RHOA Therapeutic Targeting in Hematological Cancers
Abstract
1. Introduction
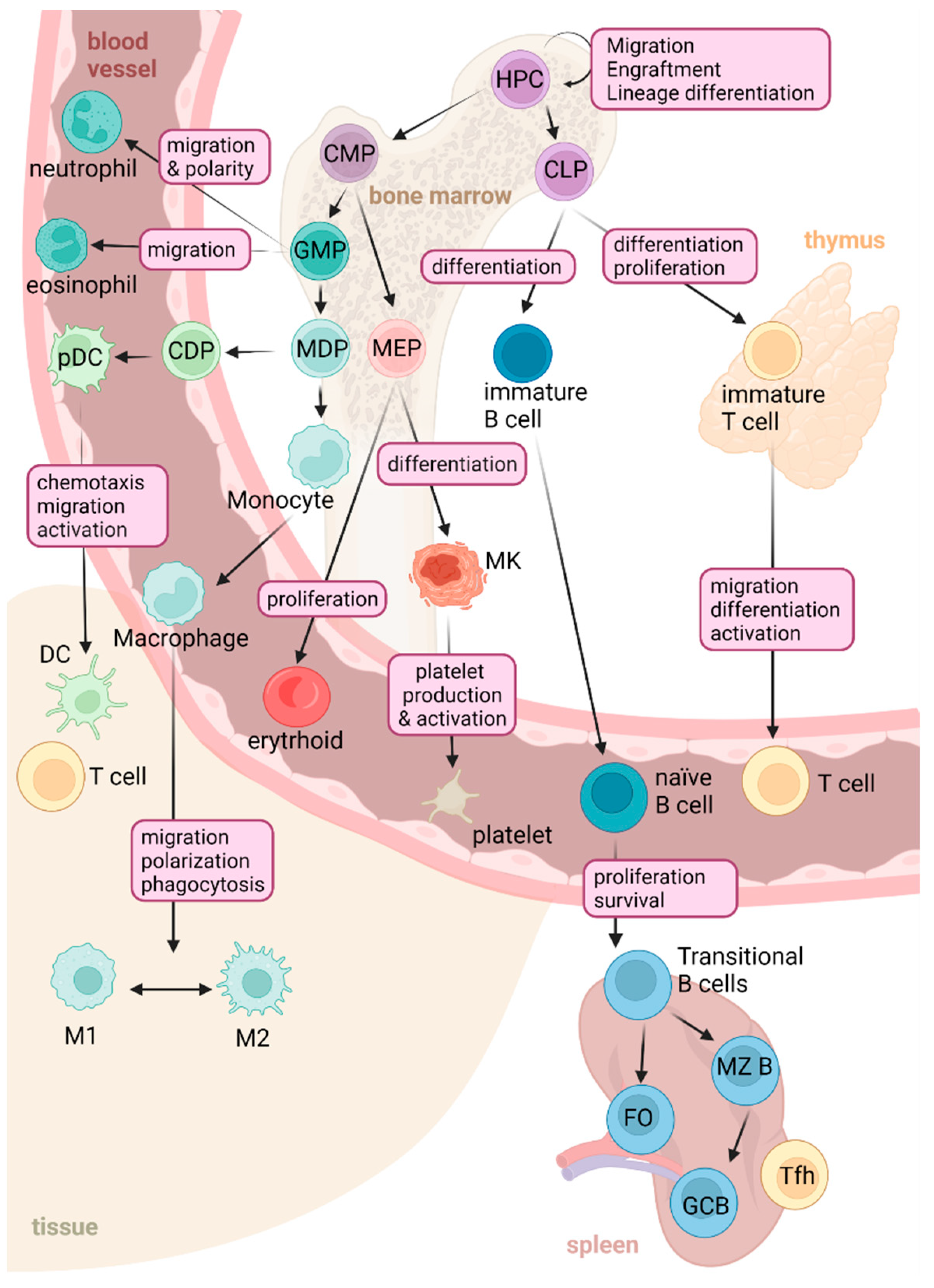
2. Physiological Functions of RHOA in the Immune System
2.1. RHOA GTPase Signaling in Myeloid Cells
2.2. RHOA GTPase in the Control of Lymphoid Lineage
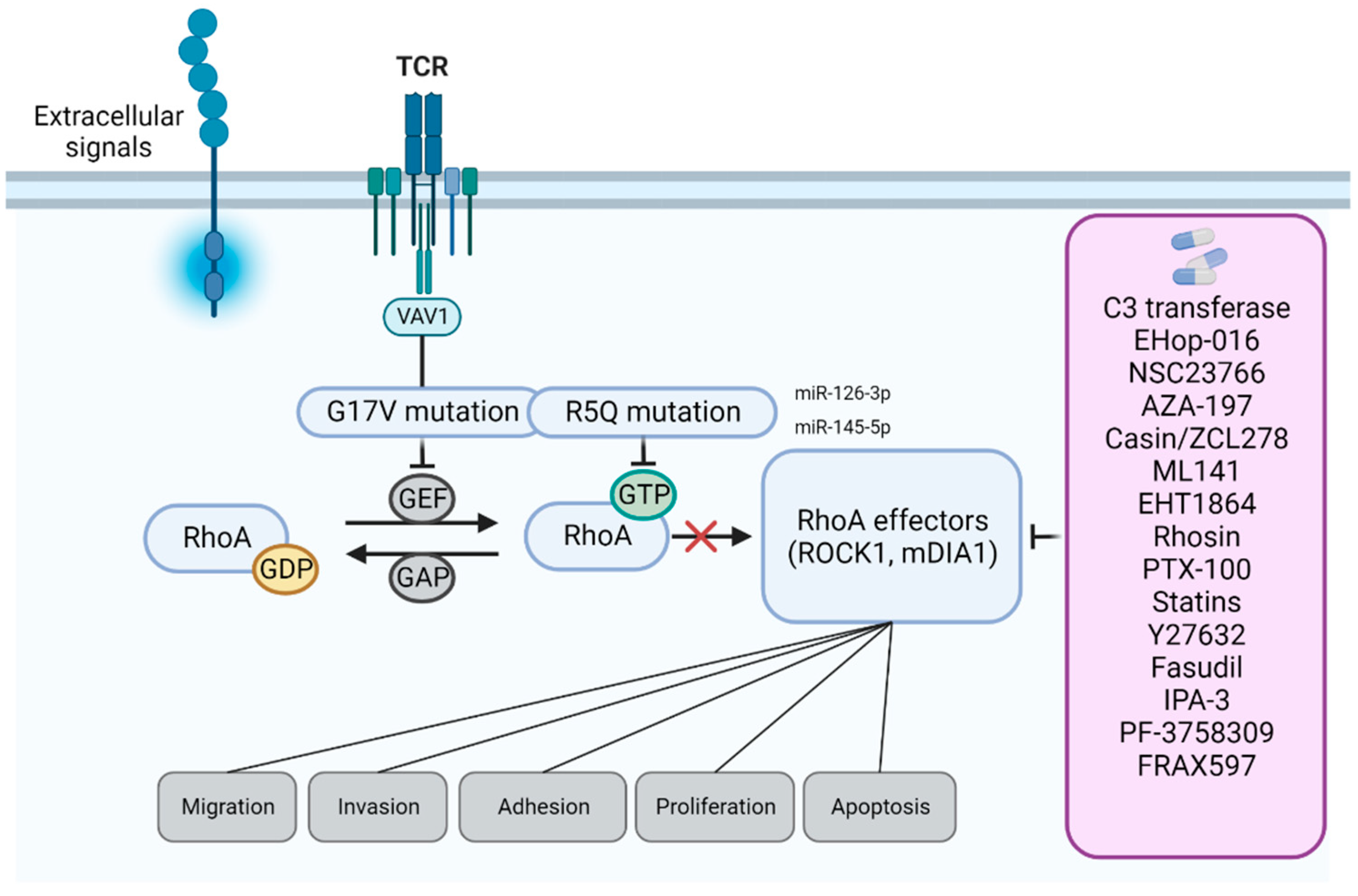
3. RhoA in Hematopoietic Tissues: Oncogene or Tumor Suppressor?
4. Bioinformatic Prediction of RHOA Therapeutic Agents
5. Approaches to Modulate RHOA Signaling in Hematological Cancers
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vega, F.M.; Ridley, A.J. Rho GTPases in cancer cell biology. FEBS Lett. 2008, 582, 2093–2101. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J. Rho GTPases and cell migration. J. Cell Sci. 2001, 114, 2713–2722. [Google Scholar] [CrossRef] [PubMed]
- Hodge, R.G.; Ridley, A.J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016, 17, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.G.; Chen, J.; Mccormick, F.; Symons, M. A role for Rho in Ras transformation. Proc. Natl. Acad. Sci. USA 1995, 92, 11781–11785. [Google Scholar] [CrossRef]
- Khosravi-Far, R.; Solski, P.A.; Clark, G.J.; Kinch, M.S.; Der, C.J. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol. Cell. Biol. 1995, 15, 6443–6453. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, R.; Pedersen, E.D.; Wang, Z.; Brakebusch, C. Rho GTPase function in tumorigenesis. Biochim. Biophys. Acta-Rev. Cancer 2009, 1796, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Murray, D.; Horgan, G.; MacMathuna, P.; Doran, P. NET1-mediated RhoA activation facilitates lysophosphatidic acid-induced cell migration and invasion in gastric cancer. Br. J. Cancer 2008, 99, 1322–1329. [Google Scholar] [CrossRef]
- Joshi, R.; Qin, L.; Cao, X.; Zhong, S.; Voss, C.; Min, W.; Li, S.S.C. DLC1 SAM domain-binding peptides inhibit cancer cell growth and migration by inactivating RhoA. J. Biol. Chem. 2020, 295, 645–656. [Google Scholar] [CrossRef]
- Kaida, T.; Nitta, H.; Kitano, Y.; Yamamura, K.; Arima, K.; Izumi, D.; Higashi, T.; Kurashige, J.; Imai, K.; Hayashi, H.; et al. C5α receptor (CD88) promotes motility and invasiveness of gastric cancer by activating RhoA. Oncotarget 2016, 7, 84798–84809. [Google Scholar] [CrossRef]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.N.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Sakata-Yanagimoto, M.; Enami, T.; Yoshida, K.; Shiraishi, Y.; Ishii, R.; Miyake, Y.; Muto, H.; Tsuyama, N.; Sato-Otsubo, A.; Okuno, Y.; et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014, 46, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.Y.; Sung, M.K.; Lee, S.H.; Kim, S.; Lee, H.; Park, S.; Kim, S.C.; Lee, B.; Rho, K.; Lee, J.E.; et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014, 46, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Kontani, K.; Enami, T.; Kataoka, K.; Ishii, R.; Totoki, Y.; Kataoka, T.R.; Hirata, M.; Aoki, K.; Nakano, K.; et al. Variegated RHOA mutations in adult T-cell leukemia/lymphoma. Blood 2016, 127, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Rohde, M.; Richter, J.; Schlesner, M.; Betts, M.J.; Claviez, A.; Bonn, B.R.; Zimmermann, M.; Damm-Welk, C.; Russell, R.B.; Borkhardt, A.; et al. Recurrent RHOA mutations in pediatric Burkitt lymphoma treated according to the NHL-BFM protocols. Genes Chromosom. Cancer 2014, 53, 911–916. [Google Scholar] [CrossRef]
- O’Hayre, M.; Inoue, A.; Kufareva, I.; Wang, Z.; Mikelis, C.M.; Drummond, R.A.; Avino, S.; Finkel, K.; Kalim, K.W.; Dipasquale, G.; et al. Inactivating mutations in GNA13 and RHOA in Burkitt’s lymphoma and diffuse large B-cell lymphoma: A tumor suppressor function for the Gα13/RhoA axis in B cells. Oncogene 2016, 35, 3771–3780. [Google Scholar] [CrossRef]
- Huang, M.; Prendergast, G.C. RhoB in cancer suppression. Histol. Histopathol. 2006, 21, 213–218. [Google Scholar] [CrossRef]
- Bhavsar, P.J.; Infante, E.; Khwaja, A.; Ridley, A.J. Analysis of Rho GTPase expression in T-ALL identifies RhoU as a target for Notch involved in T-ALL cell migration. Oncogene 2013, 32, 198–208. [Google Scholar] [CrossRef]
- Lang, S.; Busch, H.; Boerries, M.; Brummer, T.; Timme, S.; Lassmann, S.; Aktories, K.; Schmidt, G. Specific role of RhoC in tumor invasion and metastasis. Oncotarget 2017, 8, 87364–87378. [Google Scholar] [CrossRef]
- Ridley, A.J. RhoA, RhoB and RhoC have different roles in cancer cell migration. J. Microsc. 2013, 251, 242–249. [Google Scholar] [CrossRef]
- Luo, J.; Li, D.; Wei, D.; Wang, X.; Wang, L.; Zeng, X. RhoA and RhoC are involved in stromal cell-derived factor-1-induced cell migration by regulating F-actin redistribution and assembly. Mol. Cell. Biochem. 2017, 436, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Egami, Y.; Kawai, K.; Araki, N. RhoC regulates the actin remodeling required for phagosome formation during FcγR-mediated phagocytosis. J. Cell Sci. 2017, 130, 4168–4179. [Google Scholar] [CrossRef] [PubMed]
- Bros, M.; Haas, K.; Moll, L.; Grabbe, S. RhoA as a Key Regulator of Innate and Adaptive Immunity. Cells 2019, 8, 733. [Google Scholar] [CrossRef]
- Williams, D.A.; Zheng, Y.; Cancelas, J.A. Rho GTPases and Regulation of Hematopoietic Stem Cell Localization. Methods Enzymol. 2008, 439, 365–393. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, A.V.; Freund, D.; Bornhäuser, M.; Corbeil, D. Polarization and migration of hematopoietic stem and progenitor cells rely on the RhoA/ROCK I pathway and an active reorganization of the microtubule network. J. Biol. Chem. 2010, 285, 31661–31671. [Google Scholar] [CrossRef]
