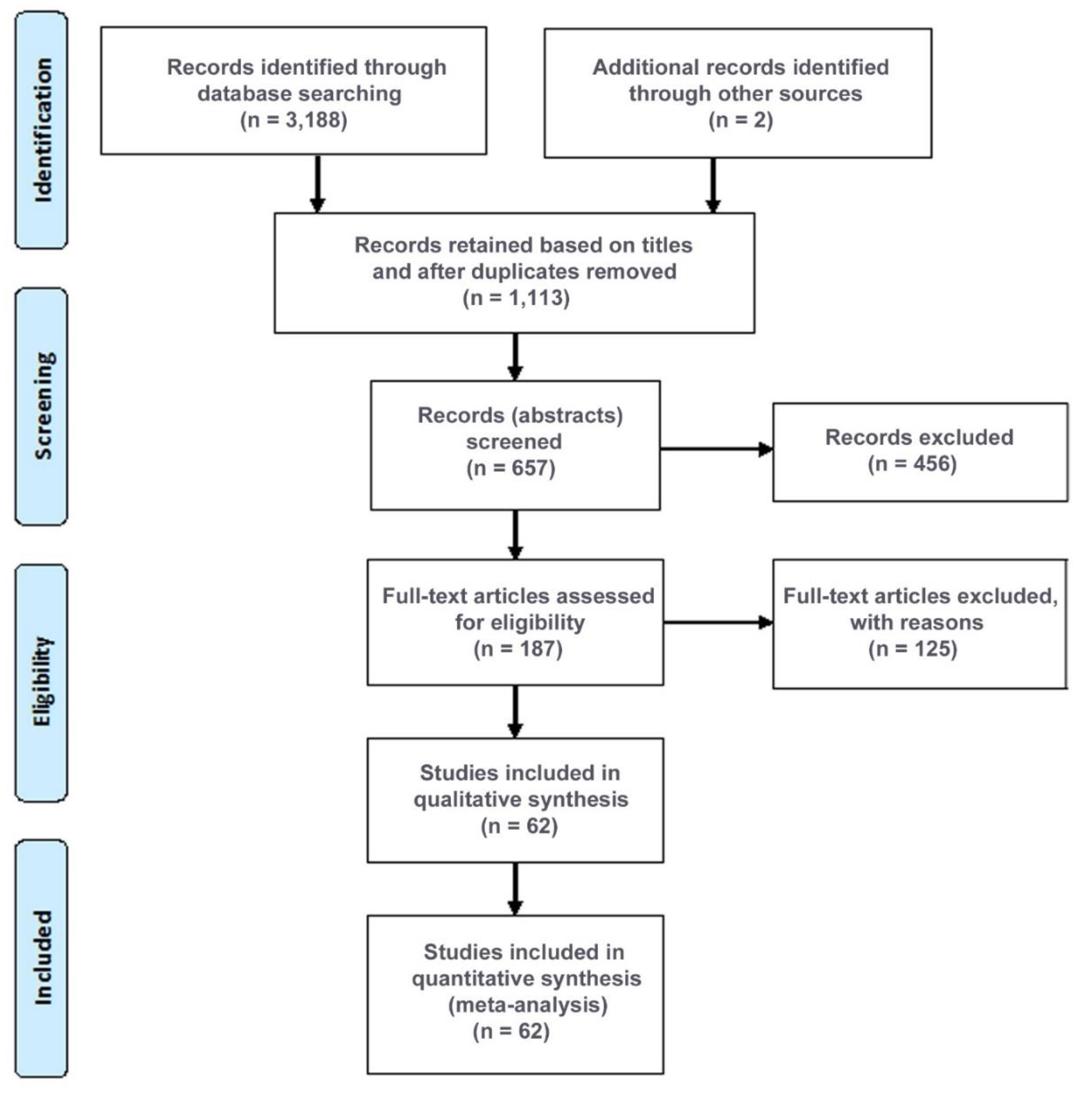
2. Methods
2.1. Search Strategy
2.2. Inclusion/Exclusion Criteria
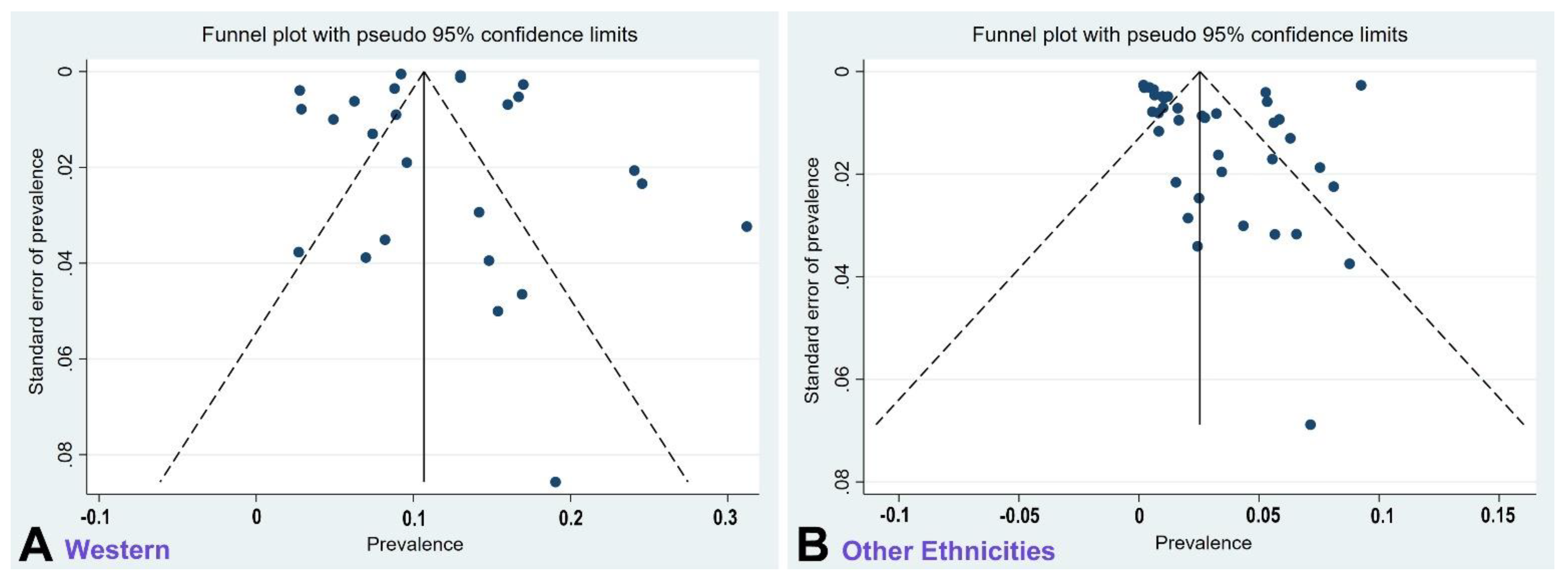
2.3. Quality Assessment and Publication Bias
2.4. Data Extraction
2.5. Subgroup Analyses and Comparisons
2.6. Data Synthesis
2.7. Statistical Analyses
3. Results
3.1. Properties of Studies
3.2. Global Prevalence of Olfactory Dysfunction
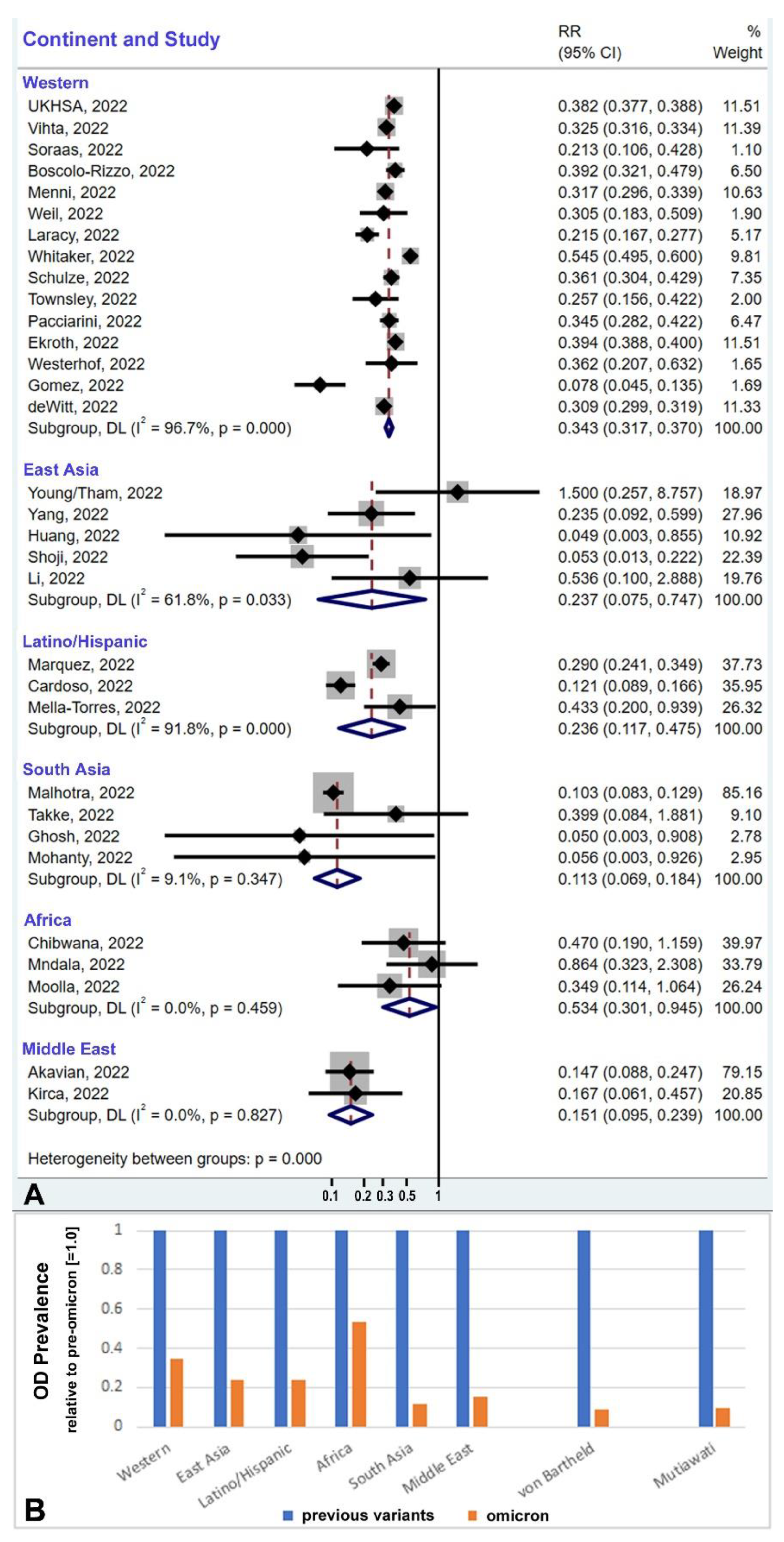
3.3. Geographic/Ethnic Differences
3.4. Global Prevalence Considering Ethnic Differences and Population Sizes
3.5. Ethnic Profiles: Omicron-Induced Hyposmia vs. UGT2A1 Risk Allele Frequency
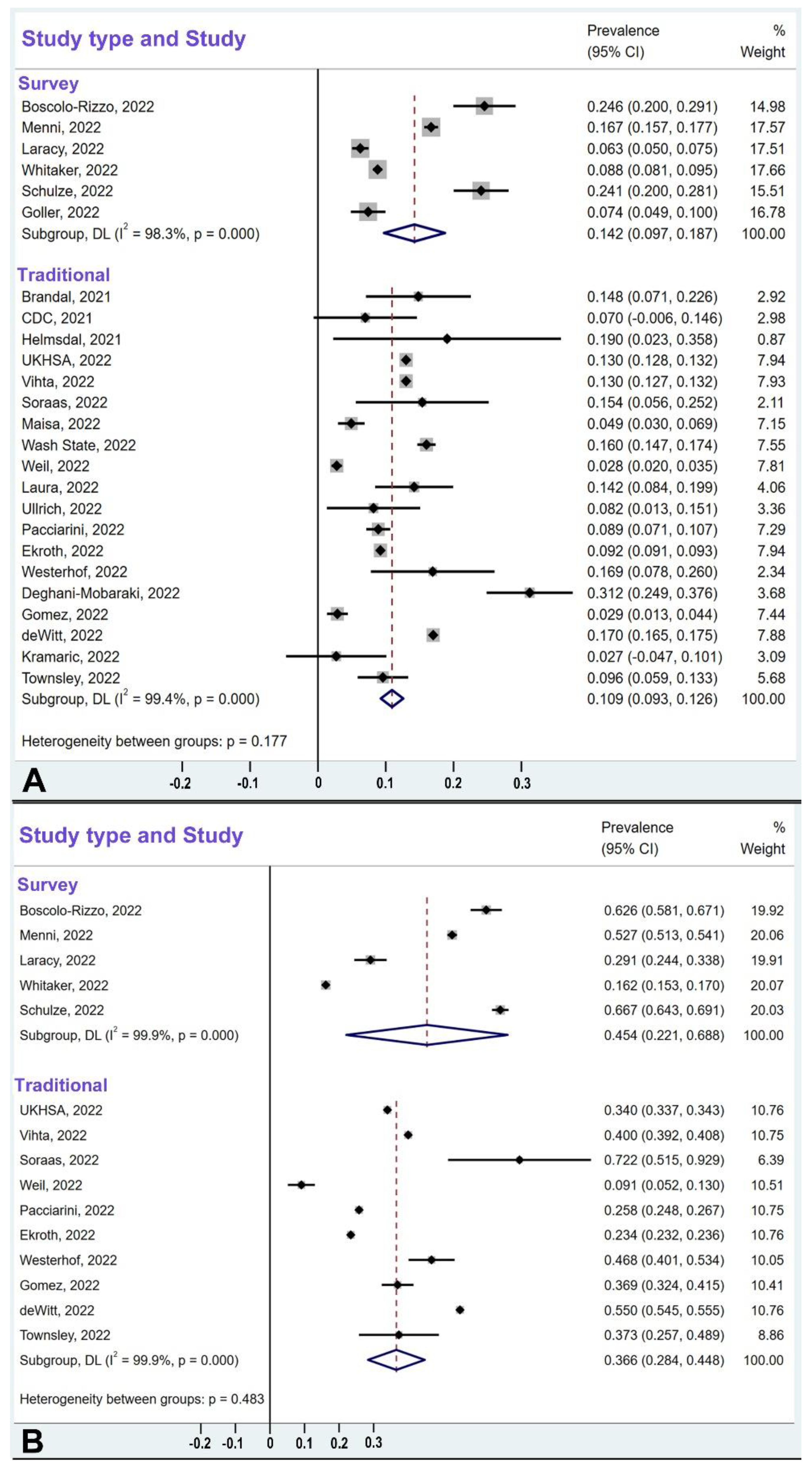
3.6. Comparison of Survey-Type Studies and Traditional-Design Studies
4. Discussion
4.1. Global Prevalence of Olfactory Dysfunction Caused by Omicron Variant Infection
4.2. Why Is Omicron’s Effect on Olfaction Different than That of Previous Variants?
4.3. Ethnic Differences in UGT2A1 Risk Allele Frequency: Implications
4.4. Technical Considerations of Methodology
4.5. Limitations of our Review
4.6. Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodriguez-Sevilla, J.J.; Güerri-Fernádez, R.; Bertran Recasens, B. Is There Less Alteration of Smell Sensation in Patients with Omicron SARS-CoV-2 Variant Infection? Front. Med. 2022, 9, 852998. [Google Scholar] [CrossRef]
