Future Perspectives of Prime Editing for the Treatment of Inherited Retinal Diseases
Abstract
1. Introduction
2. Development and Improvement of the Prime Editing Technology
2.1. Engineering the Prime Editor Protein
2.2. Improving pegRNA Stability and Structure
2.3. Suppressing DNA Repair Mechanisms
2.4. Improving Accessibility of the Target DNA
2.5. Utilizing Two pegRNAs
2.6. Additional Strategies
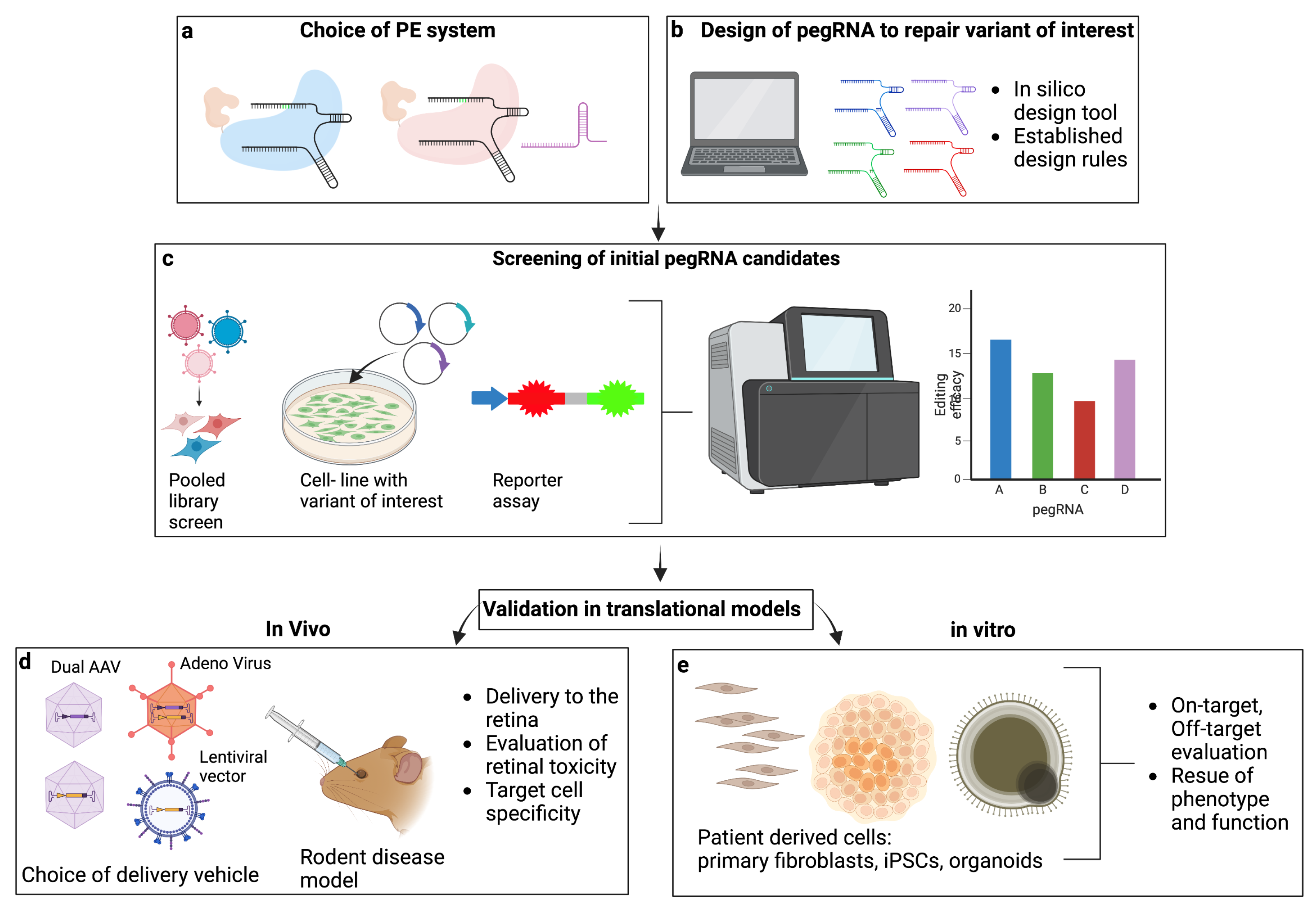
3. Identification of Successful pegRNA Candidates
3.1. Design of pegRNAs
| Name | Summary of Key Features | ON/Off Target Sore | Web Access | Ref |
|---|---|---|---|---|
| PINECONE (Prime Induced Nucleotide Engineering Creator of New Edits) | Provides reference genome of different organisms for definition of target sequence Provides cloning nucleotides, PCR, and sequencing primers for target locus | provides MIT score (UCSC browser) for pegRNAs specificity, candidates with highest score are chosen | https://github.com/xiaowanglab/PINE-CONE | [72] |
| pegFinder | Enables pegRNA design for different Cas9 orthologues (Cas9-NGG, Cas9-NG, and Cas9-SpRY) | Provides Optional sgRNA on/off-target score (based on Broad sgRNA designer [80,81] and CRISPscan [82]) | http://pegfinder.sidichenlab.org | [73] |
| PE-Designer | Enables pegRNA design for different Cas9 orthologues (SpCas9, SpCas9-VQR, SpCas9-SpRY, SpCas9-VRER, NG-Cas9, iSpyMac Cas9, and xCas9) Supports pegRNA design for 543 different organisms Provides visual interface displaying location of pegRNA and nicking sgRNA on target DNA | Provides evaluation of potential off-targets for 544 different organisms (based on Cas-OFFinder [83]) | http://www.rgenome.net/pe-designer/ | [74] |
| PrimeDesign | Supports pegRNA design for single and combination editing applications and genome-wide and saturation mutagenesis screens Provides visually display of pegRNA secondary structure Includes PrimeVar database with candidate pegRNA and nicking sgRNA combinations for > 68.500 pathogenic human genetic ClinVar variants | Provides CFD score for spacer specificity [80] | https://primedesign.pinellolab.partners.org | [75] |
| Prime Editing design Tool | Provides pegRNA design for ClinVar Variants (correction or introduction) Custom Target sequence can be defined with ClinVar variant ID or Gene ID (no need for manual entry of target sequence) Supports pegRNA design for different Cas orthologues (Cas9-NG, xCas9, xCas9-NG or Cas9SpRy), | Includes pegRNA with Hsu-Scott off-target score [84] >50 only | https://primeedit.nygenome.org | [76] |
| PnB designer | Provides pegRNA design for single or multiple targets for different organisms Accepts sequence input or genomic coordinates as input format for target sequence Additionally enables design of Base editing guide RNAs | https://fgcz-shiny.uzh.ch/PnBDesigner/ | [77] | |
| pegIT | Provides design of single and multiple pegRNAs Accepts gene name or gene ID (reference genome for different organisms is available), Clinvar ID, or manual input of WT and edited target sequence as input format for target sequence Supports pegRNA design for different Cas9 orthologues (spCas9, CjCas9, SaCas9, SaCas9KKH, SpCas9NG) Modular structure allows rapid adjustment for new prime editing variants and CRISPR scoring rules | pegRNAs are ranked by ON-target score (based on CFD score [80]), Provides off-target evolution (including 0–3 mismatches) | https://pegit.giehmlab.dk | [78] |
| Easy-Prime | Machine learning based design tool Provides predicted editing efficacy for top pegRNAs and nicking sgRNA combinations Includes prediction of RNA folding in design of pegRNA candidates Provides visual interface displaying location of pegRNA and nicking sgRNA on target DNA v1.2 includes Option for EpegRNA design Provides pegRNA design for 89.5% of 152.352 GWAS variants | Provides Spacer specificity (based on DeepSpCas9-Score) | http://easy-prime.cc | [79] |
| DeepPE | Machine learning based Provides expected prime editing efficacy for 57 pegRNA candidates per target sequence Provides pegRNA candidates to generate different types of edits (deletions, insertions, substitutions) | provides Spacer specificity (based on DeepSpCas9 Score [85]) | http://deepcrispr.info/DeepPE | [48] |
3.2. pegRNA Screening Approaches
4. Delivery of Prime Editing Components In Vivo for Treatment of IRD
4.1. Split-PE Delivery
4.2. Single PE Construct Delivery
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Cepko, C.L.; Vandenberghe, L.H. Retinal gene therapy coming of age. Hum. Gene Ther. 2013, 24, 242–244. [Google Scholar] [CrossRef]
- Diakatou, M.; Manes, G.; Bocquet, B.; Meunier, I.; Kalatzis, V. Genome Editing as a Treatment for the Most Prevalent Causative Genes of Autosomal Dominant Retinitis Pigmentosa. Int. J. Mol. Sci. 2019, 20, 2542. [Google Scholar] [CrossRef] [PubMed]