- Fonseca, A.V.; Corbeil, D. The hematopoietic stem cell polarization and migration: A dynamic link between rhoa signaling pathway, microtubule network and ganglioside-based membrane microdomains. Commun. Integr. Biol. 2011, 4, 201–204. [Google Scholar] [CrossRef]
- Ghiaur, G.; Lee, A.; Bailey, J.; Cancelas, J.A.; Zheng, Y.; Williams, D.A. Inhibition of RhoA GTPase activity enhances hematopoietic stem and progenitor cell proliferation and engraftment. Blood 2006, 108, 2087–2094. [Google Scholar] [CrossRef]
- Jaganathan, B.G.; Anjos-Afonso, F.; Kumar, A.; Bonnet, D. Active RHOA favors retention of human hematopoietic stem/progenitor cells in their niche. J. Biomed. Sci. 2013, 20, 1–8. [Google Scholar] [CrossRef]
- Zhou, X.; Florian, M.C.; Arumugam, P.; Chen, X.; Cancelas, J.A.; Lang, R.; Malik, P.; Geiger, H.; Zheng, Y. RhoA GTPase controls cytokinesis and programmed necrosis of hematopoietic progenitors. J. Exp. Med. 2013, 210, 2371–2385. [Google Scholar] [CrossRef]
- Nayak, R.C.; Chang, K.H.; Vaitinadin, N.S.; Cancelas, J.A. Rho GTPases control specific cytoskeleton-dependent functions of hematopoietic stem cells. Immunol. Rev. 2013, 256, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhou, X.; Zheng, Y. In vivo rescue assay of rhoa-deficient hematopoietic stem and progenitor cells. Methods Mol. Biol. 2018, 1821, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Boukharov, A.A.; Cohen, C.M. Guanine nucleotide-dependent translocation of RhoA from cytosol to high affinity membrane binding sites in human erythrocytes. Biochem. J. 1998, 330, 1391–1398. [Google Scholar] [CrossRef] [PubMed]
- Thuet, K.M.; Bowles, E.A.; Ellsworth, M.L.; Sprague, R.S.; Stephenson, A.H. The Rho kinase inhibitor Y-27632 increases erythrocyte deformability and low oxygen tension-induced ATP release. Am. J. Physiol.-Hear. Circ. Physiol. 2011, 301, 1891–1896. [Google Scholar] [CrossRef]
- Kalfa, T.A.; Pushkaran, S.; Mohandas, N.; Hartwig, J.H.; Fowler, V.M.; Johnson, J.F.; Joiner, C.H.; Williams, D.A.; Zheng, Y. Rac GTPases regulate the morphology and deformability of the erythrocyte cytoskeleton. Blood 2006, 108, 3637–3645. [Google Scholar] [CrossRef] [PubMed]
- Gabet, A.S.; Coulon, S.; Fricot, A.; Vandekerckhove, J.; Chang, Y.; Ribeil, J.A.; Lordier, L.; Zermati, Y.; Asnafi, V.; Belaid, Z.; et al. Caspase-activated ROCK-1 allows erythroblast terminal maturation independently of cytokine-induced Rho signaling. Cell Death Differ. 2011, 18, 678–689. [Google Scholar] [CrossRef]
- Kalfa, T.A.; Zheng, Y. Rho GTPases in Erythroid Maturation. Curr. Opin. Hematol. 2014, 21, 165. [Google Scholar] [CrossRef] [PubMed]
- Pleines, I.; Cherpokova, D.; Bender, M. Rho GTPases and their downstream effectors in megakaryocyte biology. Platelets 2019, 30, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.C.; Teixeira, A.M.; Chen, R.C.; Wang, L.; Gao, Y.; Hahn, K.L.; Krause, D.S. Induction of megakaryocyte differentiation drives nuclear accumulation and transcriptional function of MKL1 via actin polymerization and RhoA activation. Blood 2013, 121, 1094–1101. [Google Scholar] [CrossRef]
- Gao, Y.; Smith, E.; Ker, E.; Campbell, P.; Cheng, E.C.; Zou, S.; Lin, S.; Wang, L.; Halene, S.; Krause, D.S. Role of RhoA-Specific Guanine Exchange Factors in Regulation of Endomitosis in Megakaryocytes. Dev. Cell 2012, 22, 573–584. [Google Scholar] [CrossRef]
- Chang, Y.; Auradé, F.; Larbret, F.; Zhang, Y.; Le Couedic, J.P.; Momeux, L.; Larghero, J.; Bertoglio, J.; Louache, F.; Cramer, E.; et al. Proplatelet formation is regulated by the Rho/ROCK pathway. Blood 2007, 109, 4229–4236. [Google Scholar] [CrossRef]
- Pleines, I.; Hagedorn, I.; Gupta, S.; May, F.; Chakarova, L.; Van Hengel, J.; Offermanns, S.; Krohne, G.; Kleinschnitz, C.; Brakebusch, C.; et al. Megakaryocyte-specific RhoA deficiency causes macrothrombocytopenia and defective platelet activation in hemostasis and thrombosis. Blood 2012, 119, 1054–1063. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Shin, J.W.; Wang, Y.; Min, S.H.; Poncz, M.; Choi, J.K.; Discher, D.E.; Carpenter, C.L.; Lian, L.; Zhao, L.; et al. RhoA Is Essential for Maintaining Normal Megakaryocyte Ploidy and Platelet Generation. PLoS ONE 2013, 8, e69315. [Google Scholar] [CrossRef] [PubMed]
- Mulloy, J.C.; Cancelas, J.A.; Filippi, M.D.; Kalfa, T.A.; Guo, F.; Zheng, Y. Rho GTPases in hematopoiesis and hemopathies. Blood 2010, 115, 936–947. [Google Scholar] [CrossRef]
- Xu, J.; Wang, F.; Van Keymeulen, A.; Herzmark, P.; Straight, A.; Kelly, K.; Takuwa, Y.; Sugimoto, N.; Mitchison, T.; Bourne, H.R. Divergent Signals and Cytoskeletal Assemblies Regulate Self-Organizing Polarity in Neutrophils. Cell 2003, 114, 201–214. [Google Scholar] [CrossRef]
- Van Keymeulen, A.; Wong, K.; Knight, Z.A.; Govaerts, C.; Hahn, K.M.; Shokat, K.M.; Bourne, H.R. To stabilize neutrophil polarity, PIP3 and Cdc42 augment RhoA activity at the back as well as signals at the front. J. Cell Biol. 2006, 174, 437–445. [Google Scholar] [CrossRef]
- Kilian, L.S.; Frank, D.; Rangrez, A.Y. RhoA Signaling in Immune Cell Response and Cardiac Disease. Cells 2021, 10, 1681. [Google Scholar] [CrossRef]
- Filippi, M.D.; Szczur, K.; Harris, C.E.; Berclaz, P.Y. Rho GTPase Rac1 is critical for neutrophil migration into the lung. Blood 2007, 109, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Francis, S.A.; Shen, X.; Young, J.B.; Kaul, P.; Lerner, D.J. Rho GEF Lsc is required for normal polarization, migration, and adhesion of formyl-peptide–stimulated neutrophils. Blood 2006, 107, 1627–1635. [Google Scholar] [CrossRef]
