Molecular Influence of Resiniferatoxin on the Urinary Bladder Wall Based on Differential Gene Expression Profiling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Laboratory Animals
2.2. RNA Extraction, Library Construction, and Sequencing
2.3. Transcriptome Sequencing, Differential Expression Profiles, and Functional Annotations
2.4. Real-Time PCR
3. Results
3.1. Transcript Assembly, Quantification and Transcriptionally Active Region (TAR) Analysis
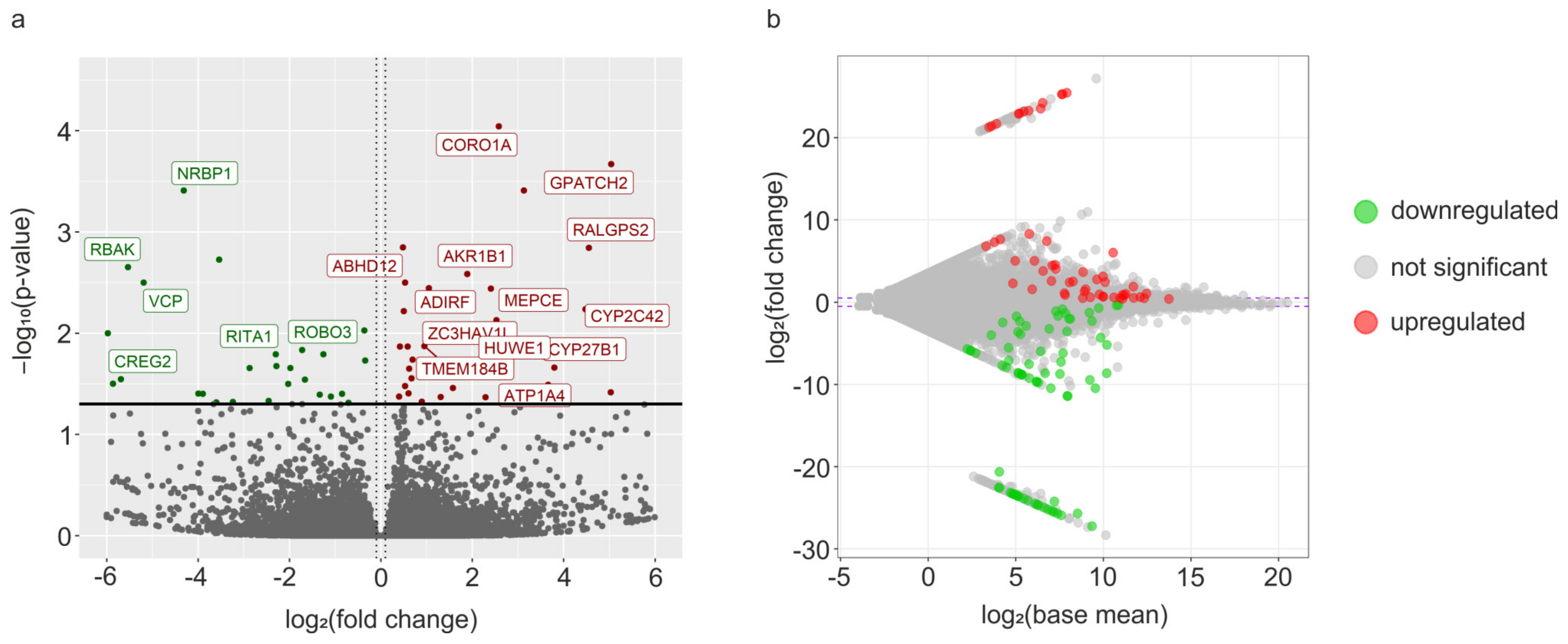
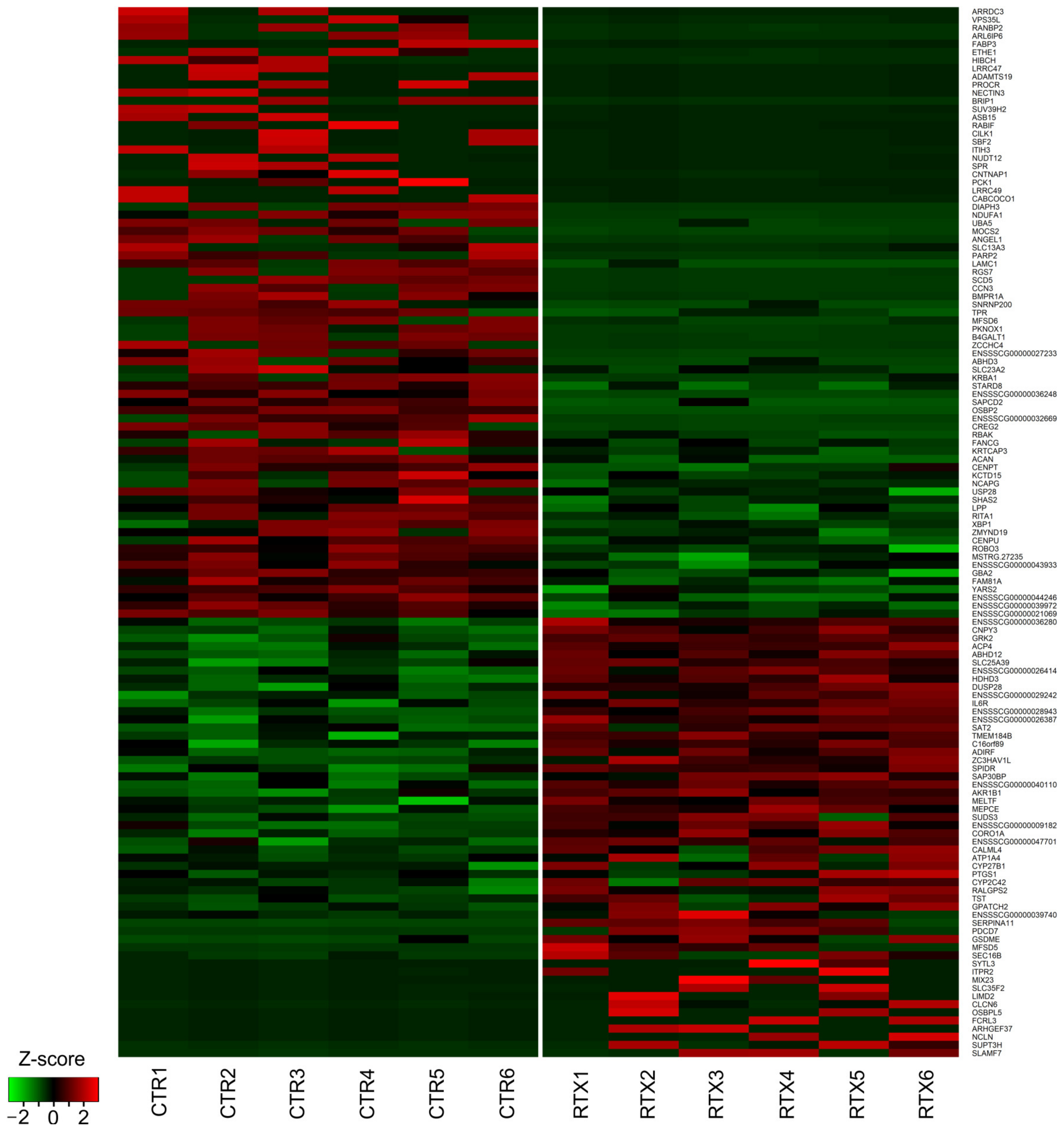
3.2. Transcriptomic Signatures of Differentially Expressed Genes (DEGs) and Functional Annotations
3.3. Validation of the Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Szallasi, A.; Blumberg, P.M. Resiniferatoxin, a Phorbol-Related Diterpene, Acts as an Ultrapotent Analog of Capsaicin, the Irritant Constituent in Red Pepper. Neuroscience 1989, 30, 515–520. [Google Scholar] [CrossRef] [Green Version]
- Cavanaugh, D.J.; Chesler, A.T.; Bráz, J.M.; Shah, N.M.; Julius, D.; Basbaum, A.I. Restriction of Transient Receptor Potential Vanilloid-1 to the Peptidergic Subset of Primary Afferent Neurons Follows Its Developmental Downregulation in Nonpeptidergic Neurons. J. Neurosci. 2011, 31, 10119–10127. [Google Scholar] [CrossRef] [PubMed]
- Bates, B.D.; Mitchell, K.; Keller, J.M.; Chan, C.-C.; Swaim, W.D.; Yaskovich, R.; Mannes, A.J.; Iadarola, M.J. Prolonged Analgesic Response of Cornea to Topical Resiniferatoxin, a Potent TRPV1 Agonist. Pain 2010, 149, 522–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelhamid, R.E.; Kovács, K.J.; Honda, C.N.; Nunez, M.G.; Larson, A.A. Resiniferatoxin (RTX) Causes a Uniquely Protracted Musculoskeletal Hyperalgesia in Mice by Activation of TRPV1 Receptors. J. Pain 2013, 14, 1629–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, B.; Li, X.; Chen, J.; Su, B.; Li, X.; Yang, S.; Guan, Z.; Wang, R. Resiniferatoxin for Treatment of Lifelong Premature Ejaculation: A Preliminary Study. Int. J. Urol. 2014, 21, 923–926. [Google Scholar] [CrossRef]
- Winter, J.; Dray, A.; Wood, J.N.; Yeats, J.C.; Bevan, S. Cellular Mechanism of Action of Resiniferatoxin: A Potent Sensory Neuron Excitotoxin. Brain Res. 1990, 520, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Mannes, A.; Hughes, M.; Quezado, Z.; Berger, A.; Fojo, T.; Smith, R.; Butman, J.; Lonser, R.; Iadarola, M. Resiniferatoxin, a Potent TRPV1 Agonist: Intrathecal Administration to Treat Severe Pain Associated with Advanced Cancer—Case Report. J. Pain 2010, 11, S43. [Google Scholar] [CrossRef]
- Leiman, D.; Minkowitz, H.; Levitt, R.C.; Solanki, D.; Horn, D.; Janfaza, D.; Sarno, D.; Albores-Ibarra, N.; Bai, X.; Takeshita, K.; et al. Preliminary Results from a Phase 1b Double-Blind Study to Assess the Safety, Tolerability and Efficacy of Intra-Articular Administration of Resiniferatoxin or Placebo for the Treatment of Moderate to Severe Pain Due to Osteoarthritis of the Knee. Osteoarthr. Cartil. 2020, 28, S138. [Google Scholar] [CrossRef]
- Nahama, A.; Ramachandran, R.; Cisternas, A.F.; Ji, H. The Role of Afferent Pulmonary Innervation in ARDS Associated with COVID-19 and Potential Use of Resiniferatoxin to Improve Prognosis: A Review. Med. Drug Discov. 2020, 5, 100033. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Chancellor, M.B. Intravesical Neuromodulatory Drugs: Capsaicin and Resiniferatoxin to Treat the Overactive Bladder. J. Endourol. 2000, 14, 97–103. [Google Scholar] [CrossRef]
- Cruz, F.; Dinis, P. Resiniferatoxin and Botulinum Toxin Type A for Treatment of Lower Urinary Tract Symptoms. Neurourol. Urodyn. 2007, 26, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Foster, H.E.; Lake, A.G. Use of Vanilloids in Urologic Disorders. In Capsaicin as a Therapeutic Molecule; Springer: Basel, Switzerland, 2014; Volume 68, pp. 307–317. ISBN 9783034808279. [Google Scholar]
- Chancellor, M.B.; de Groat, W.C. Intravesical Capsaicin and Resiniferatoxin Therapy: Spicing up the Ways to Treat the Overactive Bladder. J. Urol. 1999, 162, 3–11. [Google Scholar] [CrossRef]