- Hafler, B.P. Clinical Progress in Inherited Retinal Degenerations: Gene Therapy Clinical Trials and Advances in Genetic Sequencing. Retina 2017, 37, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Sohocki, M.M.; Daiger, S.P.; Bowne, S.J.; Rodriquez, J.A.; Northrup, H.; Heckenlively, J.R.; Birch, D.G.; Mintz-Hittner, H.; Ruiz, R.S.; Lewis, R.A.; et al. Prevalence of Mutations Causing Retinitis Pigmentosa and Other Inherited Retinopathies. Hum. Mutat. 2001, 17, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, N.; Michaelides, M.; Smith, A.J.; Ali, R.R.; Bainbridge, J.W.B. Retinal gene therapy. Br. Med. Bull. 2018, 126, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Benhar, I.; London, A.; Schwartz, M. The privileged immunity of immune privileged organs: The case of the eye. Front. Immunol. 2012, 3, 296. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.C.; Lee, V.; Maguire, A.M. Recent advances in ocular gene therapy. Curr. Opin. Ophthalmol. 2009, 20, 377–381. [Google Scholar] [CrossRef]
- Maguire, A.M.; Russell, S.; Chung, D.C.; Yu, Z.F.; Tillman, A.; Drack, A.V.; Simonelli, F.; Leroy, B.P.; Reape, K.Z.; High, K.A.; et al. Durability of Voretigene Neparvovec for Biallelic RPE65-Mediated Inherited Retinal Disease: Phase 3 Results at 3 and 4 Years. Ophthalmology 2021, 128, P1460–P1468. [Google Scholar] [CrossRef]
- Trapani, I.; Auricchio, A. Has retinal gene therapy come of age? From bench to bedside and back to bench. Hum. Mol. Genet. 2019, 28, R108–R118. [Google Scholar] [CrossRef]
- Quinn, J.; Musa, A.; Kantor, A.; McClements, M.E.; Cehajic-Kapetanovic, J.; MacLaren, R.E.; Xue, K. Genome-Editing Strategies for Treating Human Retinal Degenerations. Hum. Gene Ther. 2021, 32, 247–259. [Google Scholar] [CrossRef]
- Maeder, M.L.; Stefanidakis, M.; Wilson, C.J.; Baral, R.; Barrera, L.A.; Bounoutas, G.S.; Bumcrot, D.; Chao, H.; Ciulla, D.M.; DaSilva, J.A.; et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019, 25, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008, 283, P1–P5. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wu, Z. In Vivo Applications of CRISPR-Based Genome Editing in the Retina. Front. Cell Dev. Biol. 2018, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A. The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, R.; Onyango, D.O.; Stark, J.M. Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet. 2016, 32, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Greene, E.C. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet. 2021, 37, 639–656. [Google Scholar] [CrossRef]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef]
- Cullot, G.; Boutin, J.; Toutain, J.; Prat, F.; Pennamen, P.; Rooryck, C.; Teichmann, M.; Rousseau, E.; Lamrissi-Garcia, I.; Guyonnet-Duperat, V.; et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019, 10, 1136. [Google Scholar] [CrossRef]
- Alanis-Lobato, G.; Zohren, J.; McCarthy, A.; Fogarty, N.M.E.; Kubikova, N.; Hardman, E.; Greco, M.; Wells, D.; Turner, J.M.A.; Niakan, K.K. Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proc. Natl. Acad. Sci. USA 2021, 118, e2004832117. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, M.L.; Papathanasiou, S.; Doerfler, P.A.; Blaine, L.J.; Sun, L.; Yao, Y.; Zhang, C.-Z.; Weiss, M.J.; Pellman, D. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet. 2021, 53, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Wang, Q.; Mendez-Dorantes, C.; Burns, K.H.; Chiarle, R. Frequency and mechanisms of LINE-1 retrotransposon insertions at CRISPR/Cas9 sites. Nat. Commun. 2022, 13, 3685. [Google Scholar] [CrossRef] [PubMed]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018, 24, 927–930. [Google Scholar] [CrossRef]
- Enache, O.M.; Rendo, V.; Abdusamad, M.; Lam, D.; Davison, D.; Pal, S.; Currimjee, N.; Hess, J.; Pantel, S.; Nag, A.; et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 2020, 52, 662–668. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353, aaf8729. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Park, J.E.; Paa, P.; Rajakumar, P.D.; Prekop, H.T.; Chew, Y.T.; Manivannan, S.N.; Chew, W.L. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 2021, 12, 1384. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Liu, Z.; Zhang, Y.; Chen, M.; Sui, T.; Lai, L.; Li, Z. Large-Fragment Deletions Induced by Cas9 Cleavage while Not in the BEs System. Mol. Ther. Nucleic Acids 2020, 21, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef] [PubMed]