- Hind, L.E.; Vincent, W.J.B.; Huttenlocher, A. Leading from the Back: The Role of the Uropod in Neutrophil Polarization and Migration. Dev. Cell 2016, 38, 161–169. [Google Scholar] [CrossRef]
- Alblas, J.; Ulfman, L.; Hordijk, P.; Koenderman, L. Activation of RhoA and ROCK Are Essential for Detachment of Migrating Leukocytes. Mol. Biol. Cell 2001, 12, 2137. [Google Scholar] [CrossRef]
- Li, S.; Dislich, B.; Brakebusch, C.H.; Lichtenthaler, S.F.; Brocker, T. Control of Homeostasis and Dendritic Cell Survival by the GTPase RhoA. J. Immunol. 2015, 195, 4244–4256. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kumamoto, Y.; Wang, P.; Gan, X.; Lehmann, D.; Smrcka, A.V.; Cohn, L.; Iwasaki, A.; Li, L.; Wu, D. Regulation of immature dendritic cell migration by RHoA guanine nucleotide exchange factor Arhgef5. J. Biol. Chem. 2009, 284, 28599–28606. [Google Scholar] [CrossRef]
- Swetman, C.A.; Leverrier, Y.; Garg, R.; Gan, C.H.V.; Ridley, A.J.; Katz, D.R.; Chain, B.M. Extension, retraction and contraction in the formation of a dendritic cell dendrite: Distinct roles for Rho GTPases. Eur. J. Immunol. 2002, 32, 2074–2083. [Google Scholar] [CrossRef] [PubMed]
- Shurin, G.V.; Tourkova, I.L.; Chatta, G.S.; Schmidt, G.; Wei, S.; Djeu, J.Y.; Shurin, M.R. Small Rho GTPases Regulate Antigen Presentation in Dendritic Cells. J. Immunol. 2005, 174, 3394–3400. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, W.; Minze, L.J.; Kubiak, J.Z.; Li, X.C.; Ghobrial, R.M.; Kloc, M. Dissonant response of M0/M2 and M1 bone-marrow-derived macrophages to RhoA pathway interference. Cell Tissue Res. 2016, 366, 707–720. [Google Scholar] [CrossRef]
- Chen, W.; Zhao, Y.; Li, X.C.; Kubiak, J.Z.; Ghobrial, R.M.; Kloc, M. Rho-specific Guanine nucleotide exchange factors (Rho-GEFs) inhibition affects macrophage phenotype and disrupts Golgi complex. Int. J. Biochem. Cell Biol. 2017, 93, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Königs, V.; Jennings, R.; Vogl, T.; Horsthemke, M.; Bachg, A.C.; Xu, Y.; Grobe, K.; Brakebusch, C.; Schwab, A.; Bähler, M.; et al. Mouse Macrophages completely lacking Rho subfamily GTPases (RhoA, RhoB, and RhoC) have severe lamellipodial retraction defects, but robust chemotactic navigation and altered motility. J. Biol. Chem. 2014, 289, 30772–30784. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, S.; Bae, D.J.; Park, S.Y.; Lee, G.Y.; Park, G.M.; Kim, I.S. Coordinated balance of Rac1 and RhoA plays key roles in determining phagocytic appetite. PLoS ONE 2017, 12, e0174603. [Google Scholar] [CrossRef]
- Kim, J.G.; Moon, M.Y.; Kim, H.J.; Li, Y.; Song, D.K.; Kim, J.S.; Lee, J.Y.; Kim, J.; Kim, S.C.; Park, J.B. Ras-related GTPases Rap1 and RhoA collectively induce the phagocytosis of serum-opsonized zymosan particles in macrophages. J. Biol. Chem. 2012, 287, 5145–5155. [Google Scholar] [CrossRef]
- Zhang, S.; Konstantinidis, D.G.; Yang, J.-Q.; Mizukawa, B.; Kalim, K.; Lang, R.A.; Kalfa, T.A.; Zheng, Y.; Guo, F. Gene Targeting RhoA Reveals Its Essential Role in Coordinating Mitochondrial Function and Thymocyte Development. J. Immunol. 2014, 193, 5973–5982. [Google Scholar] [CrossRef]
- Heasman, S.J.; Ridley, A.J. Multiple roles for RhoA during T cell transendothelial migration. Small GTPases 2010, 1, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Manzanares, M.; Rey, M.; Pérez-Martínez, M.; Yáñez-Mó, M.; Sancho, D.; Cabrero, J.R.; Barreiro, O.; de la Fuente, H.; Itoh, K.; Sánchez-Madrid, F. The RhoA Effector mDia Is Induced During T Cell Activation and Regulates Actin Polymerization and Cell Migration in T Lymphocytes. J. Immunol. 2003, 171, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Heasman, S.J.; Carlin, L.M.; Cox, S.; Ng, T.; Ridley, A.J. Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J. Cell Biol. 2010, 190, 553–563. [Google Scholar] [CrossRef]
- Delaguillaumie, A.; Lagaudrière-Gesbert, C.; Popoff, M.R.; Conjeaud, H. Rho GTPase link cytoskeletal rearrangements and activation processes induced via the tetraspanin CD82 in T lymphocytes. J. Cell Sci. 2002, 115, 433–443. [Google Scholar] [CrossRef]
- El Masri, R.; Delon, J. RHO GTPases: From new partners to complex immune syndromes. Nat. Rev. Immunol. 2021, 21, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Froehlich, J.; Versapuech, M.; Megrelis, L.; Largeteau, Q.; Meunier, S.; Tanchot, C.; Bismuth, G.; Delon, J.; Mangeney, M. FAM65B controls the proliferation of transformed and primary T cells. Oncotarget 2016, 7, 63215–63225. [Google Scholar] [CrossRef] [PubMed]
- Megrelis, L.; El Ghoul, E.; Moalli, F.; Versapuech, M.; Cassim, S.; Ruef, N.; Stein, J.V.; Mangeney, M.; Delon, J. Fam65b phosphorylation relieves tonic RhoA inhibition during T cell migration. Front. Immunol. 2018, 9, 2001. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, X.; Lang, R.A.; Guo, F. RhoA of the Rho Family Small GTPases Is Essential for B Lymphocyte Development. PLoS ONE 2012, 7, e33773. [Google Scholar] [CrossRef]
- Saci, A.; Carpenter, C.L. RhoA GTPase regulates B cell receptor signaling. Mol. Cell 2005, 17, 205–214. [Google Scholar] [CrossRef]
- Madaule, P.; Axel, R. A novel ras-related gene family. Cell 1985, 41, 31–40. [Google Scholar] [CrossRef]
- Kamai, T.; Yamanishi, T.; Shirataki, H.; Takagi, K.; Asami, H.; Ito, Y.; Yoshida, K.I. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin. Cancer Res. 2004, 10, 4799–4805. [Google Scholar] [CrossRef] [PubMed]