- Butowt, R.; Bilińska, K.; von Bartheld, C. Why Does the Omicron Variant Largely Spare Olfactory Function? Implications for the Pathogenesis of Anosmia in Coronavirus Disease 2019. J. Infect. Dis. 2022, 226, 1304–1308. [Google Scholar] [CrossRef]
- Patt, Y.S.; David, P.; Bergwerk, M.; Shoenfeld, Y. The Reduced Frequency of Olfactory Dysfunction in Patients with Omicron SARS-CoV-2 Variant Infection. Ann. Otolaryngol. Rhinol. 2022, 9, 1302. [Google Scholar]
- Chee, J.; Chern, B.; Loh, W.S.; Mullol, J.; Wang, Y. Pathophysiology of SARS-CoV-2 Infection of Nasal Respiratory and Olfactory Epithelia and Its Clinical Impact. Curr. Allergy Asthma Rep. 2023, 1–11. [Google Scholar] [CrossRef]
- COVID-19 Cumulative Infection Collaborators. Estimating global, regional, and national daily and cumulative infections with SARS-CoV-2 through Nov 14, 2021: A statistical analysis. Lancet 2022, 399, 2351–2380. [Google Scholar] [CrossRef]
- IHME (Institute for Health Metrics and Evaluation). COVID-19 Results Briefing, Global, October 21, 2022. Available online: https://www.healthdata.org/sites/default/files/files/Projects/COVID/2022/1_briefing_Global_10.pdf (accessed on 13 December 2022).