- Cruz, F. Vanilloid Receptor and Detrusor Instability. Urology 2002, 59, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, C.; Peng, L.; Lu, Y.; Luo, D. Comparative Effectiveness and Safety of Intravesical Instillation Treatment of Interstitial Cystitis/Bladder Pain Syndrome: A Systematic Review and Network Meta-Analysis of Randomized Controlled Trials. Int. Urogynecol. J. 2021, 32, 1061–1071. [Google Scholar] [CrossRef]
- Gevaert, T.; Vandepitte, J.; Ost, D.; Nilius, B.; De Ridder, D. Autonomous Contractile Activity in the Isolated Rat Bladder Is Modulated by a TRPV1 Dependent Mechanism. Neurourol. Urodyn. 2007, 26, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Lepiarczyk, E.; Majewski, M.; Bossowska, A. The Influence of Intravesical Administration of Resiniferatoxin (RTX) on the Chemical Coding of Sympathetic Chain Ganglia (SChG) Neurons Supplying the Porcine Urinary Bladder. Histochem. Cell Biol. 2015, 144, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Lepiarczyk, E.; Bossowska, A.; Kaleczyc, J.; Skowrońska, A.; Majewska, M.; Majewski, M.; Majewski, M. The Influence of Resiniferatoxin (RTX) and Tetrodotoxin (TTX) on the Distribution, Relative Frequency, and Chemical Coding of Noradrenergic and Cholinergic Nerve Fibers Supplying the Porcine Urinary Bladder Wall. Toxins 2017, 9, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothschild, M.F.; Ruvinsky, A. The Genetics of the Pig, 2nd ed.; CABI: Wallingford, UK, 2011; ISBN 9781845937560. [Google Scholar]
- Swindle, M.M. The Development of Swine Models in Drug Discovery and Development. Future Med. Chem. 2012, 4, 1771–1772. [Google Scholar] [CrossRef]
- Schook, L.B.; Collares, T.V.; Darfour-Oduro, K.A.; De, A.K.; Rund, L.A.; Schachtschneider, K.M.; Seixas, F.K. Unraveling the Swine Genome: Implications for Human Health. Annu. Rev. Anim. Biosci. 2015, 3, 219–244. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 December 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Ballgown Bridges the Gap between Transcriptome Assembly and Expression Analysis. Nat. Biotechnol. 2015, 33, 243–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Kárai, L.J.; Russell, J.T.; Iadarola, M.J.; Oláh, Z. Vanilloid Receptor 1 Regulates Multiple Calcium Compartments and Contributes to Ca2+-Induced Ca2+ Release in Sensory Neurons. J. Biol. Chem. 2004, 279, 16377–16387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olah, Z.; Szabo, T.; Karai, L.; Hough, C.; Fields, R.D.; Caudle, R.M.; Blumberg, P.M.; Iadarola, M.J. Ligand-Induced Dynamic Membrane Changes and Cell Deletion Conferred by Vanilloid Receptor 1. J. Biol. Chem. 2001, 276, 11021–11030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.-P.; Yang, Y.-P.; She, R.-X.; Xing, Z.-M.; Chen, H.-W.; Zhang, Y.-W. MicroRNA-329 Reduces Bone Cancer Pain through the LPAR1-Dependent LPAR1/ERK Signal Transduction Pathway in Mice. Ther. Adv. Med. Oncol. 2019, 11, 1758835919875319. [Google Scholar] [CrossRef]
- BRADY, C. P2X3-Immunoreactive Nerve Fibres in Neurogenic Detrusor Overactivity and the Effect of Intravesical Resiniferatoxin*1. Eur. Urol. 2004, 46, 247–253. [Google Scholar] [CrossRef]
- Guan, X.; Lu, J.; Sun, F.; Li, Q.; Pang, Y. The Molecular Evolution and Functional Divergence of Lamprey Programmed Cell Death Genes. Front. Immunol. 2019, 10, 1382. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Huang, B.; Yang, H.; Kan, S.; Yao, Y.; Liu, X.; Pu, S.; He, G.; Khan, T.-M.; Qi, G.; et al. Latexin Deficiency in Mice Up-Regulates Inflammation and Aggravates Colitis through HECTD1/Rps3/NF-ΚB Pathway. Sci. Rep. 2020, 10, 9868. [Google Scholar] [CrossRef]
- Zeng, L.; Zampetaki, A.; Margariti, A.; Pepe, A.E.; Alam, S.; Martin, D.; Xiao, Q.; Wang, W.; Jin, Z.-G.; Cockerill, G.; et al. Sustained Activation of XBP1 Splicing Leads to Endothelial Apoptosis and Atherosclerosis Development in Response to Disturbed Flow. Proc. Natl. Acad. Sci. USA 2009, 106, 8326–8331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.-J.; Liu, Y.; Zhou, J.; Luo, S.; Zhao, W.; Li, X.; Liu, C. XBP1S Protects Cells from ER Stress-Induced Apoptosis through Erk1/2 Signaling Pathway Involving CHOP. Histochem. Cell Biol. 2012, 138, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.A.; Rios, J.C.; Lu, Y.; Garcia-Fresco, G.P.; Ching, W.; Martin, M.S.; Li, J.; Einheber, S.; Chesler, M.; Rosenbluth, J.; et al. Axon-Glia Interactions and the Domain Organization of Myelinated Axons Requires Neurexin IV/Caspr/Paranodin. Neuron 2001, 30, 369–383. [Google Scholar] [CrossRef] [Green Version]