- Fritz, G.; Brachetti, C.; Bahlmann, F.; Schmidt, M.; Kaina, B. Rho GTPases in human breast tumours: Expression and mutation analyses and correlation with clinical parameters. Br. J. Cancer 2002, 87, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Fritz, G.; Just, I.; Kaina, B. Rho GTPases are over-expressed in human tumors. Int. J. Cancer 1999, 81, 682–687. [Google Scholar] [CrossRef]
- Abraham, M.T.; Kuriakose, M.A.; Sacks, P.G.; Yee, H.; Chiriboga, L.; Bearer, E.L.; Delacure, M.D. Motility-related proteins as markers for head and neck squamous cell cancer. Laryngoscope 2001, 111, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Pillé, J.Y.; Denoyelle, C.; Varet, J.; Bertrand, J.R.; Soria, J.; Opolon, P.; Lu, H.; Pritchard, L.L.; Vannier, J.P.; Malvy, C.; et al. Anti-RhoA and Anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol. Ther. 2005, 11, 267–274. [Google Scholar] [CrossRef]
- Liu, N.; Bi, F.; Pan, Y.; Sun, L.; Xue, V.; Shi, Y.; Yao, X.; Zheng, Y.; Fan, D. Reversal of the malignant phenotype of gastric cancer cells by inhibition of RhoA expression and activity. Clin. Cancer Res. 2004, 10, 6239–6247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Tang, Q.; Xu, F.; Xue, Y.; Zhen, Z.; Deng, Y.; Liu, M.; Chen, J.; Liu, S.; Qiu, M.; et al. RhoA regulates G1-S progression of gastric cancer cells by modulation of multiple INK4 family tumor suppressors. Mol. Cancer Res. 2009, 7, 570–580. [Google Scholar] [CrossRef]
- Zandvakili, I.; Davis, A.K.; Hu, G.; Zheng, Y. Loss of RhoA exacerbates, rather than dampens, oncogenic K-Ras induced lung adenoma formation in mice. PLoS ONE 2015, 10, e0127923. [Google Scholar] [CrossRef]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Kakiuchi, M.; Nishizawa, T.; Ueda, H.; Gotoh, K.; Tanaka, A.; Hayashi, A.; Yamamoto, S.; Tatsuno, K.; Katoh, H.; Watanabe, Y.; et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat. Genet. 2014, 46, 583–587. [Google Scholar] [CrossRef]
- Zhang, H.; Schaefer, A.; Wang, Y.; Hodge, R.G.; Blake, D.R.; Diehl, J.N.; Papageorge, A.G.; Stachler, M.D.; Liao, J.; Zhou, J.; et al. Gain-of-function RHOA mutations promote focal adhesion kinase activation and dependency in diffuse gastric cancer. Cancer Discov. 2020, 10, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Palomero, T.; Couronné, L.; Khiabanian, H.; Kim, M.Y.; Ambesi-Impiombato, A.; Perez-Garcia, A.; Carpenter, Z.; Abate, F.; Allegretta, M.; Haydu, J.E.; et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 2014, 46, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Manso, R.; Rojo, F.; Rodríguez-Pinilla, S.M.; Sánchez-Beato, M.; Gómez, S.; Monsalvo, S.; Llamas, P.; Cereceda, L.; Piris, M.A.; Mollejo, M.; et al. The RHOA G17V gene mutation occurs frequently in peripheral T-cell lymphoma and is associated with a characteristic molecular signature. Blood 2014, 123, 2893–2894. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.R.; Ambesi-Impiombato, A.; Couronné, L.; Quinn, S.A.; Kim, C.S.; da Silva Almeida, A.C.; West, Z.; Belver, L.; Martin, M.S.; Scourzic, L.; et al. RHOA G17V Induces T Follicular Helper Cell Specification and Promotes Lymphomagenesis. Cancer Cell 2018, 33, 259–273.e7. [Google Scholar] [CrossRef]
- Abate, F.; Da Silva-Almeida, A.C.; Zairis, S.; Robles-Valero, J.; Couronne, L.; Khiabanian, H.; Quinn, S.A.; Kim, M.Y.; Laginestra, M.A.; Kim, C.; et al. Activating mutations and translocations in the guanine exchange factor VAV1 in peripheral T-cell lymphomas. Proc. Natl. Acad. Sci. USA 2017, 114, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Vallois, D.; Dobay, M.P.D.; Morin, R.D.; Lemonnier, F.; Missiaglia, E.; Juilland, M.; Iwaszkiewicz, J.; Fataccioli, V.; Bisig, B.; Roberti, A.; et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood 2016, 128, 1490–1502. [Google Scholar] [CrossRef]
- Fujisawa, M.; Sakata-Yanagimoto, M.; Nishizawa, S.; Komori, D.; Gershon, P.; Kiryu, M.; Tanzima, S.; Fukumoto, K.; Enami, T.; Muratani, M.; et al. Activation of RHOA-VAV1 signaling in angioimmunoblastic T-cell lymphoma. Leukemia 2018, 32, 694–702. [Google Scholar] [CrossRef]
- Karvonen, H.; Perttilä, R.; Niininen, W.; Hautanen, V.; Barker, H.; Murumägi, A.; Heckman, C.A.; Ungureanu, D. Wnt5a and ROR1 activate non-canonical Wnt signaling via RhoA in TCF3-PBX1 acute lymphoblastic leukemia and highlight new treatment strategies via Bcl-2 co-targeting. Oncogene 2019, 38, 3288–3300. [Google Scholar] [CrossRef]
- Li, F.; Jiang, Q.; Shi, K.J.; Luo, H.; Yang, Y.; Xu, C.M. RhoA modulates functional and physical interaction between ROCK1 and Erk1/2 in selenite-induced apoptosis of leukaemia cells. Cell Death Dis. 2013, 4, e708. [Google Scholar] [CrossRef]
- Cleverley, S.C.; Costello, P.S.; Henning, S.W.; Cantrell, D.A. Loss of Rho function in the thymus is accompanied by the development of thymic lymphoma. Oncogene 2000, 19, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Kalim, K.W.; Yang, J.Q.; Modur, V.; Nguyen, P.; Li, Y.; Zheng, Y.; Guo, F. Graded RhoA GTPase Expression in Treg Cells Distinguishes Tumor Immunity From Autoimmunity. Front. Immunol. 2021, 12, 726393. [Google Scholar] [CrossRef] [PubMed]
- Lone, W.; Bouska, A.; Sharma, S.; Amador, C.; Saumyaranjan, M.; Herek, T.A.; Heavican, T.B.; Yu, J.; Lim, S.T.; Ong, C.K.; et al. Genome-wide miRNA expression profiling of molecular subgroups of peripheral T-cell lymphoma. Clin. Cancer Res. 2021, 27, 6039–6053. [Google Scholar] [CrossRef]