- Von Bartheld, C.S.; Hagen, M.M.; Butowt, R. The D614G Virus Mutation Enhances Anosmia in COVID-19 Patients: Evidence from a Systematic Review and Meta-analysis of Studies from South Asia. ACS Chem. Neurosci. 2021, 12, 3535–3549. [Google Scholar] [CrossRef]
- See, A.; Ko, K.K.K.; Toh, S.T. Epidemiological analysis in support of hypothesis that D614G virus mutation is a major contributing factor to chemosensory dysfunction in COVID-19 patients. Eur. Arch. Otorhinolaryngol. 2021, 278, 3595–3596. [Google Scholar] [CrossRef]
- Boscolo-Rizzo, P.; Tirelli, G.; Meloni, P.; Hopkins, C.; Madeddu, G.; De Vito, A.; Gardenal, N.; Valentinotti, R.; Tofanelli, M.; Borsetto, D.; et al. Coronavirus disease 2019 (COVID-19)-related smell and taste impairment with widespread diffusion of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) Omicron variant. Int. Forum Allergy Rhinol. 2022, 12, 1273–1281. [Google Scholar] [CrossRef]
- Tan, B.K.J.; Han, R.; Zhao, J.J.; Tan, N.K.W.; Quah, E.S.H.; Tan, C.J.; Chan, Y.H.; Teo, N.W.Y.; Charn, T.C.; See, A.; et al. Prognosis and persistence of smell and taste dysfunction in patients with COVID-19: Meta-analysis with parametric cure modelling of recovery curves. BMJ 2022, 378, e069503. [Google Scholar] [CrossRef]
- Butowt, R.; Bilinska, K.; von Bartheld, C.S. Chemosensory Dysfunction in COVID-19: Integration of Genetic and Epidemiological Data Points to D614G Spike Protein Variant as a Contributing Factor. ACS Chem. Neurosci. 2020, 11, 3180–3184. [Google Scholar] [CrossRef]
- Kumar, A.A.; Lee, S.W.Y.; Lock, C.; Keong, N.C. Geographical Variations in Host Predisposition to COVID-19 Related Anosmia, Ageusia, and Neurological Syndromes. Front. Med. 2021, 8, 661359. [Google Scholar] [CrossRef]
- Shelton, J.F.; Shastri, A.J.; Fletez-Brant, K.; 23andMe COVID-19 Team; Aslibekyan, S.; Auton, A. The UGT2A1/UGT2A2 locus is associated with COVID-19-related loss of smell or taste. Nat. Genet. 2022, 54, 121–124. [Google Scholar] [CrossRef]
- Braga-Paz, I.; Ferreira de Araújo, J.L.; Alves, H.J.; de Ávila, R.E.; Resende, G.G.; Teixeira, M.M.; de Aguiar, R.S.; de Souza, R.P.; Bahia, D. Negative correlation between ACE2 gene expression levels and loss of taste in a cohort of COVID-19 hospitalized patients: New clues to long-term cognitive disorders. Front. Cell. Infect. Microbiol. 2022, 12, 905757. [Google Scholar] [CrossRef]
- Heydel, J.; Leclerc, S.; Bernard, P.; Pelczar, H.; Gradinaru, D.; Magdalou, J.; Minn, A.; Artur, Y.; Goudonnet, H. Rat olfactory bulb and epithelium UDP-glucuronosyltransferase 2A1 (UGT2A1) expression: In situ mRNA localization and quantitative analysis. Brain Res. Mol. Brain Res. 2001, 90, 83–92. [Google Scholar] [CrossRef]
- Heydel, J.M.; Coelho, A.; Thiebaud, N.; Legendre, A.; Le Bon, A.M.; Faure, P.; Neiers, F.; Artur, Y.; Golebiowski, J.; Briand, L. Odorant-binding proteins and xenobiotic metabolizing enzymes: Implications in olfactory perireceptor events. Anat. Rec. 2013, 296, 1333–1345. [Google Scholar] [CrossRef]
- Lazard, D.; Zupko, K.; Poria, Y.; Nef, P.; Lazarovits, J.; Horn, S.; Khen, M.; Lancet, D. Odorant signal termination by olfactory UDP glucuronosyl transferase. Nature 1991, 349, 790–793. [Google Scholar] [CrossRef]
- Schwartz, M.; Menetrier, F.; Heydel, J.M.; Chavanne, E.; Faure, P.; Labrousse, M.; Lirussi, F.; Canon, F.; Mannervik, B.; Briand, L.; et al. Interactions between Odorants and Glutathione Transferases in the Human Olfactory Cleft. Chem. Senses 2020, 45, 645–654. [Google Scholar] [CrossRef]
- Khan, M.; Yoo, S.J.; Clijsters, M.; Backaert, W.; Vanstapel, A.; Speleman, K.; Lietaer, C.; Choi, S.; Hether, T.D.; Marcelis, L.; et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell 2021, 184, 5932–5949.e15. [Google Scholar] [CrossRef] [PubMed]
- Finlay, J.B.; Brann, D.H.; Abi-Hachem, R.; Jang, D.W.; Oliva, A.D.; Ko, T.; Gupta, R.; Wellford, S.A.; Moseman, E.A.; Jang, S.S.; et al. Persistent post-COVID-19 smell loss is associated with immune cell infiltration and altered gene expression in olfactory epithelium. Sci. Transl. Med. 2022, 14, eadd0484. [Google Scholar] [CrossRef]
- Hintschich, C.A.; Niv, M.Y.; Hummel, T. The taste of the pandemic-contemporary review on the current state of research on gustation in coronavirus disease 2019 (COVID-19). Int. Forum Allergy Rhinol. 2022, 12, 210–216. [Google Scholar] [CrossRef]
- Langstaff, L.; Clark, A.; Salam, M.; Philpott, C.M. Cultural Adaptation and Validity of the Sniffin’ Sticks Psychophysical Test for the UK Setting. Chem. Percept. 2021, 14, 102–108. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, C.; Perofsky, A.C.; Burstein, R.; Famulare, M.; Boyle, S.; Prentice, R.; Marshall, C.; McCormick, B.J.J.; Reinhart, D.; Capodanno, B.; et al. Trends in Risk Factors and Symptoms Associated with SARS-CoV-2 and Rhinovirus Test Positivity in King County, Washington, June 2020 to July 2022. JAMA Netw. Open. 2022, 5, e2245861. [Google Scholar] [CrossRef]
- Yadav, M.; Singh, A.; Meena, J.; Sankar, J.M. A systematic review and meta-analysis of otorhinolaryngological manifestations of coronavirus disease 2019 in paediatric patients. J. Laryngol. Otol. 2022, 136, 588–603. [Google Scholar] [CrossRef]
- Coelho, D.H.; Reiter, E.R.; French, E.; Costanzo, R.M. Decreasing Incidence of Chemosensory Changes by COVID-19 Variant. Otolaryngol. Head Neck Surg. 2022, 1945998221097656. [Google Scholar] [CrossRef] [PubMed]
- Boscutti, A.; Delvecchio, G.; Pigoni, A.; Cereda, G.; Ciappolino, V.; Bellani, M.; Fusar-Poli, P.; Brambilla, P. Olfactory and gustatory dysfunctions in SARS-CoV-2 infection: A systematic review. Brain Behav. Immun. Health 2021, 15, 100268. [Google Scholar] [CrossRef]
- Lerner, D.K.; Garvey, K.L.; Arrighi-Allisan, A.E.; Filimonov, A.; Filip, P.; Shah, J.; Tweel, B.; Del Signore, A.; Schaberg, M.; Colley, P.; et al. Clinical Features of Parosmia Associated with COVID-19 Infection. Laryngoscope 2022, 132, 633–639. [Google Scholar] [CrossRef]
- Ohla, K.; Veldhuizen, M.G.; Green, T.; Hannum, M.E.; Bakke, A.J.; Moein, S.T.; Tognetti, A.; Postma, E.M.; Pellegrino, R.; Hwang, D.L.D.; et al. A follow-up on quantitative and qualitative olfactory dysfunction and other symptoms in patients recovering from COVID-19 smell loss. Rhinology 2022. Epub ahead of print. [Google Scholar] [CrossRef]
- Schulze, H.; Bayer, W. Changes in Symptoms Experienced by SARS-CoV-2-Infected Individuals—From the First Wave to the Omicron Variant. Front. Virol. 2022, 2, 880707. [Google Scholar] [CrossRef]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [Green Version]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control Clin. Trials. 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thomas, J.; Cochrane Collaboration. Cochrane Handbook for Systematic Reviews of Interventions, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2020. [Google Scholar]
- Hood, K.; Robling, M.; Ingledew, D.; Gillespie, D.; Greene, G.; Ivins, R.; Russell, I.; Sayers, A.; Shaw, C.; Williams, J. Mode of data elicitation, acquisition and response to surveys: A systematic review. Health Technol. Assess. 2012, 16, 1–162. [Google Scholar] [CrossRef] [Green Version]
- Egger, M.; Smith, G.D.; Phillips, A.N. Meta-analysis: Principles and procedures. BMJ 1997, 315, 1533–1537. [Google Scholar] [CrossRef] [Green Version]
- Brandal, L.T.; MacDonald, E.; Veneti, L.; Ravlo, T.; Lange, H.; Naseer, U.; Feruglio, S.; Bragstad, K.; Hungnes, O.; Ødeskaug, L.E.; et al. Outbreak caused by the SARS-CoV-2 Omicron variant in Norway, November to December 2021. Eurosurveillance 2021, 26, 2101147. [Google Scholar] [CrossRef] [PubMed]
- CDC COVID-19 Response Team. SARS-CoV-2 B.1.1.529 (Omicron) Variant—United States, December 1–8, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1731–1734. [Google Scholar] [CrossRef] [PubMed]
- Helmsdal, G.; Hansen, O.K.; Møller, L.F.; Christiansen, D.H.; Petersen, M.S.; Kristiansen, M.F. Omicron Outbreak at a Private Gathering in the Faroe Islands, Infecting 21 of 33 Triple-Vaccinated Healthcare Workers. Clin. Infect. Dis. 2022, 75, 893–896. [Google Scholar] [CrossRef]
- UK Health Security Agency. SARS-CoV-2 Variants of Concern and Variants under Investigation in England. Technical Briefing 34. 2022. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1046853/technical-briefing-34-14-january-2022.pdf (accessed on 13 December 2022).
- Vihta, K.D.; Pouwels, K.B.; Peto, T.E.; Pritchard, E.; House, T.; Studley, R.; Rourke, E.; Cook, D.; Diamond, I.; Crook, D.; et al. Omicron-associated changes in SARS-CoV-2 symptoms in the United Kingdom. Clin. Infect. Dis. 2022, ciac613. [Google Scholar] [CrossRef]
- Søraas, A.; Grødeland, G.; Granerud, B.K.; Ueland, T.; Lind, A.; Fevang, B.; Murphy, S.L.; Huse, C.; Nygaard, A.B.; Steffensen, A.K.; et al. Breakthrough infections with the omicron and delta variants of SARS-CoV-2 result in similar re-activation of vaccine-induced immunity. Front. Immunol. 2022, 13, 964525. [Google Scholar] [CrossRef]
- Maisa, A.; Spaccaferri, G.; Fournier, L.; Schaeffer, J.; Deniau, J.; Rolland, P.; Coignard, B.; regional COVID-19 investigation team; EMERGEN consortium. First cases of Omicron in France are exhibiting mild symptoms, November 2021–January 2022. Infect. Dis. Now. 2022, 52, 160–164. [Google Scholar] [CrossRef]
- Kramarič, J.; Ješe, R.; Tomšič, M.; Rotar, Ž.; Hočevar, A. COVID-19 among patients with giant cell arteritis: A single-centre observational study from Slovenia. Clin. Rheumatol. 2022, 41, 2449–2456. [Google Scholar] [CrossRef]
- Menni, C.; Valdes, A.M.; Polidori, L.; Antonelli, M.; Penamakuri, S.; Nogal, A.; Louca, P.; May, A.; Figueiredo, J.C.; Hu, C.; et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: A prospective observational study from the ZOE COVID Study. Lancet 2022, 399, 1618–1624. [Google Scholar] [CrossRef]
- Washington State Department of Health, SARS-CoV-2 Sequencing and Variants in Washington State April 13, 2022. Available online: https://doh.wa.gov/sites/default/files/2022-02/420-316-SequencingAndVariantsReport.pdf (accessed on 8 May 2022).