- Gordon, A.; Adamsky, K.; Vainshtein, A.; Frechter, S.; Dupree, J.L.; Rosenbluth, J.; Peles, E. Caspr and Caspr2 Are Required for Both Radial and Longitudinal Organization of Myelinated Axons. J. Neurosci. 2014, 34, 14820–14826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hengel, H.; Magee, A.; Mahanjah, M.; Vallat, J.-M.; Ouvrier, R.; Abu-Rashid, M.; Mahamid, J.; Schüle, R.; Schulze, M.; Krägeloh-Mann, I.; et al. CNTNAP1 Mutations Cause CNS Hypomyelination and Neuropathy with or without Arthrogryposis. Neurol. Genet. 2017, 3, e144. [Google Scholar] [CrossRef] [Green Version]
- Mehta, P.; Küspert, M.; Bale, T.; Brownstein, C.A.; Towne, M.C.; De Girolami, U.; Shi, J.; Beggs, A.H.; Darras, B.T.; Wegner, M.; et al. Novel Mutation in CNTNAP1 Results in Congenital Hypomyelinating Neuropathy. Muscle Nerve 2017, 55, 761–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarroel-Campos, D.; Bronfman, F.C.; Gonzalez-Billault, C. Rab GTPase Signaling in Neurite Outgrowth and Axon Specification. Cytoskeleton 2016, 73, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Li, Y.; Wang, J.; Huo, Y.; Huang, D.; Li, S.; Liu, J.; Li, X.; Liu, R.; Chen, X.; et al. A Functional Missense Variant in ITIH3 Affects Protein Expression and Neurodevelopment and Confers Schizophrenia Risk in the Han Chinese Population. J. Genet. Genomics 2020, 47, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Llavero Hurtado, M.; Fuller, H.R.; Wong, A.M.S.; Eaton, S.L.; Gillingwater, T.H.; Pennetta, G.; Cooper, J.D.; Wishart, T.M. Proteomic Mapping of Differentially Vulnerable Pre-Synaptic Populations Identifies Regulators of Neuronal Stability in Vivo. Sci. Rep. 2017, 7, 12412. [Google Scholar] [CrossRef] [Green Version]
- Loupatty, F.J.; Clayton, P.T.; Ruiter, J.P.N.; Ofman, R.; IJlst, L.; Brown, G.K.; Thorburn, D.R.; Harris, R.A.; Duran, M.; DeSousa, C.; et al. Mutations in the Gene Encoding 3-Hydroxyisobutyryl-CoA Hydrolase Results in Progressive Infantile Neurodegeneration. Am. J. Hum. Genet. 2007, 80, 195–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera-Serrano, M.; Coote, D.J.; Azmanov, D.; Goullee, H.; Andersen, E.; McLean, C.; Davis, M.; Ishimura, R.; Stark, Z.; Vallat, J.-M.; et al. A Homozygous UBA5 Pathogenic Variant Causes a Fatal Congenital Neuropathy. J. Med. Genet. 2020, 57, 835–842. [Google Scholar] [CrossRef]
- Al-Saady, M.L.; Kaiser, C.S.; Wakasuqui, F.; Korenke, G.C.; Waisfisz, Q.; Polstra, A.; Pouwels, P.J.W.; Bugiani, M.; van der Knaap, M.S.; Lunsing, R.J.; et al. Homozygous UBA5 Variant Leads to Hypomyelination with Thalamic Involvement and Axonal Neuropathy. Neuropediatrics 2021, 52, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Lombet, A.; Planque, N.; Bleau, A.M.; Li, C.L.; Perbal, B. CCN3 and Calcium Signaling. Cell Commun. Signal. 2003, 1, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Vega Gallardo, N.; Dittmer, M.; Dombrowski, Y.; Fitzgerald, D.C. Regenerating CNS Myelin: Emerging Roles of Regulatory T Cells and CCN Proteins. Neurochem. Int. 2019, 130, 104349. [Google Scholar] [CrossRef] [Green Version]
- Naughton, M.; Moffat, J.; Eleftheriadis, G.; de la Vega Gallardo, N.; Young, A.; Falconer, J.; Hawkins, K.; Pearson, B.; Perbal, B.; Hogan, A.; et al. CCN3 Is Dynamically Regulated by Treatment and Disease State in Multiple Sclerosis. J. Neuroinflammation 2020, 17, 349. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Baek, I.J.; Kim, H.; Woo, D.K.; Park, Y.J.; Shim, S. CCN3 Overexpression Inhibits Growth of Callosal Projections via Upregulation of RAB25. Biochem. Biophys. Res. Commun. 2015, 461, 456–462. [Google Scholar] [CrossRef]
- Röhr, D.; Halfter, H.; Schulz, J.B.; Young, P.; Gess, B. Sodium-Dependent Vitamin C Transporter 2 Deficiency Impairs Myelination and Remyelination after Injury: Roles of Collagen and Demethylation. Glia 2017, 65, 1186–1200. [Google Scholar] [CrossRef] [PubMed]
- Senderek, J. Mutation of the SBF2 Gene, Encoding a Novel Member of the Myotubularin Family, in Charcot-Marie-Tooth Neuropathy Type 4B2/11p15. Hum. Mol. Genet. 2003, 12, 349–356. [Google Scholar] [CrossRef]