- Zuo, E.; Sun, Y.; Wei, W.; Yuan, T.; Ying, W.; Sun, H.; Yuan, L.; Steinmetz, L.M.; Li, Y.; Yang, H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019, 364, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zong, Y.; Gao, Q.; Zhu, Z.; Wang, Y.; Qin, P.; Liang, C.; Wang, D.; Qiu, J.L.; Zhang, F.; et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 2019, 364, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Zhi, S.; Chen, Y.; Wu, G.; Wen, J.; Wu, J.; Liu, Q.; Li, Y.; Kang, R.; Hu, S.; Wang, J.; et al. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 2022, 30, 283–294. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Liu, Y.; Kao, H.I.; Bambara, R.A. Flap endonuclease 1: A central component of DNA metabolism. Annu. Rev. Biochem. 2004, 73, 589–615. [Google Scholar] [CrossRef]
- Gao, P.; Lyu, Q.; Ghanam, A.R.; Lazzarotto, C.R.; Newby, G.A.; Zhang, W.; Choi, M.; Slivano, O.J.; Holden, K.; Walker, J.A., II; et al. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol. 2021, 22, 83. [Google Scholar] [CrossRef]
- Schene, I.F.; Joore, I.P.; Oka, R.; Mokry, M.; van Vugt, A.H.M.; van Boxtel, R.; van der Doef, H.P.J.; van der Laan, L.J.W.; Verstegen, M.M.A.; van Hasselt, P.M.; et al. Prime editing for functional repair in patient-derived disease models. Nat. Commun. 2020, 11, 5352. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Lin, Q.; Jin, S.; Zong, Y.; Yu, H.; Zhu, Z.; Liu, G.; Kou, L.; Wang, Y.; Qiu, J.-L.; Li, J.; et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021, 39, 923–927. [Google Scholar] [CrossRef]
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of Plant Prime-Editing Systems for Precise Genome Editing. Plant Commun. 2020, 1, 100043. [Google Scholar] [CrossRef] [PubMed]
- Koblan, L.W.; Doman, J.L.; Wilson, C.; Levy, J.M.; Tay, T.; Newby, G.A.; Maianti, J.P.; Raguram, A.; Liu, D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018, 36, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liang, S.-Q.; Zheng, C.; Mintzer, E.; Zhao, Y.G.; Ponnienselvan, K.; Mir, A.; Sontheimer, E.J.; Gao, G.; Flotte, T.R.; et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 2021, 12, 2121. [Google Scholar] [CrossRef]
- Song, M.; Lim, J.M.; Min, S.; Oh, J.-S.; Kim, D.Y.; Woo, J.-S.; Nishimasu, H.; Cho, S.-R.; Yoon, S.; Kim, H.H. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nat. Commun. 2021, 12, 5617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, L.; Zhu, B.; Wang, L.; Chen, C.; Hong, M.; Huang, Y.; Li, H.; Han, H.; Cai, B.; et al. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell Biol. 2020, 22, 740–750. [Google Scholar] [CrossRef]
- Velimirovic, M.; Zanetti, L.C.; Shen, M.W.; Fife, J.D.; Lin, L.; Cha, M.; Akinci, E.; Barnum, D.; Yu, T.; Sherwood, R.I. Peptide fusion improves prime editing efficiency. Nat. Commun. 2022, 13, 3512. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Liu, Y.; Xue, C.; Li, B.; Li, X.; Wang, Y.; Li, J.; Liu, G.; Huang, X.; Cao, X.; et al. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 2022, 40, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Yu, G.; Park, J.; Min, S.; Lee, S.; Yoon, S.; Kim, H.H. Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 2021, 39, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022, 40, 402–410. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, Y.; Huang, S.; Qu, S.; Cheng, D.; Yao, Y.; Ji, Q.; Wang, X.; Huang, X.; Liu, J. Enhancement of prime editing via xrRNA motif-joined pegRNA. Nat. Commun. 2022, 13, 1856. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Sun, W.; Huang, S.; Zhong, M.; Yao, Y.; Ji, Q.; Huang, X. Enhancing prime editing efficiency by modified pegRNA with RNA G-quadruplexes. J. Mol. Cell Biol. 2022, 14, mjac022. [Google Scholar] [CrossRef]
- Li, X.; Zhou, L.; Gao, B.Q.; Li, G.; Wang, X.; Wang, Y.; Wei, J.; Han, W.; Wang, Z.; Li, J.; et al. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure. Nat. Commun. 2022, 13, 1669. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, G.; Huang, S.; Li, X.; Wang, X.; Li, G.; Chi, T.; Chen, Y.; Huang, X.; Wang, X. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 2021, 31, 1134–1136. [Google Scholar] [CrossRef] [PubMed]
- Petri, K.; Zhang, W.; Ma, J.; Schmidts, A.; Lee, H.; Horng, J.E.; Kim, D.Y.; Kurt, I.C.; Clement, K.; Hsu, J.Y.; et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 2022, 40, 189–193. [Google Scholar] [CrossRef]
- Chen, P.J.; Hussmann, J.A.; Yan, J.; Knipping, F.; Ravisankar, P.; Chen, P.F.; Chen, C.; Nelson, J.W.; Newby, G.A.; Sahin, M.; et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 2021, 184, 5635–5652.e29. [Google Scholar] [CrossRef] [PubMed]
- Ferreira da Silva, J.; Oliveira, G.P.; Arasa-Verge, E.A.; Kagiou, C.; Moretton, A.; Timelthaler, G.; Jiricny, J.; Loizou, J.I. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair. Nat. Commun. 2022, 13, 760. [Google Scholar] [CrossRef]