- Weil, A.A.; Luiten, K.G.; Casto, A.M.; Bennett, J.C.; O’Hanlon, J.; Han, P.D.; Gamboa, L.S.; McDermot, E.; Truong, M.; Gottlieb, G.S.; et al. Genomic surveillance of SARS-CoV-2 Omicron variants on a university campus. Nat. Commun. 2022, 13, 5240. [Google Scholar] [CrossRef] [PubMed]
- Laracy, J.C.; Robilotti, E.V.; Yan, J.; Lucca, A.; Aslam, A.; Babady, N.E.; Kamboj, M. Comparison of coronavirus disease 2019 (COVID-19) symptoms at diagnosis among healthcare personnel before and after the emergence of the omicron variant. Infect. Control Hosp. Epidemiol. 2022, 1–3. [Google Scholar] [CrossRef]
- Whitaker, M.; Elliott, J.; Bodinier, B.; Barclay, W.; Ward, H.; Cooke, G.; Donnelly, C.A.; Chadeau-Hyam, M.; Elliott, P. Variant-specific symptoms of COVID-19 in a study of 1,542,510 adults in England. Nat. Commun. 2022, 13, 6856. [Google Scholar] [CrossRef]
- Laura, L.; Dalmatin-Dragišić, M.; Martinović, K.; Tutiš, B.; Herceg, I.; Arapović, M.; Arapović, J. Does pre-existing immunity determine the course of SARS-CoV-2 infection in health-care workers? Single-center experience. Infection 2022, 13, 1–8. [Google Scholar] [CrossRef]
- Ullrich, F.; Hanoun, C.; Turki, A.T.; Liebregts, T.; Breuckmann, K.; Alashkar, F.; Reinhardt, H.C.; von Tresckow, B.; von Tresckow, J. Early report on the severity of COVID-19 in hematologic patients infected with the SARS-CoV2 omicron variant. Eur. J. Haematol. 2022, 109, 364–372. [Google Scholar] [CrossRef]
- Townsley, H.; Carr, E.C.; Russell, T.W.; Adams, L.; Mears, H.V.; Bailey, C.; Black, J.R.M.; Fowler, A.S.; Wilkinson, K.; Hutchinson, M.; et al. Non-hospitalised, vaccinated adults with COVID-19 caused by Omicron BA.1 and BA.2 present with changing symptom profiles compared to those with Delta despite similar viral kinetics. MedRxiv 2022. [Google Scholar] [CrossRef]
- Pacchiarini, N.; Sawyer, C.; Williams, C.; Sutton, D.; Roberts, C.; Simkin, F.; King, G.; McClure, V.; Cottrell, S.; Clayton, H.; et al. Epidemiological analysis of the first 1000 cases of SARS-CoV-2 lineage BA.1 (B.1.1.529, Omicron) compared with co-circulating Delta in Wales, UK. Influenza Other Respir. Viruses 2022, 16, 986–993. [Google Scholar] [CrossRef]
- Ekroth, A.K.E.; Patrzylas, P.; Turner, C.; Hughes, G.J.; Anderson, C. Comparative symptomatology of infection with SARS-CoV-2 variants Omicron (B.1.1.529) and Delta (B.1.617.2) from routine contact tracing data in England. Epidemiol. Infect. 2022, 150, e162. [Google Scholar] [CrossRef] [PubMed]
- Westerhof, I.; de Hoog, M.; Ieven, M.; Lammens, C.; van Beek, J.; Rozhnova, G.; Eggink, D.; Euser, S.; Wildenbeest, J.; Duijts, L.; et al. The impact of variant and vaccination on SARS-CoV-2 symptomatology; three prospective household cohorts. Int. J. Infect. Dis. 2022, 128, 140–147. [Google Scholar] [CrossRef]
- Goller, K.V.; Moritz, J.; Ziemann, J.; Kohler, C.; Becker, K.; Hübner, N.O.; The CoMV-Gen Study Group. Differences in Clinical Presentations of Omicron Infections with the Lineages BA.2 and BA.5 in Mecklenburg-Western Pomerania, Germany, between April and July 2022. Viruses 2022, 14, 2033. [Google Scholar] [CrossRef]
- Dehgani-Mobaraki, P.; Patel, Z.; Zaidi, A.K.; Giannandrea, D.; Hopkins, C. The Omicron variant of SARS-CoV-2 and its effect on the olfactory system. Int. Forum Allergy Rhinol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.; Kelly, M.; Sloan-Gardner, T.S.; Voo, T.V.; Kirk, M.D. Severity and Symptom Characteristics between Omicron and Delta SARS-CoV-2 Variant Infections in the Australian Capital Territory: A Cross-Sectional Study. Res. Sq. 2022. [Google Scholar] [CrossRef]
- DeWitt, M.E.; Tjaden, A.H.; Herrington, D.; Schieffelin, J.; Gibbs, M.; Weintraub, W.S.; Sanders, J.W.; Edelstein, S.L.; COVID-19 Community Research Partnership. COVID-19 Symptoms by Variant Period in the North Carolina COVID-19 Community Research Partnership, North Carolina, USA. Emerg. Infect. Dis. 2023, 29, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Lee, B.; Choi, Y.Y.; Um, J.; Lee, K.S.; Sung, H.K.; Kim, Y.; Park, J.S.; Lee, M.; Jang, H.C.; et al. Clinical Characteristics of 40 Patients Infected with the SARS-CoV-2 Omicron Variant in Korea. J. Korean Med. Sci. 2022, 37, e31. [Google Scholar] [CrossRef] [PubMed]
- Tham, S.M.; Fong, S.-W.; Chang, Z.-W.; Tan, K.S.; Rouers, A.; Goh, Y.S.; Tay, D.J.W.; Ong, S.W.X.; Hao, Y.; Chua, S.L.; et al. Comparison of the clinical features, viral shedding and immune response in vaccine breakthrough infection by the Omicron and Delta variants. SSRN 2022. Available online: https://ssrn.com/abstract=4142078 (accessed on 24 June 2022). [CrossRef]
- Lee, H.Y.; Lee, J.J.; Park, H.; Yu, M.; Kim, J.M.; Lee, S.-E.; Park, Y.-J.; Kim, M.; Kim, S.; Yoo, H.; et al. Importation and community transmission of SARS-CoV-2 B.1.1.529 (Omicron) variant of concern, the Republic of Korea, December 2021. Public Health Wkly. Rep. (PHWR) 2022, 14, 338–343. [Google Scholar]
- Ren, Y.; Shi, L.; Xie, Y.; Wang, C.; Zhang, W.; Wang, F.; Sun, H.; Huang, L.; Wu, Y.; Xing, Z.; et al. Course and clinical severity of the SARS-CoV-2 Omicron variant infection in Tianjin, China. MedRxiv 2022. [Google Scholar] [CrossRef]
- Sohn, Y.J.; Shin, P.J.; Oh, W.S.; Kim, E.; Kim, Y.; Kim, Y.K. Clinical Characteristics of Patients Who Contracted the SARS-CoV-2 Omicron Variant from an Outbreak in a Single Hospital. Yonsei Med. J. 2022, 63, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Mao, X.; Kuang, M.; Zhi, J.; Zhang, Z.; Bo, M.; Zhang, G.; Lin, P.; Wang, W.; Shen, Z. Interleukin-6 affects the severity of olfactory disorder: A cross-sectional survey of 148 patients who recovered from Omicron infection using the Sniffin’ Sticks test in Tianjin, China. Int. J. Infect. Dis. 2022, 123, 17–24. [Google Scholar] [CrossRef]
- Ao, Y.; Li, J.; Wei, Z.; Wang, Z.; Tian, H.; Qiu, Y.; Fu, X.; Ma, W.; Li, L.; Zeng, M.; et al. Clinical and virological characteristics of SARS-CoV-2 Omicron BA.2.2 variant outbreaks during April to May, 2022, Shanghai, China. J. Infect. 2022, 85, 573–607. [Google Scholar] [CrossRef]
- Yang, W.; Yang, S.; Wang, L.; Zhou, Y.; Xin, Y.; Li, H.; Mu, W.; Wu, Q.; Xu, L.; Zhao, M.; et al. Clinical characteristics of 310 SARS-CoV-2 Omicron variant patients and comparison with Delta and Beta variant patients in China. Virol. Sin. 2022, 37, 704–715. [Google Scholar] [CrossRef]
- Zee, S.T.; Kwok, L.F.; Kee, K.M.; Fung, L.H.; Luk, W.P.; Chan, T.L.; Leung, C.P.; Yu, P.W.; Hung, J.; SzeTo, K.Y.; et al. Impact of COVID-19 Vaccination on Healthcare Worker Infection Rate and Outcome during SARS-CoV-2 Omicron Variant Outbreak in Hong Kong. Vaccines 2022, 10, 1322. [Google Scholar] [CrossRef]
- Huang, R.C.; Chiu, C.H.; Shang, H.S.; Perng, C.L.; Chiang, T.T.; Tsai, C.C.; Wang, C.H. Clinical characteristics analysis of COVID-19 patients from the first significant community outbreak by SARS-CoV-2 variant B.1.1.7 in Taiwan as experienced from a single northern medical center. J. Microbiol. Immunol. Infect. 2022, 55, 1036–1043. [Google Scholar] [CrossRef]
- Shoji, K.; Tsuzuki, S.; Akiyama, T.; Matsunaga, N.; Asai, Y.; Suzuki, S.; Iwamoto, N.; Funaki, T.; Yamada, M.; Ozawa, N.; et al. Comparison of clinical characteristics of COVID-19 in pregnant women between the Delta and Omicron variants of concern predominant periods. J. Infect. Chemother. 2023, 29, 33–38. [Google Scholar] [CrossRef]
- Li, H.; Zhu, M.; Zhang, P.; Yan, X.; Niu, J.; Wang, Z.; Cao, J. Milder symptoms and shorter course in patients with re-positive COVID-19: A cohort of 180 patients from Northeast China. Front. Microbiol. 2022, 13, 989879. [Google Scholar] [CrossRef]
- Li, Q.; Liu, X.; Li, L.; Hu, X.; Cui, G.; Sun, R.; Zhang, D.; Li, J.; Li, Y.; Zhang, Y.; et al. Comparison of clinical characteristics between SARS-CoV-2 Omicron variant and Delta variant infections in China. Front. Med. 2022, 9, 944909. [Google Scholar] [CrossRef]
- Shen, J.; Wu, L.; Wang, P.; Shen, X.; Jiang, Y.; Liu, J.; Chen, W. Clinical characteristics and short-term recovery of hyposmia in hospitalized non-severe COVID-19 patients with Omicron variant in Shanghai, China. Front. Med. 2022, 9, 1038938. [Google Scholar] [CrossRef]
- Shen, X.; Wang, P.; Shen, J.; Jiang, Y.; Wu, L.; Nie, X.; Liu, J.; Chen, W. Neurological Manifestations of hospitalized patients with mild to moderate infection with SARS-CoV-2 Omicron variant in Shanghai, China. J. Infect. Public Health 2022, 16, 155–162. [Google Scholar] [CrossRef]
- Haruta, M.; Otsubo, S.; Otsubo, Y. Characteristics of the 6th Japanese wave of COVID-19 in hemodialysis patients. Ren. Replace. Ther. 2022, 8, 61. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, W.; Ye, X.; Zhou, Y.; Zheng, Y.; Weng, Z.; Xie, J.; Zheng, K.; Su, Z.; Zhuang, X.; et al. Clinical characteristics of patients infected with novel coronavirus wild strain, Delta variant strain and Omicron variant strain in Quanzhou: A real world study. Exp. Ther. Med. 2022, 25, 62. [Google Scholar] [CrossRef]
- Sheng, W.H.; Chang, H.C.; Chang, S.Y.; Hsieh, M.J.; Chen, Y.C.; Wu, Y.Y.; Pan, S.C.; Wang, J.T.; Chen, Y.C. SARS-CoV-2 infection among healthcare workers whom already received booster vaccination during epidemic outbreak of omicron variant in Taiwan. J. Formos. Med. Assoc. 2022, S0929-6646(22)00442-9. [Google Scholar] [CrossRef]
- Debroy, S. Omicron-Hit Had Sore Throat, Body Ache, Didn’t Lose Smell. Times of India, 17 December 2021. Available online: https://timesofindia.indiatimes.com/city/mumbai/mumbai-omicron-hit-had-sore-throat-body-ache-didnt-lose-smell/articleshow/88327372.cms (accessed on 30 November 2022).