- Cunningham, G.M.; Shen, F.; Wu, X.; Cantor, E.L.; Gardner, L.; Philips, S.; Jiang, G.; Bales, C.L.; Tan, Z.; Liu, Y.; et al. The Impact of SBF2 on Taxane-Induced Peripheral Neuropathy. PLoS Genet. 2022, 18, e1009968. [Google Scholar] [CrossRef]
- Long, J.Z.; Cisar, J.S.; Milliken, D.; Niessen, S.; Wang, C.; Trauger, S.A.; Siuzdak, G.; Cravatt, B.F. Metabolomics Annotates ABHD3 as a Physiologic Regulator of Medium-Chain Phospholipids. Nat. Chem. Biol. 2011, 7, 763–765. [Google Scholar] [CrossRef] [Green Version]
- Calderon, R.O.; Attema, B.; DeVries, G.H. Lipid Composition of Neuronal Cell Bodies and Neurites from Cultured Dorsal Root Ganglia. J. Neurochem. 2002, 64, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Lauwers, E.; Goodchild, R.; Verstreken, P. Membrane Lipids in Presynaptic Function and Disease. Neuron 2016, 90, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Yang, L.; Liu, K.; Ji, X.; Tang, C.; Li, W.; Ma, L.; Mei, Y.; Peng, T.; Feng, B.; et al. Transcriptional Networks Identify Synaptotagmin-like 3 as a Regulator of Cortical Neuronal Migration during Early Neurodevelopment. Cell Rep. 2021, 34, 108802. [Google Scholar] [CrossRef]
- Johnson, J.L.; Monfregola, J.; Napolitano, G.; Kiosses, W.B.; Catz, S.D. Vesicular Trafficking through Cortical Actin during Exocytosis Is Regulated by the Rab27a Effector JFC1/Slp1 and the RhoA-GTPase–Activating Protein Gem-Interacting Protein. Mol. Biol. Cell 2012, 23, 1902–1916. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. Mitochondrial Transporters of the SLC25 Family and Associated Diseases: A Review. J. Inherit. Metab. Dis. 2014, 37, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Slabbaert, J.R.; Kuenen, S.; Swerts, J.; Maes, I.; Uytterhoeven, V.; Kasprowicz, J.; Fernandes, A.C.; Blust, R.; Verstreken, P. Shawn, the Drosophila Homolog of SLC25A39/40, Is a Mitochondrial Carrier That Promotes Neuronal Survival. J. Neurosci. 2016, 36, 1914–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holick, M.F. Vitamin D Deficiency. N. Engl. J. Med. 2007, 357, 266–281. [Google Scholar] [CrossRef] [PubMed]
- AlJohri, R.; AlOkail, M.; Haq, S.H. Neuroprotective Role of Vitamin D in Primary Neuronal Cortical Culture. eNeurologicalSci 2019, 14, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Poët, M.; Kornak, U.; Schweizer, M.; Zdebik, A.A.; Scheel, O.; Hoelter, S.; Wurst, W.; Schmitt, A.; Fuhrmann, J.C.; Planells-Cases, R.; et al. Lysosomal Storage Disease upon Disruption of the Neuronal Chloride Transport Protein ClC-6. Proc. Natl. Acad. Sci. USA 2006, 103, 13854–13859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jentsch, T.J.; Pusch, M. CLC Chloride Channels and Transporters: Structure, Function, Physiology, and Disease. Physiol. Rev. 2018, 98, 1493–1590. [Google Scholar] [CrossRef] [PubMed]
- Polovitskaya, M.M.; Barbini, C.; Martinelli, D.; Harms, F.L.; Cole, F.S.; Calligari, P.; Bocchinfuso, G.; Stella, L.; Ciolfi, A.; Niceta, M.; et al. A Recurrent Gain-of-Function Mutation in CLCN6, Encoding the ClC-6 Cl−/H+-Exchanger, Causes Early-Onset Neurodegeneration. Am. J. Hum. Genet. 2020, 107, 1062–1077. [Google Scholar] [CrossRef] [PubMed]
- Frias, B.; Merighi, A. Capsaicin, Nociception and Pain. Molecules 2016, 21, 797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evron, T.; Daigle, T.L.; Caron, M.G. GRK2: Multiple Roles beyond G Protein-Coupled Receptor Desensitization. Trends Pharmacol. Sci. 2012, 33, 154–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavelaars, A.; Eijkelkamp, N.; Willemen, H.L.D.M.; Wang, H.; Carbajal, A.G.; Heijnen, C.J. Microglial GRK2: A Novel Regulator of Transition from Acute to Chronic Pain. Brain. Behav. Immun. 2011, 25, 1055–1060. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Heijnen, C.J.; Eijkelkamp, N.; Carbajal, A.G.; Schedlowski, M.; Kelley, K.W.; Dantzer, R.; Kavelaars, A. GRK2 in Sensory Neurons Regulates Epinephrine-Induced Signalling and Duration of Mechanical Hyperalgesia. Pain 2011, 152, 1649–1658. [Google Scholar] [CrossRef]