- Kitai, Y.; Ishiura, M.; Saitoh, K.; Matsumoto, N.; Owashi, K.; Yamada, S.; Muromoto, R.; Kashiwakura, J.I.; Oritani, K.; Matsuda, T. CD47 promotes T-cell lymphoma metastasis by up-regulating AKAP13-mediated RhoA activation. Int. Immunol. 2021, 33, 273–280. [Google Scholar] [CrossRef]
- Pan, Y.R.; Chen, C.C.; Chan, Y.T.; Wang, H.J.; Chien, F.T.; Chen, Y.L.; Liu, J.L.; Yang, M.H. STAT3-coordinated migration facilitates the dissemination of diffuse large B-cell lymphomas. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, G.; Zhang, L.; Wang, Q.; Li, H.; Han, Y.; Xie, L.; Yan, Z.; Li, Y.; An, Y.; et al. Comprehensive Review of Web Servers and Bioinformatics Tools for Cancer Prognosis Analysis. Front. Oncol. 2020, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Bayo, J.; Fiore, E.J.; Dominguez, L.M.; Cantero, M.J.; Ciarlantini, M.S.; Malvicini, M.; Atorrasagasti, C.; Garcia, M.G.; Rossi, M.; Cavasotto, C.; et al. Bioinformatic analysis of RHO family of GTPases identifies RAC1 pharmacological inhibition as a new therapeutic strategy for hepatocellular carcinoma. Gut 2021, 70, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Aspenström, P. Activated Rho GTPases in Cancer—The Beginning of a New Paradigm. Int. J. Mol. Sci. 2018, 19, 3949. [Google Scholar] [CrossRef]
- Jakobsen, N.A.; Vyas, P. From genomics to targeted treatment in haematological malignancies: A focus on acute myeloid leukaemia. Clin. Med. 2018, 18, s47–s53. [Google Scholar] [CrossRef]
- Hardison, R.C. Comparative Genomics. PLoS Biol. 2003, 1, e58. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506. [Google Scholar] [CrossRef]
- Madeddu, L.; Stilo, G. Deep learning methods for network biology. In Deep Learning in Biology and Medicine; Bacciu, D., Lisboa, P.J.G., Vellido, A., Eds.; World Scientific Publishing Group: London, UK, 2022. [Google Scholar] [CrossRef]
- Rocca, A.; Kholodenko, B.N. Can Systems Biology Advance Clinical Precision Oncology? Cancers 2021, 13, 6312. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Lin, Y.; Gao, F.; Zhang, C.T.; Zhang, R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014, 42, D574. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; Del-Toro, N.; et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.D.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2020, 48, D148. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Hartline, E.; et al. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234. [Google Scholar] [CrossRef]
- Farkas, I.J.; Szántó-Várnagy, Á.; Korcsmáros, T. Linking proteins to signaling pathways for experiment design and evaluation. PLoS ONE 2012, 7, e36202. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019, 20, 1085. [Google Scholar] [CrossRef]
- Chun, K.H. Discovery of Cellular RhoA Functions by the Integrated Application of Gene Set Enrichment Analysis. Biomol. Ther. 2022, 30, 98–116. [Google Scholar] [CrossRef] [PubMed]
- Fathima, S.; Sinha, S.; Donakonda, S. Network analysis identifies drug targets and small molecules to modulate apoptosis resistant cancers. Cancers 2021, 13, 851. [Google Scholar] [CrossRef]
- Maldonado, M.d.M.; Dharmawardhane, S. Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res. 2018, 78, 3101–3111. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.; Gosens, R.; Wieland, T.; Schmidt, M. Paving the Rho in cancer metastasis: Rho GTPases and beyond. Pharmacol. Ther. 2018, 183, 1–21. [Google Scholar] [CrossRef]
- Vogelsgesang, M.; Pautsch, A.; Aktories, K. C3 exoenzymes, novel insights into structure and action of Rho-ADP-ribosylating toxins. Naunyn. Schmiedebergs. Arch. Pharmacol. 2007, 374, 347–360. [Google Scholar] [CrossRef]
- Lord-Fontaine, S.; Yang, F.; Diep, Q.; Dergham, P.; Munzer, S.; Tremblay, P.; McKerracher, L. Local Inhibition of Rho Signaling by Cell-Permeable Recombinant Protein BA-210 Prevents Secondary Damage and Promotes Functional Recovery following Acute Spinal Cord Injury. J. Neurotrauma 2008, 25, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Schwartz, B.R.; Lin, N.; Winn, R.K.; Harlan, J.M. Requirement for RhoA kinase activation in leukocyte de-adhesion. J. Immunol. 2002, 169, 2330–2336. [Google Scholar] [CrossRef]
- Montalvo-Ortiz, B.L.; Castillo-Pichardo, L.; Hernández, E.; Humphries-Bickley, T.; De La Mota-Peynado, A.; Cubano, L.A.; Vlaar, C.P.; Dharmawardhane, S. Characterization of EHop-016, Novel Small Molecule Inhibitor of Rac GTPase. J. Biol. Chem. 2012, 287, 13228–13238. [Google Scholar] [CrossRef]
- Zeng, R.-J.; Zheng, C.-W.; Gu, J.-E.; Zhang, H.-X.; Xie, L.; Xu, L.-Y.; Li, E.-M. RAC1 inhibition reverses cisplatin resistance in esophageal squamous cell carcinoma and induces downregulation of glycolytic enzymes. Mol. Oncol. 2019, 13, 2010–2030. [Google Scholar] [CrossRef]
- Ito, M.A.; Kojima, E.; Yanagihara, Y.; Yoshida, K.; Matsuoka, I. Differential Effects of Gq Protein-Coupled Uridine Receptor Stimulation on IL-8 Production in 1321N1 Human Astrocytoma Cells. Biol. Pharm. Bull. 2022, 45, 691–697. [Google Scholar] [CrossRef]
- Hemsing, A.L.; Rye, K.P.; Hatfield, K.J.; Reikvam, H. NPM1-Mutated Patient-Derived AML Cells Are More Vulnerable to Rac1 Inhibition. Biomedicines 2022, 10, 1881. [Google Scholar] [CrossRef] [PubMed]
- Akbar, H.; Cancelas, J.; Williams, D.A.; Zheng, J.; Zheng, Y. Rational design and applications of a Rac GTPase-specific small molecule inhibitor. Methods Enzymol. 2006, 406, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Dickerson, J.B.; Guo, F.; Zheng, J.; Zheng, Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA 2004, 101, 7618–7623. [Google Scholar] [CrossRef]