- Thirunahari, P.S.; Moluguri, A.; Rajamouli, J.; Patruni, M.; Gurnule, S.R. Alarming symptoms in COVID-19 omicron variant, Karimnagar, Telangana. Rev. Geintec 2022, 12, 73–80. Available online: https://revistageintec.net/wp-content/uploads/2022/09/REVISTA-GEINTEC-6001.pdf (accessed on 10 January 2023).
- Malhotra, S.; Mani, K.; Lodha, R.; Bakhshi, S.; Mathur, V.P.; Gupta, P.; Kedia, S.; Sankar, M.J.; Kumar, P.; Kumar, A.H.V.; et al. COVID-19 infection, and reinfection, and vaccine effectiveness against symptomatic infection among health care workers in the setting of omicron variant transmission in New Delhi, India. Lancet Reg. Health Southeast Asia 2022, 3, 100023. [Google Scholar] [CrossRef]
- Gulzar, B.; Rishi, S.; Farhana, A.; Sheikh, A.A.; Dewani, S. Clinico-Demographic characteristics of Third Wave by Omicron (B.1.1.529) variant of SARS-CoV-2 at a Tertiary Care Center, J&K India. J. Adv. Med. Dent. Sci. Res. 2022, 10, 5–9. [Google Scholar] [CrossRef]
- Takke, A.; Zarekar, M.; Muthuraman, V.; Ashar, A.; Patil, K.; Badhavkar, A.; Trivedi, J.; Khargekar, N.; Madkaikar, M.; Banerjee, A. Comparative study of clinical features and vaccination status in Omicron and non-Omicron infected patients during the third wave in Mumbai, India. J. Fam. Med. Prim. Care 2022, 11, 6135–6142. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Landt, O.; Yeasmin, M.; Sharif, M.; Ratul, R.H.; Molla, M.A.; Nafisa, T.; Mosaddeque, M.B.; Hosen, N.; Bulbul, M.R.H.; et al. Clinical Presentation of COVID-19 and Antibody Responses in Bangladeshi Patients Infected with the Delta or Omicron Variants of SARS-CoV-2. Vaccines 2022, 10, 1959. [Google Scholar] [CrossRef]
- Mohanty, M.; Mishra, B.; Singh, A.K.; Mohapatra, P.R.; Gupta, K.; Patro, B.K.; Sahu, D.P.; Kar, P.; Purushotham, P.; Saha, S.; et al. Comparison of Clinical Presentation and Vaccine Effectiveness Among Omicron and Non-omicron SARS Coronavirus-2 Patients. Cureus 2022, 14, e32354. [Google Scholar] [CrossRef]
- Karyakarte, R.; Das, R.; Dudhate, S.; Agarasen, J.; Pillai, P.; Chandankhede, P.; Labshetwar, R.; Gadiyal, Y.; Rajmane, M.; Kulkarni, P.; et al. Clinical Characteristics and Outcomes of Laboratory-Confirmed SARS-CoV-2 Cases Infected with Omicron subvariants and XBB recombinant variant. MedRxiv 2023. [Google Scholar] [CrossRef]
- Sgorlon-Oliveira, G.; Queiroz, J.A.D.S.; Gasparelo, N.W.F.; Roca, T.P.; Passos-Silva, A.M.; Teixeira, K.S.; Oliveira, A.A.D.S.; Souza, P.R.F.D.; Silva, E.D.S.; Silva, C.C.D.; et al. The SARS-CoV-2 Omicron Variant of Concern and its Rapid Spread throughout the Western Brazilian Amazon. Preprints 2022, 2022040266. [Google Scholar] [CrossRef]
- Marquez, C.; Kerkhoff, A.D.; Schrom, J.; Rojas, S.; Black, D.; Mitchell, A.; Wang, C.Y.; Pilarowski, G.; Ribeiro, S.; Jones, D.; et al. COVID-19 Symptoms and Duration of Rapid Antigen Test Positivity at a Community Testing and Surveillance Site During Pre-Delta, Delta, and Omicron BA.1 Periods. JAMA Netw. Open 2022, 5, e2235844. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.C.; Rossi, Á.D.; Galliez, R.M.; Faffe, D.S.; Tanuri, A.; Castiñeiras, T.M.P.P. Olfactory Dysfunction in Patients with Mild COVID-19 During Gamma, Delta, and Omicron Waves in Rio de Janeiro, Brazil. JAMA 2022, 328, 582–583. [Google Scholar] [CrossRef] [PubMed]
- Mella-Torres, A.; Escobar, A.; Barrera-Avalos, C.; Vargas-Salas, S.; Pirazzoli, M.; Gonzalez, U.; Valdes, D.; Rojas, P.; Luraschi, R.; Vallejos-Vidal, E.; et al. Epidemiological characteristics of Omicron and Delta SARS-CoV-2 variant infection in Santiago, Chile. Front. Public Health 2022, 10, 984433. [Google Scholar] [CrossRef] [PubMed]
- Thornycroft, P.; Brown, W. South African doctor who raised alarm about omicron variant says symptoms are ‘unusual but mild’. Telegraph, 27 November 2021. Available online: https://www.telegraph.co.uk/global-health/science-and-disease/south-african-doctor-raised-alarm-omicron-variant-says-symptoms/ (accessed on 30 November 2022).