- Wang, H.; Heijnen, C.J.; van Velthoven, C.T.J.; Willemen, H.L.D.M.; Ishikawa, Y.; Zhang, X.; Sood, A.K.; Vroon, A.; Eijkelkamp, N.; Kavelaars, A. Balancing GRK2 and EPAC1 Levels Prevents and Relieves Chronic Pain. J. Clin. Invest. 2013, 123, 5023–5034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eijkelkamp, N.; Heijnen, C.J.; Willemen, H.L.D.M.; Deumens, R.; Joosten, E.A.J.; Kleibeuker, W.; den Hartog, I.J.M.; van Velthoven, C.T.J.; Nijboer, C.; Nassar, M.A.; et al. GRK2: A Novel Cell-Specific Regulator of Severity and Duration of Inflammatory Pain. J. Neurosci. 2010, 30, 2138–2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latremoliere, A.; Latini, A.; Andrews, N.; Cronin, S.J.; Fujita, M.; Gorska, K.; Hovius, R.; Romero, C.; Chuaiphichai, S.; Painter, M.; et al. Reduction of Neuropathic and Inflammatory Pain through Inhibition of the Tetrahydrobiopterin Pathway. Neuron 2015, 86, 1393–1406. [Google Scholar] [CrossRef]
- Moore, B.J.R.; Islam, B.; Ward, S.; Jackson, O.; Armitage, R.; Blackburn, J.; Haider, S.; McHugh, P.C. Repurposing of Tranilast for Potential Neuropathic Pain Treatment by Inhibition of Sepiapterin Reductase in the BH4 Pathway. ACS Omega 2019, 4, 11960–11972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHugh, P.C.; Joyce, P.R.; Deng, X.; Kennedy, M.A. A Polymorphism of the GTP-Cyclohydrolase i Feedback Regulator Gene Alters Transcriptional Activity and May Affect Response to SSRI Antidepressants. Pharmacogenomics J. 2011, 11, 207–213. [Google Scholar] [CrossRef]
- Werner, E.R.; Blau, N.; Thöny, B. Tetrahydrobiopterin: Biochemistry and Pathophysiology. Biochem. J. 2011, 438, 397–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallingford, J.B.; Niswander, L.A.; Shaw, G.M.; Finnell, R.H. The Continuing Challenge of Understanding, Preventing, and Treating Neural Tube Defects. Science 2013, 339, 1222002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattson, M.P.; Shea, T.B. Folate and Homocysteine Metabolism in Neural Plasticity and Neurodegenerative Disorders. Trends Neurosci. 2003, 26, 137–146. [Google Scholar] [CrossRef]
- Iskandar, B.J.; Nelson, A.; Resnick, D.; Pate Skene, J.H.; Gao, P.; Johnson, C.; Cook, T.D.; Hariharan, N. Folic Acid Supplementation Enhances Repair of the Adult Central Nervous System. Ann. Neurol. 2004, 56, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Miranpuri, G.S.; Schomberg, D.T.; Alrfaei, B.; King, K.C.; Rynearson, B.; Wesley, V.S.; Khan, N.; Obiakor, K.; Wesley, U.V.; Resnick, D.K. Role of Matrix Metalloproteinases 2 in Spinal Cord Injury-Induced Neuropathic Pain. Ann. Neurosci. 2016, 23, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Apostolidis, A.; Gonzales, G.E.; Fowler, C.J. Effect of Intravesical Resiniferatoxin (RTX) on Lower Urinary Tract Symptoms, Urodynamic Parameters, and Quality of Life of Patients with Urodynamic Increased Bladder Sensation. Eur. Urol. 2006, 50, 1299–1305. [Google Scholar] [CrossRef]
- Giordano, J.; Schultea, T. Serotonin 5-HT(3) Receptor Mediation of Pain and Anti-Nociception: Implications for Clinical Therapeutics. Pain Physician 2004, 7, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Ramage, A.G. The Role of Central 5-Hydroxytryptamine (5-HT, Serotonin) Receptors in the Control of Micturition. Br. J. Pharmacol. 2006, 147, S120–S131. [Google Scholar] [CrossRef] [PubMed]
- Conley, R.K.; Williams, T.J.; Ford, A.P.D.W.; Ramage, A.G. The Role of α 1 -Adrenoceptors and 5-HT 1A Receptors in the Control of the Micturition Reflex in Male Anaesthetized Rats. Br. J. Pharmacol. 2001, 133, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Mbaki, Y.; Ramage, A.G. Investigation of the Role of 5-HT 2 Receptor Subtypes in the Control of the Bladder and the Urethra in the Anaesthetized Female Rat. Br. J. Pharmacol. 2008, 155, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Islas, E.; Vidal-Cantú, G.C.; Bravo-Hernández, M.; Cervantes-Durán, C.; Quiñonez-Bastidas, G.N.; Pineda-Farias, J.B.; Barragán-Iglesias, P.; Granados-Soto, V. Spinal 5-HT5A Receptors Mediate 5-HT-Induced Antinociception in Several Pain Models in Rats. Pharmacol. Biochem. Behav. 2014, 120, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Kruithof, P.D.; Lunev, S.; Aguilar Lozano, S.P.; de Assis Batista, F.; Al-dahmani, Z.M.; Joles, J.A.; Dolga, A.M.; Groves, M.R.; van Goor, H. Unraveling the Role of Thiosulfate Sulfurtransferase in Metabolic Diseases. Biochim. Biophys. Acta - Mol. Basis Dis. 2020, 1866, 165716. [Google Scholar] [CrossRef] [PubMed]