- Thomas, E.K.; Cancelas, J.A.; Chae, H.-D.; Cox, A.D.; Keller, P.J.; Perrotti, D.; Neviani, P.; Druker, B.J.; Setchell, K.D.R.; Zheng, Y.; et al. Rac Guanosine Triphosphatases Represent Integrating Molecular Therapeutic Targets for BCR-ABL-Induced Myeloproliferative Disease. Cancer Cell 2007, 12, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Zins, K.; Lucas, T.; Reichl, P.; Abraham, D.; Aharinejad, S. A Rac1/Cdc42 GTPase-Specific Small Molecule Inhibitor Suppresses Growth of Primary Human Prostate Cancer Xenografts and Prolongs Survival in Mice. PLoS ONE 2013, 8, e74924. [Google Scholar] [CrossRef]
- Zins, K.; Gunawardhana, S.; Lucas, T.; Abraham, D.; Aharinejad, S. Targeting Cdc42 with the small molecule drug AZA197 suppresses primary colon cancer growth and prolongs survival in a preclinical mouse xenograft model by downregulation of PAK1 activity. J. Transl. Med. 2013, 11, 295. [Google Scholar] [CrossRef]
- Florian, M.C.; Dörr, K.; Niebel, A.; Daria, D.; Schrezenmeier, H.; Rojewski, M.; Filippi, M.-D.; Hasenberg, A.; Gunzer, M.; Scharffetter-Kochanek, K.; et al. Cdc42 Activity Regulates Hematopoietic Stem Cell Aging and Rejuvenation. Cell Stem Cell 2012, 10, 520–530. [Google Scholar] [CrossRef]
- Friesland, A.; Zhao, Y.; Chen, Y.-H.; Wang, L.; Zhou, H.; Lu, Q. Small molecule targeting Cdc42–intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. USA 2013, 110, 1261–1266. [Google Scholar] [CrossRef]
- Aguilar, B.J.; Zhao, Y.; Zhou, H.; Huo, S.; Chen, Y.-H.; Lu, Q. Inhibition of Cdc42–intersectin interaction by small molecule ZCL367 impedes cancer cell cycle progression, proliferation, migration, and tumor growth. Cancer Biol. Ther. 2019, 20, 740–749. [Google Scholar] [CrossRef]
- Leins, H.; Mulaw, M.; Eiwen, K.; Sakk, V.; Liang, Y.; Denkinger, M.; Geiger, H.; Schirmbeck, R. Aged murine hematopoietic stem cells drive aging-associated immune remodeling. Blood 2018, 132, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Shutes, A.; Onesto, C.; Picard, V.; Leblond, B.; Schweighoffer, F.; Der, C.J. Specificity and Mechanism of Action of EHT 1864, a Novel Small Molecule Inhibitor of Rac Family Small GTPases. J. Biol. Chem. 2007, 282, 35666–35678. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Kenney, S.R.; Phillips, G.K.; Simpson, D.; Schroeder, C.E.; Nöth, J.; Romero, E.; Swanson, S.; Waller, A.; Strouse, J.J.; et al. Characterization of a Cdc42 Protein Inhibitor and Its Use as a Molecular Probe. J. Biol. Chem. 2013, 288, 8531–8543. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Song, X.; Ma, S.; Wang, X.; Xu, J.; Zhang, H.; Wu, Q.; Zhao, K.; Cao, J.; Qiao, J.; et al. Cdc42 inhibitor ML141 enhances G-CSF-induced hematopoietic stem and progenitor cell mobilization. Int. J. Hematol. 2015, 101, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Sims, A.H.; Sproul, D.; Caldwell, H.; Michael Dixon, J.; Meehan, R.R.; Harrison, D.J. Targeting of Rac GTPases blocks the spread of intact human breast cancer. Oncotarget 2012, 3, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Ramos, D.F.V.; Mancuso, R.I.; Contieri, B.; Duarte, A.; Paiva, L.; de Melo Carrilho, J.; Saad, S.T.O.; Lazarini, M. Rac GTPases in acute myeloid leukemia cells: Expression profile and biological effects of pharmacological inhibition. Toxicol. Appl. Pharmacol. 2022, 442, 115990. [Google Scholar] [CrossRef]
- Shang, X.; Marchioni, F.; Sipes, N.; Evelyn, C.R.; Jerabek-Willemsen, M.; Duhr, S.; Seibel, W.; Wortman, M.; Zheng, Y. Rational Design of Small Molecule Inhibitors Targeting RhoA Subfamily Rho GTPases. Chem. Biol. 2012, 19, 699–710. [Google Scholar] [CrossRef]
- Yoon, C.; Cho, S.-J.; Aksoy, B.A.; Park, D.J.; Schultz, N.; Ryeom, S.W.; Yoon, S.S. Chemotherapy Resistance in Diffuse-Type Gastric Adenocarcinoma Is Mediated by RhoA Activation in Cancer Stem-Like Cells. Clin. Cancer Res. 2016, 22, 971–983. [Google Scholar] [CrossRef]
- Tsubaki, M.; Genno, S.; Takeda, T.; Matsuda, T.; Kimura, N.; Yamashita, Y.; Morii, Y.; Shimomura, K.; Nishida, S. Rhosin Suppressed Tumor Cell Metastasis through Inhibition of Rho/YAP Pathway and Expression of RHAMM and CXCR4 in Melanoma and Breast Cancer Cells. Biomedicines 2021, 9, 35. [Google Scholar] [CrossRef]
- Evelyn, C.R.; Ferng, T.; Rojas, R.J.; Larsen, M.J.; Sondek, J.; Neubig, R.R. High-Throughput Screening for Small-Molecule Inhibitors of LARG-Stimulated RhoA Nucleotide Binding via a Novel Fluorescence Polarization Assay. SLAS Discov. 2000, 14, 161–172. [Google Scholar] [CrossRef]
- Shang, X.; Marchioni, F.; Evelyn, C.R.; Sipes, N.; Zhou, X.; Seibel, W.; Wortman, M.; Zheng, Y. Small-molecule inhibitors targeting G-protein–coupled Rho guanine nucleotide exchange factors. Proc. Natl. Acad. Sci. USA 2013, 110, 3155–3160. [Google Scholar] [CrossRef] [PubMed]
- Mitin, N.; Roberts, P.J.; Chenette, E.J.; Der, C.J. Posttranslational lipid modification of rho family small GTPases. Methods Mol. Biol. 2012, 827, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Aspenström, P.; Ruusala, A.; Pacholsky, D. Taking Rho GTPases to the next level: The cellular functions of atypical Rho GTPases. Exp. Cell Res. 2007, 313, 3673–3679. [Google Scholar] [CrossRef]