- Rashid, N. Sars-CoV-2 B.1.1.529 (Omicron) variant outbreak: Case series presentations and response to treatment at the Islamic University in Uganda health facility. ScienceRise 2022, 1, 36–40. [Google Scholar] [CrossRef]
- Chibwana, M.G.; Thole, H.W.; Anscombe, C.; Ashton, P.M.; Green, E.; Barnes, K.G.; Cornick, J.; Turner, A.; Witte, D.; Nthala, S.; et al. Differential symptoms among COVID-19 outpatients before and during periods of SARS-CoV-2 Omicron variant dominance in Blantyre, Malawi: A prospective observational study. MedRxiv 2022. [Google Scholar] [CrossRef]
- Mndala, L.; Monk, E.J.M.; Phiri, D.; Riches, J.; Makuluni, R.; Gadama, L.; Kachale, F.; Bilesi, R.; Mbewe, M.; Likaka, A.; et al. Comparison of maternal and neonatal outcomes of COVID-19 before and after SARS-CoV-2 omicron emergence in maternity facilities in Malawi (MATSurvey): Data from a national maternal surveillance platform. Lancet Glob. Health 2022, 10, e1623–e1631. [Google Scholar] [CrossRef] [PubMed]
- Moolla, M.S.; Maponga, T.; Moolla, H.; Kollenberg, E.; Anie, S.; Moolla, A.; Moodley, D.; Lalla, U.; Allwood, B.W.; Schrueder, N.; et al. A tale of two waves: Characteristics and outcomes of COVID-19 admissions during the omicron-driven 4th wave in Cape Town, South Africa and implications for the future. IJID Reg. 2022, 6, 42–47. [Google Scholar] [CrossRef]
- Hajjo, R.; AbuAlSamen, M.M.; Alzoubi, H.M.; Alqutob, R. The Epidemiology of Hundreds of Individuals Infected with Omicron BA.1 in Middle Eastern Jordan. MedRxiv 2022. [Google Scholar] [CrossRef]
- Akavian, I.; Nitzan, I.; Talmy, T.; Nitecki, M.; Gendler, S.; Besor, O. SARS-CoV-2 Omicron Variant: Clinical Presentation and Occupational Implications in Young and Healthy IDF Soldiers. Mil. Med. 2022, 3, usac263. [Google Scholar] [CrossRef] [PubMed]
- Kirca, F.; Aydoğan, S.; Gözalan, A.; Kayipmaz, A.E.; Özdemir, F.A.E.; Tekçe, Y.T.; Beşer, İ.O.; Gün, P.; Ökten, R.S.; Dinç, B. Comparison of clinical characteristics of wild-type SARS-CoV-2 and Omicron. Rev. Assoc. Med. Bras. 2022, 68, 1476–1480. [Google Scholar] [CrossRef] [PubMed]
- Von Bartheld, C.S.; Hagen, M.M.; Butowt, R. Prevalence of Chemosensory Dysfunction in COVID-19 Patients: A Systematic Review and Meta-analysis Reveals Significant Ethnic Differences. ACS Chem. Neurosci. 2020, 11, 2944–2961. [Google Scholar] [CrossRef]
- Mutiawati, E.; Fahriani, M.; Mamada, S.S.; Fajar, J.K.; Frediansyah, A.; Maliga, H.A.; Ilmawan, M.; Emran, T.B.; Ophinni, Y.; Ichsan, I.; et al. Anosmia and dysgeusia in SARS-CoV-2 infection: Incidence and effects on COVID-19 severity and mortality, and the possible pathobiology mechanisms—A systematic review and meta-analysis. F1000Research 2021, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef]
- Butowt, R.; Bilinska, K.; von Bartheld, C.S. Olfactory dysfunction in COVID-19: New insights into the underlying mechanisms. Trends Neurosci. 2023, 46, 75–90. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell. Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Bilinska, K.; Jakubowska, P.; von Bartheld, C.S.; Butowt, R. Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age. ACS Chem. Neurosci. 2020, 11, 1555–1562. [Google Scholar] [CrossRef]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; Van den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.J.; Fletcher, R.B.; et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020, 6, eabc5801. [Google Scholar] [CrossRef] [PubMed]
- Karimian, A.; Behjati, M.; Karimian, M. Molecular mechanisms involved in anosmia induced by SARS-CoV-2, with a focus on the transmembrane serine protease TMPRSS2. Arch. Virol. 2022, 167, 1931–1946. [Google Scholar] [CrossRef]
- Majdoul, S.; Compton, A.A. Lessons in self-defence: Inhibition of virus entry by intrinsic immunity. Nat. Rev. Immunol. 2022, 22, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Zazhytska, M.; Kodra, A.; Hoagland, D.A.; Frere, J.; Fullard, J.F.; Shayya, H.; McArthur, N.G.; Moeller, R.; Uhl, S.; Omer, A.D.; et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell 2022, 185, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Armando, F.; Beythien, G.; Kaiser, F.K.; Allnoch, L.; Heydemann, L.; Rosiak, M.; Becker, S.; Gonzalez-Hernandez, M.; Lamers, M.M.; Haagmans, B.L.; et al. SARS-CoV-2 Omicron variant causes mild pathology in the upper and lower respiratory tract of hamsters. Nat. Commun. 2022, 13, 3519. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, S.; Heydel, J.M.; Amossé, V.; Gradinaru, D.; Cattarelli, M.; Artur, Y.; Goudonnet, H.; Magdalou, J.; Netter, P.; Pelczar, H.; et al. Glucuronidation of odorant molecules in the rat olfactory system: Activity, expression and age-linked modifications of UDP-glucuronosyltransferase isoforms, UGT1A6 and UGT2A1, and relation to mitral cell activity. Brain Res. Mol. Brain Res. 2002, 107, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.B.; Klaassen, C.D. Tissue- and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab. Dispos. 2007, 35, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gözen, E.D.; Aliyeva, C.; Tevetoğlu, F.; Karaali, R.; Balkan, İ.İ.; Yener, H.M.; Özdoğan, H.A. Evaluation of Olfactory Function with Objective Tests in COVID-19-Positive Patients: A Cross-Sectional Study. Ear Nose Throat J. 2021, 100, 169S–173S. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.A.; Silva, M.T.T.; Oliveira, R.V.; Soares, C.N.; Takano, C.L.; Azevedo, A.E.; Moraes, R.L.; Rezende, R.B.; Chagas, I.T.; Espíndola, O.; et al. Smell dysfunction in COVID-19 patients: More than a yes-no question. J. Neurol. Sci. 2020, 418, 117107. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.M.; Larsen, K.D.; Homøe, A.S.; Simonsen, A.L.; Arndal, E.; Koch, A.; Samuelsen, G.B.; Nielsen, X.C.; Todsen, T.; Homøe, P. Subjective and psychophysical olfactory and gustatory dysfunction among COVID-19 outpatients; short- and long-term results. PLoS ONE 2022, 17, e0275518. [Google Scholar] [CrossRef]
- Kaya, A.; Altıparmak, S.; Yaşar, M.; Özcan, İ.; Çelik, İ. Objective Evaluation of Smell and Taste Senses in COVID-19 Patients. Turk. Arch. Otorhinolaryngol. 2022, 60, 128–133. [Google Scholar] [CrossRef]
- Lechien, J.R.; Cabaraux, P.; Chiesa-Estomba, C.M.; Khalife, M.; Hans, S.; Calvo-Henriquez, C.; Martiny, D.; Journe, F.; Sowerby, L.; Saussez, S. Objective olfactory evaluation of self-reported loss of smell in a case series of 86 COVID-19 patients. Head Neck 2020, 42, 1583–1590. [Google Scholar] [CrossRef]
- Lechien, J.R.; Chiesa-Estomba, C.M.; Hans, S.; Barillari, M.R.; Jouffe, L.; Saussez, S. Loss of Smell and Taste in 2013 European Patients with Mild to Moderate COVID-19. Ann. Intern. Med. 2020, 173, 672–675. [Google Scholar] [CrossRef]
- Romero-Gameros, C.A.; Waizel-Haiat, S.; Mendoza-Zubieta, V.; Anaya-Dyck, A.; López-Moreno, M.A.; Colin-Martinez, T.; Martínez-Ordaz, J.L.; Ferat-Osorio, E.; Vivar-Acevedo, E.; Vargas-Ortega, G.; et al. Evaluation of predictive value of olfactory dysfunction, as a screening tool for COVID-19. Laryngoscope Investig. Otolaryngol. 2020, 5, 983–991. [Google Scholar] [CrossRef]