- Tiranti, V.; Viscomi, C.; Hildebrandt, T.; Di Meo, I.; Mineri, R.; Tiveron, C.; Levitt, M.D.; Prelle, A.; Fagiolari, G.; Rimoldi, M.; et al. Loss of ETHE1, a Mitochondrial Dioxygenase, Causes Fatal Sulfide Toxicity in Ethylmalonic Encephalopathy. Nat. Med. 2009, 15, 200–205. [Google Scholar] [CrossRef]
- Nicholas, S.; Yuan, S.Y.; Brookes, S.J.H.; Spencer, N.J.; Zagorodnyuk, V.P. Hydrogen Peroxide Preferentially Activates Capsaicin-Sensitive High Threshold Afferents via TRPA1 Channels in the Guinea Pig Bladder. Br. J. Pharmacol. 2017, 174, 126–138. [Google Scholar] [CrossRef] [Green Version]
- Andersson, K.-E. TRP Channels as Lower Urinary Tract Sensory Targets. Med. Sci. 2019, 7, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streng, T.; Axelsson, H.E.; Hedlund, P.; Andersson, D.A.; Jordt, S.-E.; Bevan, S.; Andersson, K.-E.; Högestätt, E.D.; Zygmunt, P.M. Distribution and Function of the Hydrogen Sulfide–Sensitive TRPA1 Ion Channel in Rat Urinary Bladder. Eur. Urol. 2008, 53, 391–400. [Google Scholar] [CrossRef]
- Bjorling, D.E.; Wang, Z.Y. Potential of Endocannabinoids to Control Bladder Pain. Front. Syst. Neurosci. 2018, 12, 17. [Google Scholar] [CrossRef] [Green Version]
- Blankman, J.L.; Long, J.Z.; Trauger, S.A.; Siuzdak, G.; Cravatt, B.F. ABHD12 Controls Brain Lysophosphatidylserine Pathways That Are Deregulated in a Murine Model of the Neurodegenerative Disease PHARC. Proc. Natl. Acad. Sci. USA 2013, 110, 1500–1505. [Google Scholar] [CrossRef]
- Jenstad, M.; Quazi, A.Z.; Zilberter, M.; Haglerød, C.; Berghuis, P.; Saddique, N.; Goiny, M.; Buntup, D.; Davanger, S.; Haug, F.-M.S.; et al. System A Transporter SAT2 Mediates Replenishment of Dendritic Glutamate Pools Controlling Retrograde Signaling by Glutamate. Cereb. Cortex 2009, 19, 1092–1106. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Dong, L.; Sun, B.; Guillon, M.A.; Burbach, L.R.; Nunn, P.A.; Liu, X.; Vilenski, O.; Ford, A.P.D.W.; Zhong, Y.; et al. The Role of Metabotropic Glutamate Receptor MGlu5 in Control of Micturition and Bladder Nociception. Neurosci. Lett. 2009, 450, 12–17. [Google Scholar] [CrossRef]
- Ostrovskaya, O.; Xie, K.; Masuho, I.; Fajardo-Serrano, A.; Lujan, R.; Wickman, K.; Martemyanov, K.A. RGS7/Gβ5/R7BP Complex Regulates Synaptic Plasticity and Memory by Modulating Hippocampal GABABR-GIRK Signaling. eLife 2014, 3, e02053. [Google Scholar] [CrossRef] [PubMed]
- Gerber, K.J.; Squires, K.E.; Hepler, J.R. Roles for Regulator of G Protein Signaling Proteins in Synaptic Signaling and Plasticity. Mol. Pharmacol. 2016, 89, 273–286. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.F.; Doyle, P.T.; Ferguson, D.R. Inhibition in the Human Urinary Bladder by Gamma-Amino-Butyric Acid. Br. J. Urol. 1994, 73, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Witherow, D.S.; Wang, Q.; Levay, K.; Cabrera, J.L.; Chen, J.; Willars, G.B.; Slepak, V.Z. Complexes of the G Protein Subunit Gβ5 with the Regulators of G Protein Signaling RGS7 and RGS9. J. Biol. Chem. 2000, 275, 24872–24880. [Google Scholar] [CrossRef] [Green Version]
- Sandiford, S.L.; Slepak, V.Z. The Gβ 5 −RGS7 Complex Selectively Inhibits Muscarinic M3 Receptor Signaling via the Interaction between the Third Intracellular Loop of the Receptor and the DEP Domain of RGS7. Biochemistry 2009, 48, 2282–2289. [Google Scholar] [CrossRef] [Green Version]
- Shuey, D.J.; Betty, M.; Jones, P.G.; Khawaja, X.Z.; Cockett, M.I. RGS7 Attenuates Signal Transduction Through the Gαq Family of Heterotrimeric G Proteins in Mammalian Cells. J. Neurochem. 2002, 70, 1964–1972. [Google Scholar] [CrossRef]
- Chess-Williams, R. Muscarinic Receptors of the Urinary Bladder: Detrusor, Urothelial and Prejunctional. Auton. Autacoid Pharmacol. 2002, 22, 133–145. [Google Scholar] [CrossRef] [PubMed]



| RNA-Seq Libraries | Raw Reads | Trimmed Reads | Mapped Reads | Uniquely Mapped Reads | Multi-Mapped Reads | Too Many Loci |
|---|---|---|---|---|---|---|
| CTR1 | 63,450,432 | 53,906,522 | 51,882,476 | 50,773,306 | 1,065,278 | 43,892 |
| CTR2 | 63,018,144 | 52,441,270 | 50,719,276 | 49,751,418 | 941,066 | 26,792 |
| CTR3 | 63,747,778 | 54,451,852 | 51,875,622 | 50,745,164 | 1,099,474 | 30,984 |