- Wu, J.; Corbett, A.H.; Berland, K.M. The intracellular mobility of nuclear import receptors and NLS cargoes. Biophys. J. 2009, 96, 3840–3849. [Google Scholar] [CrossRef]
- Dang, C.V.; Lee, W.M.F. Identification of the human c-myc protein nuclear translocation signal. Mol. Cell. Biol. 1988, 8, 4048–4054. [Google Scholar] [CrossRef]
- Spencer, J.M.; Zhang, X. Deep mutational scanning of S. pyogenes Cas9 reveals important functional domains. Sci. Rep. 2017, 7, 16836. [Google Scholar] [CrossRef]
- Chen, F.; Ding, X.; Feng, Y.; Seebeck, T.; Jiang, Y.; Davis, G.D. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat. Commun. 2017, 8, 14958. [Google Scholar] [CrossRef]
- Dahlman, J.E.; Abudayyeh, O.O.; Joung, J.; Gootenberg, J.S.; Zhang, F.; Konermann, S. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat. Biotechnol. 2015, 33, 1159–1161. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Jeong, T.Y.; Shin, S.K.; Yoon, D.E.; Lim, S.Y.; Kim, S.P.; Choi, J.; Lee, H.; Hong, J.I.; Ahn, J.; et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. 2021, 22, 170. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhou, L.; Lin, G.; Hu, Y.; Jiao, Y.; Wang, Y.; Liu, J.; Yang, S.; Yao, S. HDAC inhibitors improve CRISPR-Cas9 mediated prime editing and base editing. Mol. Ther. Nucleic Acids 2022, 29, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Gao, X.D.; Podracky, C.J.; Nelson, A.T.; Koblan, L.W.; Raguram, A.; Levy, J.M.; Mercer, J.A.M.; Liu, D.R. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 2022, 40, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, Z.; Wang, G.; Zhang, R.; Duan, J.; Gao, P.; Lei, X.; Qiu, H.; Zhang, C.; Zhang, Y.; et al. Efficient targeted insertion of large DNA fragments without DNA donors. Nat. Methods 2022, 19, 331–340. [Google Scholar] [CrossRef]
- Zhuang, Y.; Liu, J.; Wu, H.; Zhu, Q.; Yan, Y.; Meng, H.; Chen, P.R.; Yi, C. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat. Chem. Biol. 2022, 18, 29–37. [Google Scholar] [CrossRef]
- Choi, J.; Chen, W.; Suiter, C.C.; Lee, C.; Chardon, F.M.; Yang, W.; Leith, A.; Daza, R.M.; Martin, B.; Shendure, J. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 2022, 40, 218–226. [Google Scholar] [CrossRef]
- Tao, R.; Wang, Y.; Jiao, Y.; Hu, Y.; Li, L.; Jiang, L.; Zhou, L.; Qu, J.; Chen, Q.; Yao, S. Bi-PE: Bi-directional priming improves CRISPR/Cas9 prime editing in mammalian cells. Nucleic Acids Res. 2022, 50, 6423–6434. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, X.-O.; Weng, Z.; Xue, W. Deletion and replacement of long genomic sequences using prime editing. Nat. Biotechnol. 2022, 40, 227–234. [Google Scholar] [CrossRef]
- Yarnall, M.T.N.; Ioannidi, E.I.; Schmitt-Ulms, C.; Krajeski, R.N.; Lim, J.; Villiger, L.; Zhou, W.; Jiang, K.; Garushyants, S.K.; Roberts, N.; et al. Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases. Nat. Biotechnol. 2022, in press. [Google Scholar] [CrossRef]
- Doman, J.L.; Sousa, A.A.; Randolph, P.B.; Chen, P.J.; Liu, D.R. Designing and executing prime editing experiments in mammalian cells. Nat. Protoc. 2022, 17, 2431–2468. [Google Scholar] [CrossRef]
- Standage-Beier, K.; Tekel, S.J.; Brafman, D.A.; Wang, X. Prime Editing Guide RNA Design Automation Using PINE-CONE. ACS Synth. Biol. 2021, 10, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Chow, R.D.; Chen, J.S.; Shen, J.; Chen, S. A web tool for the design of prime-editing guide RNAs. Nat. Biomed. Eng. 2021, 5, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.H.; Jeong, Y.K.; Habib, O.; Hong, S.A.; Lim, K.; Kim, J.S.; Bae, S. PE-Designer and PE-Analyzer: Web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res. 2021, 49, W499–W504. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.Y.; Grünewald, J.; Szalay, R.; Shih, J.; Anzalone, A.V.; Lam, K.C.; Shen, M.W.; Petri, K.; Liu, D.R.; Joung, J.K.; et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 2021, 12, 1034. [Google Scholar] [CrossRef]
- Morris, J.A.; Rahman, J.A.; Guo, X.; Sanjana, N.E. Automated design of CRISPR prime editors for 56,000 human pathogenic variants. iScience 2021, 24, 103380. [Google Scholar] [CrossRef] [PubMed]
- Siegner, S.M.; Karasu, M.E.; Schröder, M.S.; Kontarakis, Z.; Corn, J.E. PnB Designer: A web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinform. 2021, 22, 101. [Google Scholar] [CrossRef]
- Anderson, M.V.; Haldrup, J.; Thomsen, E.A.; Wolff, J.H.; Mikkelsen, J.G. pegIT—A web-based design tool for prime editing. Nucleic Acids Res. 2021, 49, W505–W509. [Google Scholar] [CrossRef]