- Hintschich, C.A.; Wenzel, J.J.; Hummel, T.; Hankir, M.K.; Kühnel, T.; Vielsmeier, V.; Bohr, C. Psychophysical tests reveal impaired olfaction but preserved gustation in COVID-19 patients. Int. Forum Allergy Rhinol. 2020, 10, 1105–1107. [Google Scholar] [CrossRef] [PubMed]
- Le Bon, S.D.; Pisarski, N.; Verbeke, J.; Prunier, L.; Cavelier, G.; Thill, M.P.; Rodriguez, A.; Dequanter, D.; Lechien, J.R.; Le Bon, O.; et al. Psychophysical evaluation of chemosensory functions 5 weeks after olfactory loss due to COVID-19: A prospective cohort study on 72 patients. Eur. Arch Otorhinolaryngol. 2021, 278, 101–108. [Google Scholar] [CrossRef]
- Hummel, T.; Whitcroft, K.L.; Andrews, P.; Altundag, A.; Cinghi, C.; Costanzo, R.M.; Damm, M.; Frasnelli, J.; Gudziol, H.; Gupta, N.; et al. Position paper on olfactory dysfunction. Rhinology 2017, 54 (Suppl. S25), 7–35. [Google Scholar] [CrossRef] [Green Version]
- Fahmy, M.; Whitcroft, K. Psychophysical Testing in Chemosensory Disorders. Curr Otorhinolaryngol. Rep. 2022, 10, 393–404. [Google Scholar] [CrossRef]
- Killingley, B.; Mann, A.J.; Kalinova, M.; Boyers, A.; Goonawardane, N.; Zhou, J.; Lindsell, K.; Hare, S.S.; Brown, J.; Frise, R.; et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat. Med. 2022, 28, 1031–1041. [Google Scholar] [CrossRef]
- Mariño-Sánchez, F.; Santamaría-Gadea, A.; de Los Santos, G.; Alobid, I.; Mullol, J. Psychophysical olfactory testing in COVID-19: Is smell function really impaired in nearly all patients? Int. Forum Allergy Rhinol. 2020, 10, 951–952. [Google Scholar] [CrossRef]
- Desiato, V.M.; Levy, D.A.; Byun, Y.J.; Nguyen, S.A.; Soler, Z.M.; Schlosser, R.J. The Prevalence of Olfactory Dysfunction in the General Population: A Systematic Review and Meta-analysis. Am. J. Rhinol. Allergy 2021, 35, 195–205. [Google Scholar] [CrossRef]
- Agyeman, A.A.; Chin, K.L.; Landersdorfer, C.B.; Liew, D.; Ofori-Asenso, R. Smell and Taste Dysfunction in Patients With COVID-19: A Systematic Review and Meta-analysis. Mayo Clin. Proc. 2020, 95, 1621–1631. [Google Scholar] [CrossRef]
- Wang, X.; Chang, H.; Tian, H.; Zhu, Y.; Li, J.; Wei, Z.; Wang, Y.; Xia, A.; Ge, Y.; Liu, G.; et al. Epidemiological and clinical features of SARS-CoV-2 infection in children during the outbreak of Omicron variant in Shanghai, March 7–31, 2022. Influenza Other Respir. Viruses 2022, 16, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.R.; Harrison, C.J.; Myers, A.L.; Jackson, M.A.; Selvarangan, R. Differences in pediatric SARS-CoV-2 symptomology and Co-infection rates among COVID-19 Pandemic waves. J. Clin. Virol. 2022, 154, 105220. [Google Scholar] [CrossRef] [PubMed]
- Shoji, K.; Akiyama, T.; Tsuzuki, S.; Matsunaga, N.; Asai, Y.; Suzuki, S.; Iwamoto, N.; Funaki, T.; Ohmagari, N. Clinical characteristics of COVID-19 in hospitalized children during the Omicron variant predominant period. J. Infect. Chemother. 2022, 28, 1531–1535. [Google Scholar] [CrossRef] [PubMed]
- Stopyra, L.; Kowalik, A.; Stala, J.; Majchrzak, I.; Szebla, J.; Jakosz, M.; Grzywaczewska, K.; Kwinta, P. Characteristics of Hospitalized Pediatric Patients in the First Five Waves of the COVID-19 Pandemic in a Single Center in Poland—1407 Cases. J. Clin. Med. 2022, 11, 6806. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Ruebsteck, E.; Dewald, F.; Klein, F.; Lehmann, C.; Huenseler, C.; Weber, L.T. Clinical Aspects of the Subsequent SARS-CoV-2 Waves in Children from 2020 to 2022-Data from a Local Cohort in Cologne, Germany (n = 21,635). Viruses 2022, 14, 1607. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.; Mahon, A.; Moss, A.; Rao, S. SARS-CoV-2 infection in children evaluated in an ambulatory setting during Delta and Omicron time periods. J. Med. Virol. 2023, 95, e28318. [Google Scholar] [CrossRef]
- Pieniak, M.; Oleszkiewicz, A.; Avaro, V.; Calegari, F.; Hummel, T. Olfactory training—Thirteen years of research reviewed. Neurosci. Biobehav. Rev. 2022, 141, 104853. [Google Scholar] [CrossRef] [PubMed]
- Lechien, J.R.; Chiesa-Estomba, C.M.; Radulesco, T.; Michel, J.; Vaira, L.A.; Le Bon, S.D.; Horoi, M.; Falanga, C.; Barillari, M.R.; Hans, S.; et al. Clinical features of patients who had two COVID-19 episodes: A European multicentre case series. J. Intern. Med. 2021, 290, 421–429. [Google Scholar] [CrossRef]
- Lechien, J.R.; Chiesa-Estomba, C.M.; Vaira, L.A.; Saussez, S.; Hans, S. COVID-19 Reinfection and Second Episodes of Olfactory and Gustatory Dysfunctions: Report of First Cases. Ear Nose Throat J. 2022, 101, 499–500. [Google Scholar] [CrossRef] [PubMed]
- Boulware, D.R.; Murray, T.A.; Proper, J.L.; Tignanelli, C.J.; Buse, J.B.; Liebovitz, D.M.; Nicklas, J.M.; Cohen, K.; Puskarich, M.A.; Belani, H.K.; et al. Impact of SARS-CoV-2 vaccination and booster on COVID-19 symptom severity over time in the COVID-OUT trial. Clin. Infect. Dis. 2022, 17, ciac772, Epub ahead of print. [Google Scholar] [CrossRef]
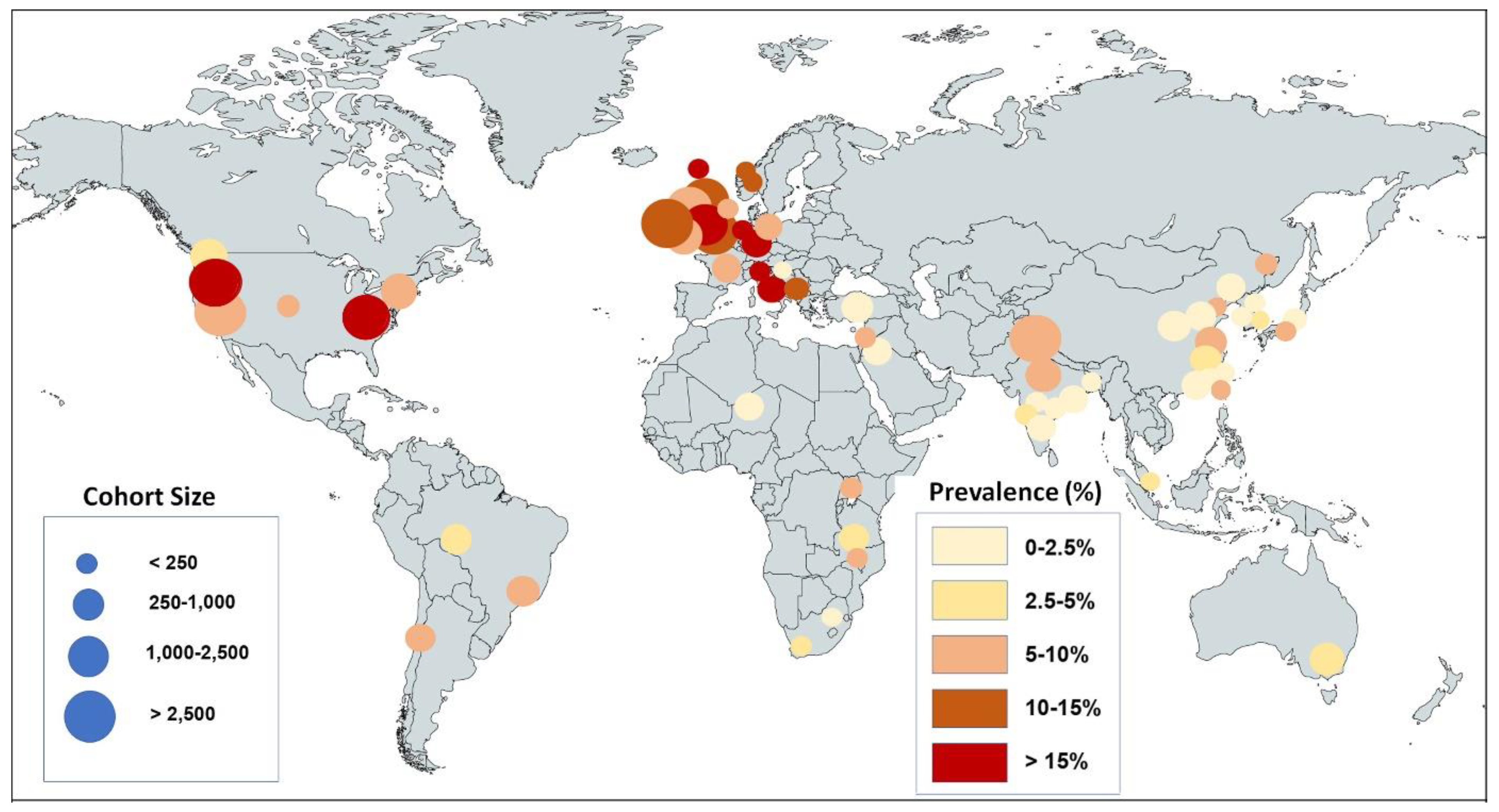



| Date First Published | Ref # | Author and First Publication Date | Cohort Country or Region | Cohort Size | Cases with OD | OD % | Quality Scores |
|---|---|---|---|---|---|---|---|
| 27 November 2021 | 91 | Thornycroft | South Africa | 24 | 0 | 0.0% | L |
| 16 December 2021 | 38 | Brandal | Norway | 81 | 12 | 14.8% | M |
| 17 December 2021 | 39 | CDC | USA | 43 | 3 | 7% | M |
| 17 December 2021 | 79 | Debroy | India | 32 | 0 | 0.0% | L |
| 31 December 2021 | 40 | Helmsdal | Denmark | 21 | 4 | 19% | L |
| 14 January 2022 | 41 | UKHSA | UK | 182,133 | 23,677 | 13% | M |
| 17 January 2022 | 61 | Kim | Korea | 40 | 1 | 2.5% | M |
| 18 January 2022 | 42 | Vihta | UK | 69,372 | 9018 | 13% | M |
| 25 January 2022 | 96 | Hajjo | Jordan | 500 | 6 | 1.2% | M |
| 27 January 2022 | 43 | Soraas | Norway | 52 | 8 | 15% | M |
| 27 January 2022 | 62 | Young/Tham | Singapore | 87 | 3 | 3.44% | M |
| January 2022 | 80 | Thirunahari | Telangana, India | 60 | 0 | 0% | H |
| 10 February 2022 | 63 | Lee | Korea | 123 | 1 | 0.8% | M |
| 12 February 2022 | 44 | Maisa | France | 468 | 23 | 4.9% | H |
| 18 February 2022 | 9 | Boscolo-Rizzo | Italy | 338 | 83 | 24.6% | H |
| 28 February 2022 | 92 | Rashid | Uganda, Africa | 14 | 1 | 7.1% | H |
| 2 April 2022 | 45 | Kramaric | Slovenia | 18 | 0 | 0.0% | M |
| 6 April 2022 | 46 | Menni | UK | 4990 | 833 | 16.7% | H |
| 13 April 2022 | 47 | Washington State | WA, USA | 2830 | 453 | 16% | L |
| 28 April 2022 | 87 | Sgorlon-Oliveira | Rondonia, Brazil | 343 | 9 | 2.6% | M |
| 28 April 2022 | 48 | Weil | WA, USA | 1730 | 48 | 2.8% | H |
| 4 May 2022 | 49 | Laracy | NY, USA | 1520 | 95 | 6.3% | M |
| 23 May 2022 | 88 | Marquez | CA, USA | 3032 | 160 | 5.3% | H |
| 23 May 2022 | 50 | Whitaker | UK | 6395 | 563 | 8.8% | H |
| 6 June 2022 | 81 | Malhotra | New Delhi, India | 1461 | 78 | 5.3% | H |