| CTR4 | 63,650,872 | 53,024,696 | 50,606,392 | 49,420,902 | 1,156,814 | 28,676 |
| CTR5 | 64,447,908 | 53,525,868 | 52,020,672 | 51,016,528 | 974,894 | 29,250 |
| CTR6 | 61,433,064 | 51,078,746 | 49,682,244 | 48,575,512 | 1,074,524 | 32,208 |
| RTX1 | 62,806,456 | 51,157,178 | 49,658,774 | 48,627,582 | 996,796 | 34,396 |
| RTX2 | 61,901,726 | 51,796,642 | 48,881,448 | 47,799,714 | 1,050,198 | 31,536 |
| RTX3 | 62,638,642 | 52,315,264 | 49,903,750 | 48,713,644 | 1,162,622 | 27,484 |
| RTX4 | 62,536,662 | 51,887,614 | 50,170,978 | 48,818,308 | 1,313,042 | 39,628 |
| RTX5 | 62,688,462 | 51,465,770 | 50,012,444 | 48,808,958 | 1,171,510 | 31,976 |
| RTX6 | 61,475,200 | 50,497,818 | 47,752,176 | 46,484,026 | 1,229,930 | 38,220 |
| Gene ID | Gene Name | log2FoldChange | p-Adjusted |
|---|---|---|---|
| MSTRG.18558 | SLAMF7 | 25.47 | 7.96 × 10−14 |
| MSTRG.24634 | SUPT3H | 25.30 | 9.68 × 10−14 |
| MSTRG.14735 | NCLN | 25.29 | 9.68 × 10−14 |
| MSTRG.15606 | ARHGEF37 | 24.25 | 1.13 × 10−12 |
| MSTRG.18613 | FCRL3 | 23.54 | 5.68 × 10−12 |
| MSTRG.13448 | OSBPL5 | 23.30 | 9.30 × 10−12 |
| MSTRG.22604 | CLCN6 | 23.18 | 9.71 × 10−18 |
| MSTRG.4591 | LIMD2 | 22.96 | 2.08 × 10−11 |
| MSTRG.27341 | SLC35F2 | 22.92 | 2.22 × 10−11 |
| MSTRG.7236 | MIX23 | 21.70 | 4.42 × 10−10 |
| MSTRG.20205 | ITPR2 | 21.42 | 8.65 × 10−10 |
| MSTRG.137 | SYTL3 | 21.28 | 1.17 × 10−9 |
| MSTRG.28195 | SEC16B | 8.31 | 0.0036 |
| MSTRG.19812 | MFSD5 | 7.62 | 0.0403 |
| MSTRG.13163 | GSDME | 7.43 | 0.0105 |
| MSTRG.1415 | PDCD7 | 7.30 | 0.0436 |
| ENSSSCG00000002479 | SERPINA11 | 6.81 | 0.0006 |
| MSTRG.13644 | ENSSSCG00000039740 | 6.03 | 0.0003 |
| MSTRG.2732 | GPATCH2 | 5.04 | 0.0002 |
| MSTRG.19650 | TST | 5.03 | 0.0389 |
| MSTRG.28205 | RALGPS2 | 4.55 | 0.0015 |
| MSTRG.9190 | CYP2C42 | 4.48 | 0.0059 |
| MSTRG.2418 | PTGS1 | 4.06 | 0.0356 |
| MSTRG.19993 | CYP27B1 | 3.79 | 0.0221 |
| MSTRG.18574 | ATP1A4 | 3.66 | 0.0328 |
| MSTRG.1486 | CALML4 | 3.13 | 0.0004 |
| MSTRG.29522 | ENSSSCG00000047701 | 2.78 | 0.0214 |
| MSTRG.16166 | CORO1A | 2.58 | 9.19 × 10−5 |
| MSTRG.26707 | ENSSSCG00000009182 | 2.53 | 0.0075 |
| MSTRG.8288 | SUDS3 | 2.46 | 0.0018 |
| MSTRG.15899 | MEPCE | 2.41 | 0.0037 |
| MSTRG.7148 | MELTF | 2.29 | 0.0434 |
| MSTRG.12780 | AKR1B1 | 1.89 | 0.0026 |
| MSTRG.4949 | ENSSSCG00000040110 | 1.58 | 0.0015 |
| MSTRG.4430 | SAP30BP | 1.58 | 0.0352 |
| MSTRG.18428 | SPIDR | 1.31 | 0.0434 |
| MSTRG.12716 | ZC3HAV1L | 1.09 | 0.0101 |
| MSTRG.9033 | ADIRF | 1.05 | 0.0036 |
| MSTRG.16386 | C16orf89 | 1.02 | 0.0221 |
| MSTRG.19607 | TMEM184B | 0.95 | 0.0136 |
| MSTRG.5521 | SAT2 | 0.89 | 0.0483 |
| MSTRG.14799 | ENSSSCG00000026387 | 0.89 | 0.0224 |
| MSTRG.21677 | ENSSSCG00000028943 | 0.85 | 2.31 × 10−15 |
| MSTRG.18711 | IL6R | 0.69 | 0.0185 |
| MSTRG.22744 | ENSSSCG00000029242 | 0.67 | 0.0283 |
| MSTRG.10953 | DUSP28 | 0.62 | 0.0227 |
| MSTRG.2338 | HDHD3 | 0.60 | 0.0399 |
| MSTRG.11546 | ENSSSCG00000026414 | 0.59 | 0.0137 |
| MSTRG.4676 | SLC25A39 | 0.53 | 0.0337 |
| MSTRG.11916 | ABHD12 | 0.53 | 0.0032 |
| MSTRG.22082 | ACP4 | 0.50 | 0.0062 |
| MSTRG.13531 | GRK2 | 0.48 | 0.0014 |
| MSTRG.24570 | CNPY3 | 0.42 | 0.0137 |
| MSTRG.8629 | ENSSSCG00000036280 | 0.40 | 0.0428 |
| Gene ID | Gene Name | log2FoldChange | p−Adjusted |
|---|---|---|---|
| MSTRG.15089 | ARRDC3 | −27.24 | 1.70 × 10−15 |
| MSTRG.16274 | VPS35L | −25.90 | 5.74 × 10−14 |
| MSTRG.16760 | RANBP2 | −25.69 | 5.74 × 10−14 |
| MSTRG.10097 | ARL6IP6 | −25.69 | 5.74 × 10−14 |
| MSTRG.22915 | FABP3 | −25.51 | 7.96 × 10−14 |
| MSTRG.21796 | ETHE1 | −25.41 | 8.44 × 10−14 |
| MSTRG.10408 | HIBCH | −25.18 | 1.23 × 10−13 |
| MSTRG.22503 | LRRC47 | −24.99 | 1.93 × 10−13 |
| MSTRG.15319 | ADAMTS19 | −24.68 | 4.27 × 10−13 |
| MSTRG.12082 | PROCR | −24.61 | 4.90 × 10−13 |
| MSTRG.7356 | NECTIN3 | −24.51 | 5.96 × 10−13 |
| MSTRG.5103 | BRIP1 | −24.23 | 1.13 × 10−12 |
| MSTRG.3245 | SUV39H2 | −24.09 | 1.57 × 10−12 |
| MSTRG.12889 | ASB15 | −23.88 | 2.67 × 10−12 |
| MSTRG.2979 | RABIF | −23.62 | 5.12 × 10−12 |
| MSTRG.24689 | CILK1 | −23.55 | 5.68 × 10−12 |
| MSTRG.14163 | SBF2 | −23.49 | 6.31 × 10−12 |
| MSTRG.6267 | ITIH3 | −23.36 | 8.48 × 10−12 |
| MSTRG.15168 | NUDT12 | −23.34 | 8.76 × 10−12 |
| MSTRG.17031 | SPR | −23.26 | 9.96 × 10−12 |
| MSTRG.4745 | CNTNAP1 | −23.16 | 5.74 × 10−14 |