- Li, Y.; Chen, J.; Tsai, S.Q.; Cheng, Y. Easy-Prime: A machine learning-based prime editor design tool. Genome Biol. 2021, 22, 235. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Listgarten, J.; Weinstein, M.; Kleinstiver, B.P.; Sousa, A.A.; Joung, J.K.; Crawford, J.; Gao, K.; Hoang, L.; Elibol, M.; Doench, J.G.; et al. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat. Biomed. Eng. 2018, 2, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, Y.; Lee, S.; Min, S.; Bae, J.Y.; Choi, J.W.; Park, J.; Jung, D.; Yoon, S.; Kim, H.H. SpCas9 activity prediction by DeepSpCas9, a deep learning-based model with high generalization performance. Sci. Adv. 2019, 5, eaax9249. [Google Scholar] [CrossRef] [PubMed]
- Schene, I.F.; Joore, I.P.; Baijens, J.H.L.; Stevelink, R.; Kok, G.; Shehata, S.; Ilcken, E.F.; Nieuwenhuis, E.C.M.; Bolhuis, D.P.; van Rees, R.C.M.; et al. Mutation-specific reporter for optimization and enrichment of prime editing. Nat. Commun. 2022, 13, 1028. [Google Scholar] [CrossRef]
- Simon, D.A.; Tálas, A.; Kulcsár, P.I.; Biczók, Z.; Krausz, S.L.; Várady, G.; Welker, E. PEAR, a flexible fluorescent reporter for the identification and enrichment of successfully prime edited cells. Elife 2022, 11, e69504. [Google Scholar] [CrossRef]
- Chemello, F.; Chai, A.C.; Li, H.; Rodriguez-Caycedo, C.; Sanchez-Ortiz, E.; Atmanli, A.; Mireault, A.A.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 2021, 7, eabg4910. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; He, S.; Huang, S.; Li, C.; Chen, Y.; Liu, Z.; Huang, X.; Wang, X. Efficient generation of mouse models with the prime editing system. Cell Discov. 2020, 6, 27. [Google Scholar] [CrossRef]
- Liu, B.; Dong, X.; Cheng, H.; Zheng, C.; Chen, Z.; Rodríguez, T.C.; Liang, S.-Q.; Xue, W.; Sontheimer, E.J. A split prime editor with untethered reverse transcriptase and circular RNA template. Nat. Biotechnol. 2022, 40, 1388–1393. [Google Scholar] [CrossRef]
- Zheng, C.; Liang, S.Q.; Liu, B.; Liu, P.; Kwan, S.Y.; Wolfe, S.A.; Xue, W. A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver. Mol. Ther. 2022, 30, 1343–1351. [Google Scholar] [CrossRef]
- Jang, H.; Jo, D.H.; Cho, C.S.; Shin, J.H.; Seo, J.H.; Yu, G.; Gopalappa, R.; Kim, D.; Cho, S.R.; Kim, J.H.; et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 2021, 6, 181–194. [Google Scholar] [CrossRef]
- van Haasteren, J.; Li, J.; Scheideler, O.J.; Murthy, N.; Schaffer, D.V. The delivery challenge: Fulfilling the promise of therapeutic genome editing. Nat. Biotechnol. 2020, 38, 845–855. [Google Scholar] [CrossRef]
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef] [PubMed]
- McClements, M.E.; MacLaren, R.E. Adeno-associated Virus (AAV) Dual Vector Strategies for Gene Therapy Encoding Large Transgenes. Yale J. Biol. Med. 2017, 90, 611–623. [Google Scholar] [PubMed]
- Grieger, J.C.; Samulski, R.J. Packaging capacity of adeno-associated virus serotypes: Impact of larger genomes on infectivity and postentry steps. J. Virol. 2005, 79, 9933–9944. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, F.; Gao, G. CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell 2020, 181, 136–150. [Google Scholar] [CrossRef]
- Sun, L.; Li, J.; Xiao, X. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nat. Med. 2000, 6, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Pryadkina, M.; Lostal, W.; Bourg, N.; Charton, K.; Roudaut, C.; Hirsch, M.L.; Richard, I. A comparison of AAV strategies distinguishes overlapping vectors for efficient systemic delivery of the 6.2 kb Dysferlin coding sequence. Mol. Ther. Methods Clin. Dev. 2015, 2, 15009. [Google Scholar] [CrossRef]
- Trapani, I.; Colella, P.; Sommella, A.; Iodice, C.; Cesi, G.; de Simone, S.; Marrocco, E.; Rossi, S.; Giunti, M.; Palfi, A.; et al. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med. 2014, 6, 194–211. [Google Scholar] [CrossRef]
- Ryu, S.M.; Koo, T.; Kim, K.; Lim, K.; Baek, G.; Kim, S.T.; Kim, H.S.; Kim, D.E.; Lee, H.; Chung, E.; et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018, 36, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Yue, Y.; Lai, Y.; Duan, D. A Hybrid Vector System Expands Adeno-associated Viral Vector Packaging Capacity in a Transgene-independent Manner. Mol. Ther. 2008, 16, 124–130. [Google Scholar] [CrossRef]
- Yan, Z.; Zhang, Y.; Duan, D.; Engelhardt, J.F. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. USA 2000, 97, 6716–6721. [Google Scholar] [CrossRef]