- Karasic, T.B.; Chiorean, E.G.; Sebti, S.M.; O’Dwyer, P.J. A Phase I Study of GGTI-2418 (Geranylgeranyl Transferase I Inhibitor) in Patients with Advanced Solid Tumors. Target. Oncol. 2019, 14, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Denoyelle, C.; Vasse, M.; Körner, M.; Mishal, Z.; Ganné, F.; Vannier, J.-P.; Soria, J.; Soria, C. Cerivastatin, an inhibitor of HMG-CoA reductase, inhibits the signaling pathways involved in the invasiveness and metastatic properties of highly invasive breast cancer cell lines: An in vitro study. Carcinogenesis 2001, 22, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Gazzerro, P.; Proto, M.C.; Gangemi, G.; Malfitano, A.M.; Ciaglia, E.; Pisanti, S.; Santoro, A.; Laezza, C.; Bifulco, M. Pharmacological Actions of Statins: A Critical Appraisal in the Management of Cancer. Pharmacol. Rev. 2012, 64, 102–146. [Google Scholar] [CrossRef]
- Tobert, J.A. Lovastatin and beyond: The history of the HMG-CoA reductase inhibitors. Nat. Rev. Drug Discov. 2003, 2, 517–526. [Google Scholar] [CrossRef]
- Beckwitt, C.H.; Brufsky, A.; Oltvai, Z.N.; Wells, A. Statin drugs to reduce breast cancer recurrence and mortality. Breast Cancer Res. 2018, 20, 144. [Google Scholar] [CrossRef]
- Markowska, A.; Antoszczak, M.; Markowska, J.; Huczyński, A. Statins: HMG-CoA Reductase Inhibitors as Potential Anticancer Agents against Malignant Neoplasms in Women. Pharmaceuticals 2020, 13, 422. [Google Scholar] [CrossRef]
- Afshari, A.R.; Mollazadeh, H.; Henney, N.C.; Jamialahmad, T.; Sahebkar, A. Effects of statins on brain tumors: A review. Semin. Cancer Biol. 2021, 73, 116–133. [Google Scholar] [CrossRef]
- Gachpazan, M.; Kashani, H.; Khazaei, M.; Hassanian, S.M.; Rezayi, M.; Asgharzadeh, F.; Ghayour-Mobarhan, M.; Ferns, G.A.; Avan, A. The Impact of Statin Therapy on the Survival of Patients with Gastrointestinal Cancer. Curr. Drug Targets 2018, 20, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, G.; Delakas, D.; Nakopoulou, L.; Kassimatis, T. Statins and prostate cancer: Molecular and clinical aspects. Eur. J. Cancer 2011, 47, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Bonovas, S.; Filioussi, K.; Tsantes, A.; Sitaras, N.M. Use of statins and risk of haematological malignancies: A meta-analysis of six randomized clinical trials and eight observational studies. Br. J. Clin. Pharmacol. 2007, 64, 255. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Xiao, W.; Li, D.; Mu, S. Combination of simvastatin and imatinib sensitizes the CD34+ cells in K562 to cell death. Med. Oncol. 2011, 28, 528–531. [Google Scholar] [CrossRef]
- Yang, Y.C.; Huang, W.F.; Chuan, L.M.; Xiao, D.W.; Zeng, Y.L.; Zhou, D.A.; Xu, G.Q.; Liu, W.; Huang, B.; Hu, Q. In vitro and in vivo study of cell growth inhibition of simvastatin on chronic myelogenous leukemia cells. Chemotherapy 2008, 54, 438–446. [Google Scholar] [CrossRef]
- Winiarska, M.; Bil, J.; Wilczek, E.; Wilczynski, G.M.; Lekka, M.; Engelberts, P.J.; Mackus, W.J.M.; Gorska, E.; Bojarski, L.; Stoklosa, T.; et al. Statins Impair Antitumor Effects of Rituximab by Inducing Conformational Changes of CD20. PLoS Med. 2008, 5, e64. [Google Scholar] [CrossRef]
- Chapman-Shimshoni, D.; Yuklea, M.; Radnay, J.; Shapiro, H.; Lishner, M. Simvastatin induces apoptosis of B-CLL cells by activation of mitochondrial caspase 9. Exp. Hematol. 2003, 31, 779–783. [Google Scholar] [CrossRef]
- Vitols, S.; Angelin, B.; Juliusson, G. Simvastatin impairs mitogen-induced proliferation of malignant B-lymphocytes from humans—In vitro and in vivo studies. Lipids 1997, 32, 255–262. [Google Scholar] [CrossRef]
- Shanafelt, T.D.; Rabe, K.G.; Kay, N.E.; Zent, C.S.; Call, T.G.; Slager, S.L.; Bowen, D.A.; Schwager, S.M.; Nowakowski, G.S. Statin and non-steroidal anti-inflammatory drug use in relation to clinical outcome among patients with Rai stage 0 chronic lymphocytic leukemia. Leuk. Lymphoma 2010, 51, 1233–1240. [Google Scholar] [CrossRef]
- Friedman, D.R.; Magura, L.A.; Warren, H.A.C.; Harrison, J.D.; Diehl, L.F.; Weinberg, J.B. Statin use and need for therapy in chronic lymphocytic leukemia. Leuk. Lymphoma 2010, 51, 2295–2298. [Google Scholar] [CrossRef]
- Gouni, S.; Strati, P.; Toruner, G.; Aradhya, A.; Landgraf, R.; Bilbao, D.; Vega, F.; Agarwal, N.K. Statins enhance the chemosensitivity of R-CHOP in diffuse large B-cell lymphoma. Leuk. Lymphoma 2022, 63, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, H.; Zou, L. Influence of Statins on the Survival Outcomes of Patients with Diffuse Large B Cell Lymphoma: A Systematic Review and Meta-Analysis. Int. J. Clin. Pract. 2022, 2022, 5618290. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.; Profitós-Pelejà, N.; Ribeiro, M.L.; Roué, G. Antitumor Activity of Simvastatin in Preclinical Models of Mantle Cell Lymphoma. Cancers 2022, 14, 5601. [Google Scholar] [CrossRef] [PubMed]
- Van Besien, H.; Sassano, A.; Altman, J.K.; Platanias, L.C. Antileukemic properties of 3-hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors. Leuk. Lymphoma 2013, 54, 2601–2605. [Google Scholar] [CrossRef]
- Kornblau, S.M.; Banker, D.E.; Stirewalt, D.; Shen, D.; Lemker, E.; Verstovsek, S.; Estrov, Z.; Faderl, S.; Cortes, J.; Beran, M.; et al. Blockade of adaptive defensive changes in cholesterol uptake and synthesis in AML by the addition of pravastatin to idarubicin + high-dose Ara-C: A phase 1 study. Blood 2007, 109, 2999–3006. [Google Scholar] [CrossRef] [PubMed]