| MSTRG.12395 | PCK1 | −22.59 | 5.18 × 10−11 |
| MSTRG.1522 | LRRC49 | −22.53 | 5.88 × 10−11 |
| MSTRG.8812 | CABCOCO1 | −20.62 | 5.39 × 10−9 |
| MSTRG.3953 | DIAPH3 | −11.41 | 2.52 × 10−5 |
| MSTRG.29959 | NDUFA1 | −11.39 | 9.13 × 10−7 |
| MSTRG.6637 | UBA5 | −10.47 | 7.95 × 10−6 |
| MSTRG.11212 | MOCS2 | −10.45 | 7.86 × 10−8 |
| MSTRG.25491 | ANGEL1 | −9.75 | 0.0013 |
| MSTRG.12280 | SLC13A3 | −9.71 | 0.0151 |
| MSTRG.25217 | PARP2 | −9.64 | 0.0018 |
| MSTRG.28269 | LAMC1 | −9.45 | 2.65 × 10−7 |
| MSTRG.2788 | RGS7 | −9.23 | 0.0018 |
| MSTRG.26818 | SCD5 | −8.80 | 0.0032 |
| MSTRG.17952 | CCN3 | −8.80 | 0.0033 |
| MSTRG.9028 | BMPR1A | −8.78 | 0.0092 |
| MSTRG.16749 | SNRNP200 | −8.72 | 7.95 × 10−6 |
| MSTRG.28308 | TPR | −8.64 | 9.15 × 10−7 |
| MSTRG.10410 | MFSD6 | −8.63 | 1.35 × 10−5 |
| MSTRG.7732 | PKNOX1 | −8.61 | 0.0003 |
| MSTRG.3103 | B4GALT1 | −8.59 | 0.0037 |
| MSTRG.25925 | ZCCHC4 | −7.93 | 0.0150 |
| MSTRG.5125 | ENSSSCG00000027233 | −7.67 | 0.0005 |
| MSTRG.23226 | ABHD3 | −7.52 | 0.0021 |
| MSTRG.11786 | SLC23A2 | −7.27 | 0.0007 |
| MSTRG.13311 | KRBA1 | −7.08 | 0.0293 |
| MSTRG.29606 | STARD8 | −6.20 | 7.95 × 10−6 |
| MSTRG.4433 | ENSSSCG00000036248 | −6.17 | 0.0004 |
| MSTRG.28999 | SAPCD2 | −5.98 | 0.0101 |
| MSTRG.8518 | OSBP2 | −5.88 | 1.08 × 10−5 |
| ENSSSCG00000032669 | ENSSSCG00000032669 | −5.86 | 0.0319 |
| ENSSSCG00000008166 | CREG2 | −5.69 | 0.0289 |
| MSTRG.15782 | RBAK | −5.53 | 0.0023 |
| MSTRG.2091 | FANCG | −5.19 | 0.0032 |
| MSTRG.17449 | KRTCAP3 | −4.32 | 0.0004 |
| MSTRG.24821 | ACAN | −4.00 | 0.0400 |
| MSTRG.21369 | CENPT | −3.90 | 0.0403 |
| MSTRG.21518 | KCTD15 | −3.60 | 0.0488 |
| MSTRG.25897 | NCAPG | −3.54 | 0.0019 |
| MSTRG.27421 | USP28 | −3.24 | 0.0485 |
| MSTRG.17940 | SHAS2 | −2.88 | 0.0224 |
| MSTRG.7109 | LPP | −2.46 | 0.0473 |
| MSTRG.8347 | RITA1 | −2.30 | 0.0164 |
| MSTRG.8476 | XBP1 | −2.29 | 0.0214 |
| MSTRG.28939 | ZMYND19 | −2.03 | 0.0321 |
| MSTRG.9945 | CENPU | −1.98 | 0.0224 |
| MSTRG.27587 | ROBO3 | −1.73 | 0.0149 |
| MSTRG.27235 | NA | −1.66 | 0.0291 |
| MSTRG.11646 | ENSSSCG00000043933 | −1.34 | 0.0410 |
| MSTRG.2104 | GBA2 | −1.26 | 0.0164 |
| MSTRG.924 | FAM81A | −1.09 | 0.0429 |
| MSTRG.20134 | YARS2 | −0.85 | 0.0403 |
| MSTRG.3339 | ENSSSCG00000044246 | −0.71 | 0.0494 |
| MSTRG.2389 | ENSSSCG00000039972 | −0.36 | 0.0095 |
| MSTRG.24246 | ENSSSCG00000021069 | −0.34 | 0.0189 |
| Term ID | KEGG Name | p-Adjusted * | Gene Number | Gene Symbol |
|---|---|---|---|---|
| KEGG:00790 | Folate biosynthesis | 0.0224 | 3 | SPR, MOCS2, AKR1B1 |
| KEGG:01100 | Metabolic pathways | 0.0621 | 18 | ETHE1, HIBCH, SUV39H2, NUDT12, SPR, PCK1, NDUFA1, MOCS2, SCD5, B4GALT1, GBA2, SAT2, AKR1B1, ENSSSCG00000009182, CYP27B1, PTGS1, CYP2C42, TST |
| KEGG:04122 | Sulfur relay system | 0.0845 | 2 | MOCS2,TST |
| KEGG:00920 | Sulfur metabolism | 0.1052 | 2 | ETHE1,TST |
| KEGG:04726 | Serotonergic synapse | 0.2291 | 4 | ENSSSCG00000040110, PTGS1, CYP2C42, ITPR2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lepiarczyk, E.; Paukszto, Ł.; Wiszpolska, M.; Łopieńska-Biernat, E.; Bossowska, A.; Majewski, M.K.; Majewska, M. Molecular Influence of Resiniferatoxin on the Urinary Bladder Wall Based on Differential Gene Expression Profiling. Cells 2023, 12, 462. https://doi.org/10.3390/cells12030462
Lepiarczyk E, Paukszto Ł, Wiszpolska M, Łopieńska-Biernat E, Bossowska A, Majewski MK, Majewska M. Molecular Influence of Resiniferatoxin on the Urinary Bladder Wall Based on Differential Gene Expression Profiling. Cells. 2023; 12(3):462. https://doi.org/10.3390/cells12030462
Chicago/Turabian StyleLepiarczyk, Ewa, Łukasz Paukszto, Marta Wiszpolska, Elżbieta Łopieńska-Biernat, Agnieszka Bossowska, Mariusz Krzysztof Majewski, and Marta Majewska. 2023. "Molecular Influence of Resiniferatoxin on the Urinary Bladder Wall Based on Differential Gene Expression Profiling" Cells 12, no. 3: 462. https://doi.org/10.3390/cells12030462
APA StyleLepiarczyk, E., Paukszto, Ł., Wiszpolska, M., Łopieńska-Biernat, E., Bossowska, A., Majewski, M. K., & Majewska, M. (2023). Molecular Influence of Resiniferatoxin on the Urinary Bladder Wall Based on Differential Gene Expression Profiling. Cells, 12(3), 462. https://doi.org/10.3390/cells12030462