- Pergolizzi, R.G.; Ropper, A.E.; Dragos, R.; Reid, A.C.; Nakayama, K.; Tan, Y.; Ehteshami, J.R.; Coleman, S.H.; Silver, R.B.; Hackett, N.R.; et al. In vivo trans-splicing of 5’ and 3’; segments of Pre-mRNA directed by corresponding DNA sequences delivered by gene transfer. Mol. Ther. 2003, 8, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Lou, H.H.; Boyer, J.L.; Limberis, M.P.; Vandenberghe, L.H.; Hackett, N.R.; Leopold, P.L.; Wilson, J.M.; Crystal, R.G. Functional Cystic Fibrosis Transmembrane Conductance Regulator Expression in Cystic Fibrosis Airway Epithelial Cells by AAV6.2-Mediated Segmental Trans-Splicing. Hum. Gene Ther. 2009, 20, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Aranko, A.S.; Wlodawer, A.; Iwaï, H. Nature’s recipe for splitting inteins. Protein Eng. Des. Sel. 2014, 27, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Dyka, F.M.; Boye, S.L.; Chiodo, V.A.; Hauswirth, W.W.; Boye, S.E. Dual adeno-associated virus vectors result in efficient in vitro and in vivo expression of an oversized gene, MYO7A. Hum. Gene Ther. Methods 2014, 25, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, L.S.; Turunen, H.T.; Wassmer, S.J.; Luna-Velez, M.V.; Xiao, R.; Bennett, J.; Vandenberghe, L.H. Evaluating Efficiencies of Dual AAV Approaches for Retinal Targeting. Front. Neurosci. 2017, 11, 503. [Google Scholar] [CrossRef] [PubMed]
- Tornabene, P.; Trapani, I.; Minopoli, R.; Centrulo, M.; Lupo, M.; de Simone, S.; Tiberi, P.; Dell’Aquila, F.; Marrocco, E.; Iodice, C.; et al. Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci. Transl. Med. 2019, 11, eaav4523. [Google Scholar] [CrossRef]
- McClements, M.E.; Barnard, A.R.; Singh, M.S.; Charbel Issa, P.; Jiang, Z.; Radu, R.A.; MacLaren, R.E. An AAV Dual Vector Strategy Ameliorates the Stargardt Phenotype in Adult Abca4(−/−) Mice. Hum. Gene Ther. 2019, 30, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.H.; Song, D.W.; Cho, C.S.; Kim, U.G.; Lee, K.J.; Lee, K.; Park, S.W.; Kim, D.; Kim, J.H.; Kim, J.S.; et al. CRISPR-Cas9-mediated therapeutic editing of Rpe65 ameliorates the disease phenotypes in a mouse model of Leber congenital amaurosis. Sci. Adv. 2019, 5, eaax1210. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.H.; Jang, H.-K.; Cho, C.S.; Han, J.H.; Ryu, G.; Jung, Y.; Bae, S.; Kim, J.H. Therapeutic adenine base editing corrects nonsense mutation and improves visual function in a mouse model of Leber congenital amaurosis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Suh, S.; Choi, E.H.; Leinonen, H.; Foik, A.T.; Newby, G.A.; Yeh, W.H.; Dong, Z.; Kiser, P.D.; Lyon, D.C.; Liu, D.R.; et al. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat. Biomed. Eng. 2021, 5, 169–178. [Google Scholar] [CrossRef]
- Chen, Y.; Zhi, S.; Liu, W.; Wen, J.; Hu, S.; Cao, T.; Sun, H.; Li, Y.; Huang, L.; Liu, Y.; et al. Development of Highly Efficient Dual-AAV Split Adenosine Base Editor for In Vivo Gene Therapy. Small Methods 2020, 4, 2000309. [Google Scholar] [CrossRef]
- Gao, Z.; Ravendran, S.; Mikkelsen, N.S.; Haldrup, J.; Cai, H.; Ding, X.; Paludan, S.R.; Thomsen, M.K.; Mikkelsen, J.G.; Bak, R.O. A truncated reverse transcriptase enhances prime editing by split AAV vectors. Mol. Ther. 2022, 30, 2942–2951. [Google Scholar] [CrossRef]
- Böck, D.; Rothgangl, T.; Villiger, L.; Schmidheini, L.; Matsushita, M.; Mathis, N.; Ioannidi, E.; Rimann, N.; Grisch-Chan, H.M.; Kreutzer, S.; et al. In vivo prime editing of a metabolic liver disease in mice. Sci. Transl. Med. 2022, 14, eabl9238. [Google Scholar] [CrossRef]
- Grünewald, J.; Miller, B.R.; Szalay, R.N.; Cabeceiras, P.K.; Woodilla, C.J.; Holtz, E.J.B.; Petri, K.; Joung, J.K. Engineered CRISPR prime editors with compact, untethered reverse transcriptases. Nat. Biotechnol. 2022, in press. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Janssen, J.M.; Tasca, F.; Mei, H.; Gonçalves, M. Broadening the reach and investigating the potential of prime editors through fully viral gene-deleted adenoviral vector delivery. Nucleic Acids Res. 2021, 49, 11986–12001. [Google Scholar] [CrossRef]
- Arsenijevic, Y.; Berger, A.; Udry, F.; Kostic, C. Lentiviral Vectors for Ocular Gene Therapy. Pharmaceutics 2022, 14, 1605. [Google Scholar] [CrossRef] [PubMed]
- Puppo, A.; Cesi, G.; Marrocco, E.; Piccolo, P.; Jacca, S.; Shayakhmetov, D.M.; Parks, R.J.; Davidson, B.L.; Colloca, S.; Brunetti-Pierri, N.; et al. Retinal transduction profiles by high-capacity viral vectors. Gene Ther. 2014, 21, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Banskota, S.; Raguram, A.; Suh, S.; Du, S.W.; Davis, J.R.; Choi, E.H.; Wang, X.; Nielsen, S.C.; Newby, G.A.; Randolph, P.B.; et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022, 185, 250–265.e16. [Google Scholar] [CrossRef]
- Staurenghi, F.; McClements, M.E.; Salman, A.; MacLaren, R.E. Minicircle Delivery to the Neural Retina as a Gene Therapy Approach. Int. J. Mol. Sci. 2022, 23, 11673. [Google Scholar] [CrossRef] [PubMed]