- Advani, A.S.; Mcdonough, S.; Copelan, E.; Willman, C.; Mulford, D.A.; List, A.F.; Sekeres, M.A.; Othus, M.; Appelbaum, F.R. SWOG0919: A Phase 2 study of idarubicin and cytarabine in combination with pravastatin for relapsed acute myeloid leukaemia. Br. J. Haematol. 2014, 167, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Kale, V.P.; Hengst, J.A.; Desai, D.H.; Amin, S.G.; Yun, J.K. The regulatory roles of ROCK and MRCK kinases in the plasticity of cancer cell migration. Cancer Lett. 2015, 361, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Rath, N.; Olson, M.F. Rho-associated kinases in tumorigenesis: Re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012, 13, 900–908. [Google Scholar] [CrossRef]
- Nagumo, H.; Sasaki, Y.; Ono, Y.; Okamoto, H.; Seto, M.; Takuwa, Y. Rho kinase inhibitor HA-1077 prevents Rho-mediated myosin phosphatase inhibition in smooth muscle cells. Am. J. Physiol. Physiol. 2000, 278, C57–C65. [Google Scholar] [CrossRef]
- William, B.M.; An, W.; Feng, D.; Nadeau, S.; Mohapatra, B.C.; Storck, M.A.; Band, V.; Band, H. Fasudil, a clinically safe ROCK inhibitor, decreases disease burden in a Cbl/Cbl-b deficiency-driven murine model of myeloproliferative disorders. Hematology 2016, 21, 218–224. [Google Scholar] [CrossRef]
- Narumiya, S.; Ishizaki, T.; Uehata, M. Use and properties of ROCK-specific inhibitor Y-27632. Methods Enzymol. 2000, 325, 273–284. [Google Scholar] [CrossRef]
- Linke, F.; Zaunig, S.; Nietert, M.M.; VonBonin, F.; Lutz, S.; Dullin, C.; Janovská, P.; Beissbarth, T.; Alves, F.; Klapper, W.; et al. WNT5A: A motility-promoting factor in Hodgkin lymphoma. Oncogene 2017, 36, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.C.; Liu, H.F.; Hsu, K.C.; Yang, J.M.; Chen, C.; Liu, K.K.; Hsu, T.S.; Chao, J.I. 7-Chloro-6-piperidin-1-yl-quinoline-5,8-dione (PT-262), a novel ROCK inhibitor blocks cytoskeleton function and cell migration. Biochem. Pharmacol. 2011, 81, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Vigil, D.; Kim, T.Y.; Plachco, A.; Garton, A.J.; Castaldo, L.; Pachter, J.A.; Dong, H.; Chen, X.; Tokar, B.; Campbell, S.L.; et al. ROCK1 and ROCK2 Are Required for Non-Small Cell Lung Cancer Anchorage-Independent Growth and Invasion. Cancer Res. 2012, 72, 5338–5347. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.A.; Forinash, K.D.; Pireddu, R.; Sun, Y.; Sun, N.; Martin, M.P.; Schönbrunn, E.; Lawrence, N.J.; Sebti, S.M. RKI-1447 Is a Potent Inhibitor of the Rho-Associated ROCK Kinases with Anti-Invasive and Antitumor Activities in Breast Cancer. Cancer Res. 2012, 72, 5025–5034. [Google Scholar] [CrossRef]
- Sadok, A.; McCarthy, A.; Caldwell, J.; Collins, I.; Garrett, M.D.; Yeo, M.; Hooper, S.; Sahai, E.; Kuemper, S.; Mardakheh, F.K.; et al. Rho Kinase Inhibitors Block Melanoma Cell Migration and Inhibit Metastasis. Cancer Res. 2015, 75, 2272–2284. [Google Scholar] [CrossRef]
- Semenova, G.; Chernoff, J. Targeting PAK1. Biochem. Soc. Trans. 2017, 45, 79–88. [Google Scholar] [CrossRef]
- Deacon, S.W.; Beeser, A.; Fukui, J.A.; Rennefahrt, U.E.E.; Myers, C.; Chernoff, J.; Peterson, J.R. An Isoform-Selective, Small-Molecule Inhibitor Targets the Autoregulatory Mechanism of p21-Activated Kinase. Chem. Biol. 2008, 15, 322–331. [Google Scholar] [CrossRef]
- Flis, S.; Bratek, E.; Chojnacki, T.; Piskorek, M.; Skorski, T. Simultaneous inhibition of BCR-ABL1 tyrosine kinase and PAK1/2 serine/threonine kinase exerts synergistic effect against chronic myeloid leukemia cells. Cancers 2019, 11, 1544. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Choi, C.K.; Lee, C.S.; Park, M.H.; Tian, X.; Kim, N.D.; Lee, K.I.; Choi, J.K.; Ahn, J.H.; Shin, E.Y.; et al. Small molecules that allosterically inhibit p21-activated kinase activity by binding to the regulatory p21-binding domain. Exp. Mol. Med. 2016, 48, e229. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Ghosh, J.; Ramdas, B.; Mali, R.S.; Martin, H.; Kobayashi, M.; Vemula, S.; Canela, V.H.; Waskow, E.R.; Visconte, V.; et al. Regulation of Stat5 by FAK and PAK1 in Oncogenic FLT3- and KIT-Driven Leukemogenesis. Cell Rep. 2014, 9, 1333–1348. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.W.; Guo, C.; Piraino, J.; Westwick, J.K.; Zhang, C.; Lamerdin, J.; Dagostino, E.; Knighton, D.; Loi, C.M.; Zager, M.; et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc. Natl. Acad. Sci. USA 2010, 107, 9446–9451. [Google Scholar] [CrossRef] [PubMed]
- Chow, H.Y.; Jubb, A.M.; Koch, J.N.; Jaffer, Z.M.; Stepanova, D.; Campbell, D.A.; Duron, S.G.; O’Farrell, M.; Cai, K.Q.; Klein-Szanto, A.J.P.; et al. p21-Activated kinase 1 is required for efficient tumor formation and progression in a Ras-mediated skin cancer model. Cancer Res. 2012, 72, 5966–5975. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, A.; Stanley, R.F.; Yu, Y.; Bartholdy, B.; Pendurti, G.; Gritsman, K.; Boultwood, J.; Chernoff, J.; Verma, A.; Steidl, U. PAK1 is a therapeutic target in acute myeloid leukemia and myelodysplastic syndrome. Blood 2015, 126, 1118–1127. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, J.C.; Profitós-Pelejà, N.; Sánchez-Vinces, S.; Roué, G. RHOA Therapeutic Targeting in Hematological Cancers. Cells 2023, 12, 433. https://doi.org/10.3390/cells12030433
Santos JC, Profitós-Pelejà N, Sánchez-Vinces S, Roué G. RHOA Therapeutic Targeting in Hematological Cancers. Cells. 2023; 12(3):433. https://doi.org/10.3390/cells12030433
Chicago/Turabian StyleSantos, Juliana Carvalho, Núria Profitós-Pelejà, Salvador Sánchez-Vinces, and Gaël Roué. 2023. "RHOA Therapeutic Targeting in Hematological Cancers" Cells 12, no. 3: 433. https://doi.org/10.3390/cells12030433
APA StyleSantos, J. C., Profitós-Pelejà, N., Sánchez-Vinces, S., & Roué, G. (2023). RHOA Therapeutic Targeting in Hematological Cancers. Cells, 12(3), 433. https://doi.org/10.3390/cells12030433