| 13 June 2022 | 51 | Laura | Bosnia | 141 | 20 | 14.2% | H |
| 16 June 2022 | 64 | Ren | Tianjin, China | 307 | 2 | 0.7% | L |
| 24 June 2022 | 89 | Cardoso | Brazil | 633 | 37 | 5.8% | H |
| 25 June 2022 | 52 | Ullrich | Germany | 61 | 5 | 8.2% | M |
| June 2022 | 82 | Gulzar | Kashmir, India | 11,715 | 1084 | 9.3% | M |
| 1 July 2022 | 30 | Schulze | Germany | 428 | 103 | 24.1% | M |
| 10 July 2022 | 53 | Townsley | London, UK | 240 | 23 | 9.6% | M |
| 12 July 2022 | 54 | Pacchiarini | Wales, UK | 1000 | 89 | 8.9% | H |
| 17 July 2022 | 93 | Chibwana | Malawi, Africa | 328 | 9 | 2.7% | H |
| 19 July 2022 | 65 | Sohn | Korea | 181 | 3 | 1.7% | H |
| 2 August 2022 | 66 | Liang | Tianjin, China | 148 | 12 | 8.1% | M |
| 8 August 2022 | 67 | Ao | Shanghai, China | 465 | 15 | 3.2% | M |
| 11 August 2022 | 68 | Yang | Tianjin, China | 310 | 5 | 1.6% | H |
| 15 August 2022 | 69 | Zee | Hong Kong, China | 454 | 2 | 0.4% | M |
| 17 August 2022 | 55 | Ekroth | UK | 309,912 | 28,569 | 13.4% | H |
| 17 August 2022 | 70 | Huang | Taiwan, China | 224 | 0 | 0.0% | M |
| 19 August 2022 | 56 | Westerhof | Netherlands | 65 | 11 | 16.9% | M |
| 03 September 2022 | 97 | Akavian | Israel | 199 | 15 | 9.1% | M |
| 9 September 2022 | 57 | Goller | Germany | 405 | 30 | 7.4% | M |
| 11 September 2022 | 71 | Shoji | Japan | 199 | 2 | 1.0% | H |
| 14 September 2022 | 58 | Deghani-Mobaraki | Italy | 205 | 64 | 31.2% | M |
| 22 September 2022 | 94 | Mndala | Malawi, Africa | 57 | 5 | 8.8% | M |
| 11 October 2022 | 72 | Li | Jilin, China | 180 | 10 | 5.6% | H |
| 12 October 2022 | 73 | Li | Henan, China | 384 | 4 | 1.0% | M |
| 21 October 2022 | 90 | Mella-Torres | Chile | 534 | 30 | 5.6% | M |
| 31 October 2022 | 83 | Takke | Mumbai, India | 46 | 2 | 4.3% | M |
| 7 November 2022 | 74 | Shen | Shanghai, China | 349 | 22 | 6.3% | H |
| 16 November 2022 | 59 | Gomez | Australia | 452 | 13 | 3.2% | M |
| 18 November 2022 | 84 | Ghosh | Bangladesh | 90 | 0 | 0.0% | M |
| 24 November 2022 | 98 | Kirca | Turkey | 411 | 4 | 1.0% | M |
| 24 November 2022 | 95 | Moolla | South Africa | 121 | 4 | 3.0% | M |
| 2 December 2022 | 76 | Haruta | Japan | 53 | 3 | 5.7% | M |
| 9 December 2022 | 85 | Mohanty | Odisha, India | 267 | 0 | 0.0% | H |
| 9 December 2022 | 77 | Zhang | Fujian, China | 20 | 0 | 0.0% | M |
| 15 December 2022 | 60 | deWitt | NC, USA | 19,189 | 3262 | 17% | M |
| 16 December 2022 | 78 | Sheng | Taiwan, China | 61 | 4 | 6.6% | M |
| 6 January 2023 | 86 | Karyakarte | Pune, India | 494 | 3 | 0.7% | H |
| Total: 625,945 | Total: 68,545 |
| Region | Ref # | Author | Country or Region | Cohort Size | Percentage of Hyposmia | Cohort Size | Percentage of Hyposmia | Reduction Om./Prev | Variant Name |
|---|---|---|---|---|---|---|---|---|---|
| Omicron | Previous Variants | ||||||||
| Middle East | 97 | Akavian | Israel | 199 | 9.1% | 119 | 51.3% | 17.5% | G614 |
| Middle East | 98 | Kirca | Turkey | 411 | 1% | 960 | 5.8% | 17.2% | wt |
| Africa | 95 | Moolla | South Africa | 121 | 3.3% | 116 | 9.5% | 34.7% | G614 |
| Africa | 93 | Chibwana | Malawi | 328 | 2.7% | 154 | 5.8% | 46.6% | δ? |
| Africa | 94 | Mndala | Malawi | 57 | 8.8% | 128 | 10.2% | 86.3% | δ |
| East Asia | 62 | Young/ Tham | Singapore | 87 | 3.44% | 87 | 2.3% | 149.6% | δ |
| East Asia | 68 | Yang | Tianjin, China | 310 | 1.6% | 422 | 6.9% | 23.2% | β, δ |
| East Asia | 73 | Li | Jilin, China | 384 | 1% | 103 | 2% | 50.0% | δ |
| East Asia | 71 | Shoji | Japan | 199 | 1.0% | 111 | 19% | 5.3% | δ |
| East Asia | 70 | Huang | Taiwan, China | 224 | 0.0% | 141 | 4.3% | 0.0% | α |
| South Asia | 84 | Ghosh | Bangladesh | 90 | 0.0% | 40 | 10.0% | 0.0% | δ |
| South Asia | 85 | Mohanty | Odisha, India | 267 | 0.0% | 461 | 3.2% | 0.0% | δ ? |
| South Asia | 83 | Takke | Mumbai, India | 46 | 0.43% | 55 | 10.9% | 39.4% | δ |
| South Asia | 81 | Malhotra | New Delhi, India | 1461 | 5.3% | 1907 | 51.6% | 10.3% | δ |
| Hispanic | 88 | Marquez | CA, USA | 3032 | 5.3% | 1533 | 18.2% | 29.1% | δ, prev. |
| Latino | 89 | Cardoso | Brazil | 633 | 5.8% | 5420 | 48.2% | 12.0% | wt, γ, δ |
| Latino | 90 | Mella-Torres | Chile | 534 | 5.6% | 54 | 13% | 43.1% | δ |
| Western | 41 | UKHSA | UK | 182,133 | 13% | 87,920 | 34% | 38.2% | δ |
| Western | 42 | Vihta | UK | 69,372 | 13% | 14,318 | 40% | 32.5% | δ |
| Western | 43 | Soraas | Norway | 52 | 15% | 18 | 72.2% | 20.8% | δ |
| Western | 9 | Boscolo-Rizzo | Italy | 338 | 24.6% | 441 | 62.6% | 39.3% | G614 |
| Western | 46 | Menni | UK | 4990 | 16.7% | 4990 | 52.7% | 31.7% | δ |
| Western | 48 | Weil | WA, USA | 1730 | 2.8% | 209 | 11.1% | 25.2% | δ |
| Western | 49 | Laracy | NY, USA | 1520 | 6.3% | 361 | 29% | 21.7% | α, δ |
| Western | 50 | Whitaker | UK | 6395 | 8.8% | 6739 | 16.2% | 54.3% | wt, α, δ |
| Western | 30 | Schulze | Germany | 428 | 24.1% | 1497 | 66.7% | 36.1% | G614, α, δ |
| Western | 54 | Pacciarini | Wales, UK | 1000 | 8.9% | 8,168 | 25.8% | 34.5% | δ |
| Western | 55 | Ekroth | UK | 309,912 | 13.4% | 123,529 | 33.7% | 39.8% | δ |
| Western | 56 | Westerhof | Netherlands | 65 | 16.9% | 216 | 46.7% | 36.2% | G614, α |
| Western | 59 | Gomez | Australia | 452 | 3.2% | 425 | 36.9% | 8.7% | δ |
| Western | 53 | Townsley | London, UK | 240 | 9.6% | 67 | 37.3% | 25.7% | δ |
| Western | 60 | deWitt | NC, USA | 19,189 | 17% | 37,711 | 55% | 30.9% | pre δ |
| Population | Adults Only | COVID-19 -Infected Adults * | OD Pre-Valence | Adults with OD | Weight | Prevalence × Weight | |
|---|---|---|---|---|---|---|---|
| Billion | Billion | Billion | % | Million | |||
| Western | 0.9 | 0.675 | 0.6075 | 11.7 | 71.1 | 0.11 | 1.32 |
| Latino/Hispanic | 0.7 | 0.525 | 0.4725 | 4.9 | 23.2 | 0.09 | 0.43 |
| Africa | 1.4 | 1.050 | 0.9450 | 3.1 | 29.3 | 0.18 | 0.54 |
| East Asia | 2.5 | 1.875 | 1.6875 | 1.9 | 32.1 | 0.31 | 0.59 |
| South Asia | 2.0 | 1.500 | 1.3500 | 2.8 | 37.8 | 0.25 | 0.70 |
| Middle East | 0.5 | 0.375 | 0.3375 | 2.2 | 7.4 | 0.06 | 0.14 |
| Total | 8.0 | 6.00 | 5.40 | 200.9 | 1.00 | 3.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Bartheld, C.S.; Wang, L. Prevalence of Olfactory Dysfunction with the Omicron Variant of SARS-CoV-2: A Systematic Review and Meta-Analysis. Cells 2023, 12, 430. https://doi.org/10.3390/cells12030430
von Bartheld CS, Wang L. Prevalence of Olfactory Dysfunction with the Omicron Variant of SARS-CoV-2: A Systematic Review and Meta-Analysis. Cells. 2023; 12(3):430. https://doi.org/10.3390/cells12030430
Chicago/Turabian Stylevon Bartheld, Christopher S., and Lingchen Wang. 2023. "Prevalence of Olfactory Dysfunction with the Omicron Variant of SARS-CoV-2: A Systematic Review and Meta-Analysis" Cells 12, no. 3: 430. https://doi.org/10.3390/cells12030430
APA Stylevon Bartheld, C. S., & Wang, L. (2023). Prevalence of Olfactory Dysfunction with the Omicron Variant of SARS-CoV-2: A Systematic Review and Meta-Analysis. Cells, 12(3), 430. https://doi.org/10.3390/cells12030430