- Vagni, P.; Perlini, L.E.; Chenais, N.A.L.; Marchetti, T.; Parrini, M.; Contestabile, A.; Cancedda, L.; Ghezzi, D. Gene Editing Preserves Visual Functions in a Mouse Model of Retinal Degeneration. Front. Neurosci. 2019, 13, 945. [Google Scholar] [CrossRef]
- Salman, A.; Kantor, A.; McClements, M.E.; Marfany, G.; Trigueros, S.; MacLaren, R.E. Non-Viral Delivery of CRISPR/Cas Cargo to the Retina Using Nanoparticles: Current Possibilities, Challenges, and Limitations. Pharmaceutics 2022, 14, 1842. [Google Scholar] [CrossRef] [PubMed]
- Rajala, A.; Wang, Y.; Zhu, Y.; Ranjo-Bishop, M.; Ma, J.-X.; Mao, C.; Rajala, R.V.S. Nanoparticle-Assisted Targeted Delivery of Eye-Specific Genes to Eyes Significantly Improves the Vision of Blind Mice In Vivo. Nano Lett. 2014, 14, 5257–5263. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.-J.; Yang, P.; Ban, Q.; Yang, Y.-P.; Wang, M.-L.; Chien, C.-S.; Chen, S.-J.; Sun, N.; Zhu, Y.; Liu, H.; et al. Dual Supramolecular Nanoparticle Vectors Enable CRISPR/Cas9-Mediated Knockin of Retinoschisin 1 Gene—A Potential Nonviral Therapeutic Solution for X-Linked Juvenile Retinoschisis. Adv. Sci. 2020, 7, 1903432. [Google Scholar] [CrossRef]
- Yang, T.-C.; Chang, C.-Y.; Yarmishyn, A.A.; Mao, Y.-S.; Yang, Y.-P.; Wang, M.-L.; Hsu, C.-C.; Yang, H.-Y.; Hwang, D.-K.; Chen, S.-J.; et al. Carboxylated nanodiamond-mediated CRISPR-Cas9 delivery of human retinoschisis mutation into human iPSCs and mouse retina. Acta Biomater. 2020, 101, 484–494. [Google Scholar] [CrossRef]
- Herrera-Barrera, M.; Ryals, R.C.; Gautam, M.; Jozic, A.; Landry, M.; Korzun, T.; Gupta, M.; Acosta, C.; Stoddard, J.; Reynaga, R.; et al. Peptide-guided lipid nanoparticles deliver mRNA to the neural retina of rodents and nonhuman primates. Sci. Adv. 2023, 9, eadd4623. [Google Scholar] [CrossRef] [PubMed]
- Fry, L.E.; McClements, M.E.; MacLaren, R.E. Analysis of Pathogenic Variants Correctable With CRISPR Base Editing Among Patients With Recessive Inherited Retinal Degeneration. JAMA Ophthalmol. 2021, 139, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Kaukonen, M.; McClements, M.E.; MacLaren, R.E. CRISPR DNA Base Editing Strategies for Treating Retinitis Pigmentosa Caused by Mutations in Rhodopsin. Genes 2022, 13, 1327. [Google Scholar] [CrossRef] [PubMed]

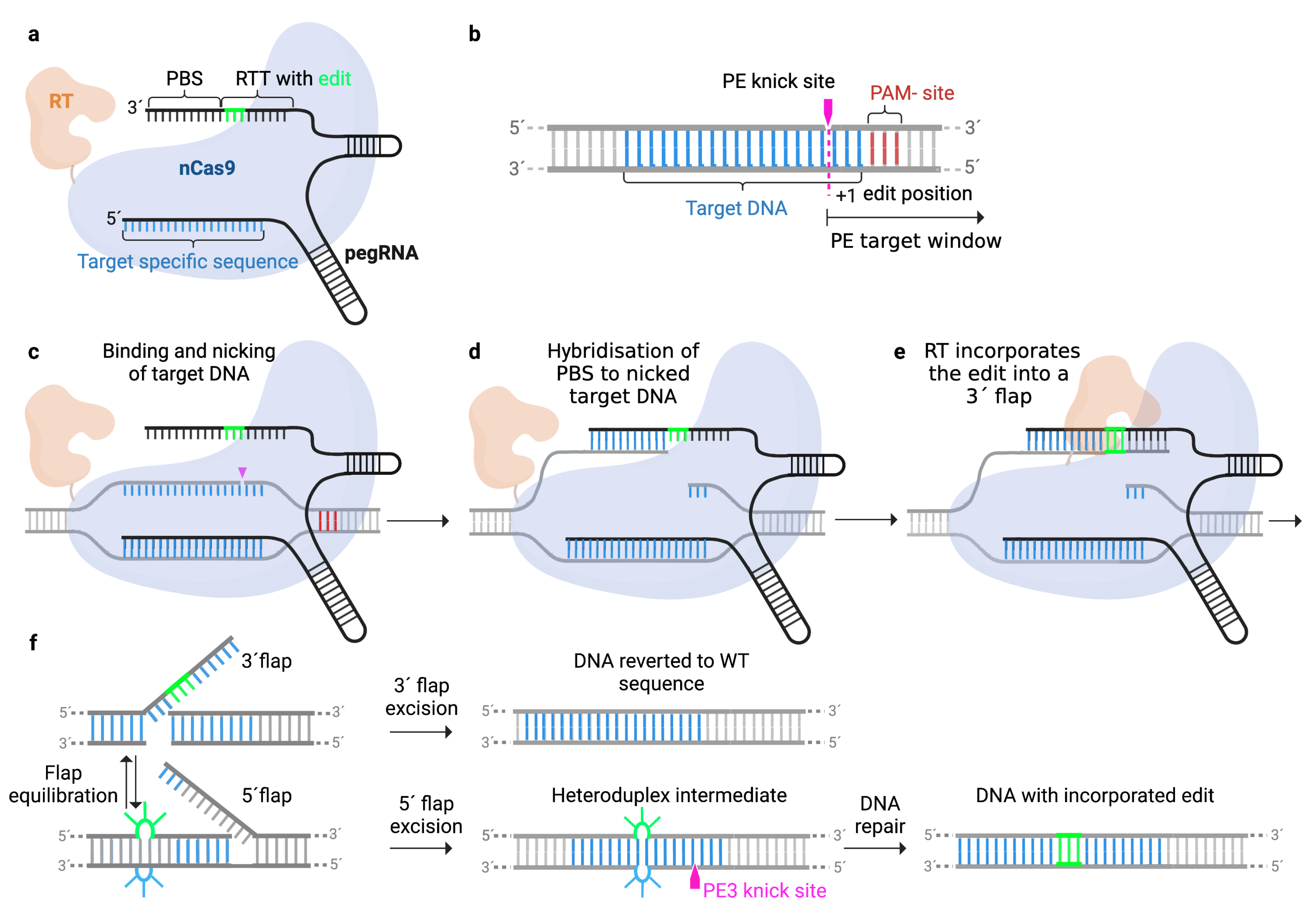

| Name | Mechanism/Improvement | Components | Editing Efficacy | Undesired Indels | Ref |
|---|---|---|---|---|---|
| PE1 | Cas9 Nickase (H840A) + Wildtype M-MLV RT | PE1 + pegRNA | 0.7–17% | 0.2% +/- 0.1% | [35] |
| PE2 | Modified M-MLV RT (D200N/LL603W/T330P/T306K/W313F) | PE2 + pegRNA | 1.6–5.1 fold increase compared to PE1 | [35] | |
| PE3 | Additional sgRNA nicking the non-edited strand -> favors incorporation of edited strand | PE2 + pegRNA+ nicking sgRNA | 1.5–4.2 fold increase compared to PE2, up to 55% | Increased compared to PE2 | [35] |
| PE3b | Nicking sgRNA with a spacer that matches the edited strand only | PE3 + pegRNA + nicking sgRNA with protospacer that only matches the edited strand | Similar to PE3 | 13-fold decrease compared to PE3 | [35] |
| PE* | Enhanced nuclear localization of the Prime editor | PE3/PE2 (with optimized NLS) + pegRNA | 1.3 to 3.4 fold increase compared to PE2 | Same or slightly higher levels than conventional PE2 and PE3 | [43] |
| hyPE2 | Addition of a Rad51 DNA-bin binding protein domain to stabilize the ssDNA that is used as a primer for reverse transcription | PE2 (with Rad51 DNA-binding domain) + pegRNA | 1.4 or 1.5 fold increase compared to PE2 | Slightly decreased compared to PE2 | [44] |
| IN-PE | Fusion of a dual peptide (IGFpm1-NFATC2IPp1) to PE 2 increases translation efficiency | IGFpm1-NFATC2IPp1 + pegRNA | Increased compared to PE2 (median in different cell-lines: 1.35X in A549, 1.92X in HCT-116, 1.17X in HEK293T, 1.78X in U2OS) | [46] | |
| ePPE | Engineered M-MLV RT (removal of ribonuclease H domain + addition of viral nucleocapsid protein with chaperon activity) | PPE (with modified M-MLV RT) + pegRNA | 5.8-fold increase compared to PPE | Similar to PPE | [47] |
| Prime editing with epegRNA | Stabilization of the 3´terminus of pegRNA with structured RNA motifs (evopreQ1 or mpknot) | PE3 + pegRNA (with evopreQ1/mpknot) | 3–4 fold increase compared to PE3 | Similar to PE3 | [49] |
| xrPE | Stabilization of 3´terminus of pegRNA with viral exoribonuclease-resistant RNA motif (xRNA) | PE3 + pegRNA (with xrRNA) | 2.5–4.5 fold increase compared to PE3 | Similar to PE3 | [50] |
| G-PE | Stabilization of 3´terminus of pegRNA with a structured RNA motif (human telomerase RNA (hTR) G-quadruplex modification) | PE + pegRNA (with hTR G-quadruplex formation) | Similar to xrPE and prime editing with epegRNA, up to 1.9 fold increase compared to conventional prime editing | Higher indel frequency than conventional prime editing | [51] |
| sPE | Introduction of same-sense mutations in RTT | PE3/PE5 + pegRNA (with SSM in RTT) | 353-fold increase (for base conversions) compared to PE3 | Similar to PE3 | [52] |
| aPE | Stabilization of pegRNA secondary structure | PE3/PE5 + pegRNA (modified) | 2.77-fold increase (for indel-editing) compared to PE3 | Similar to PE3 | [52] |
| Enhanced PE (ePE) | Prevention of pegRNA circulation by a Csy4 recognition site | PE3 (Csy4-T2A) + pegRNA (with Cys recognition site) | 1.9 fold increased compared to PE3 | Increased compared to PE3 | [53] |
| PE4 | Co-delivery of engineered MMR-inhibiting protein (MLH1d) | PE2 + co-delivered MLH1dn + pegRNA | 7-fold increase compared to PE2 | 3.4-fold improvement of edit:indel ratio in MMR proficient cells | [55] |
| PE5 | Co-delivery of engineered MMR-inhibiting protein (MLH1d) | PE3 + co-delivered MLH1dn + pegRNA | 2.0-fold increase compared to PE3 | 3.4-fold improvement of edit:indel ratio in MMR proficient cells | [55] |
| PE4 MAX | Co-delivery of engineered MMR-inhibiting protein (MLH1d)+ optimized PE architecture (human codan optimized RT, c-Myc NLS, linker, R221K, N394K mutation of SpCas)) | PE2 (optimized) + co-delivered MLH1dn + pegRNA /epegRNA | 2.8 fold improvement on average in HeLa cells compared to PE2 architecture | Decreased compared to PE2 | [55] |
| PE5 MAX | Co-delivery of engineered MMR-inhibiting protein (MLH1d)+ optimized PE architecture (human codan optimized RT, c-Myc NLS, linker, R221K, N394K mutation of SpCas)) | PE3 (optimized) + co-delivered MLH1dn + pegRNA (or epegRNA) + nicking sgRNA | 2.8 fold improvement on average in HeLa cells compared to PE2 architecture | 1.2 fold decrease compared to PE5 | [55] |
| Enhanced PE | Opening chromatin structure at target site using proximal dead sgRNA (dsgRNA) and chromatin-modulating peptides | PE3 + pegRNA + dsgRNA + CMP | 2.55–5.11-fold increase compared to PE3 | [62] | |
| Prime editing with HDACi | Opening chromatin structure at target site using HDAC inhibitors | PE3 + pegRNA + HDAC inhibitor | Compared to PE3: Increased for insertions (1.37-fold) and deletions (1.40-fold), decreased for installation of point mutations. | [63] | |
| twinPE | Two pegRNAs, each targeting the opposite strand and encoding strands complementary to each other | PE2 + 2 pegRNAs | For 108-bp insertion: 20-fold increase compared to PE3 | Fewer indels compared to paired Cas9 nuclease strategy | [64] |
| GRAND | Two pegRNAs, each targeting the opposite strand and encoding strands complementary to each other | PE2 + 2 pegRNAs | up to 63% for 150 bp insertion | [65] | |
| HOPE | Two pegRNAs, each targeting the opposite strand and encoding strands complementary to each other | PE2 + 2 pegRNAs | higher editing efficacy compared to PE2/PE3 | Fewer indels compared to PE3 | [66] |
| PRIME-Del | Two pegRNAs, targeting the opposite strand, encoding for strands complementary to each other and the DNA sequence upstream of the nick on opposite strand | PE2 + 2 pegRNAs | 1–30% editing efficacy | Fewer undesired indels compared to paired Cas9 nuclease strategy | [67] |
| Bi-PE | Two pegRNAs, targeting the opposite strand, encoding for strands complementary to each other and the DNA sequence upstream of the nick on opposite strand | PE2 + 2 pegRNAs | Compared to PE3: increased for conversions of multiple bases, no improved for single base conversions | [68] | [68] |
| PEDAR | Two pegRNAs, targeting opposite strands, Cas9 nuclease | PE-Cas9 (nuclease) + pegRNA | similar to PRIME-Del | Increased compared to PRIME-del | [69] |
| PASTE | CRISPR-Cas9 nickase fused to a reverse transcriptase and an additional serine integrase | Cas9nickase+ RT + serine integrase | 5–50% efficacy for different inserts | [70] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hansen, S.; McClements, M.E.; Corydon, T.J.; MacLaren, R.E. Future Perspectives of Prime Editing for the Treatment of Inherited Retinal Diseases. Cells 2023, 12, 440. https://doi.org/10.3390/cells12030440
Hansen S, McClements ME, Corydon TJ, MacLaren RE. Future Perspectives of Prime Editing for the Treatment of Inherited Retinal Diseases. Cells. 2023; 12(3):440. https://doi.org/10.3390/cells12030440
Chicago/Turabian StyleHansen, Silja, Michelle E. McClements, Thomas J. Corydon, and Robert E. MacLaren. 2023. "Future Perspectives of Prime Editing for the Treatment of Inherited Retinal Diseases" Cells 12, no. 3: 440. https://doi.org/10.3390/cells12030440
APA StyleHansen, S., McClements, M. E., Corydon, T. J., & MacLaren, R. E. (2023). Future Perspectives of Prime Editing for the Treatment of Inherited Retinal Diseases. Cells, 12(3), 440. https://doi.org/10.3390/cells12030440






