Djck1α Is Required for Proper Regeneration and Maintenance of the Medial Tissues in Planarians
Abstract
:1. Introduction
2. Materials and Methods
2.1. Planarian Culture
2.2. Gene Identification and Cloning
2.3. RNAi Experiments
2.4. Whole-Mount Immunostaining
2.5. In Situ Hybridization
2.6. Quantitative RT-qPCR
2.7. Statistical Analysis
3. Results
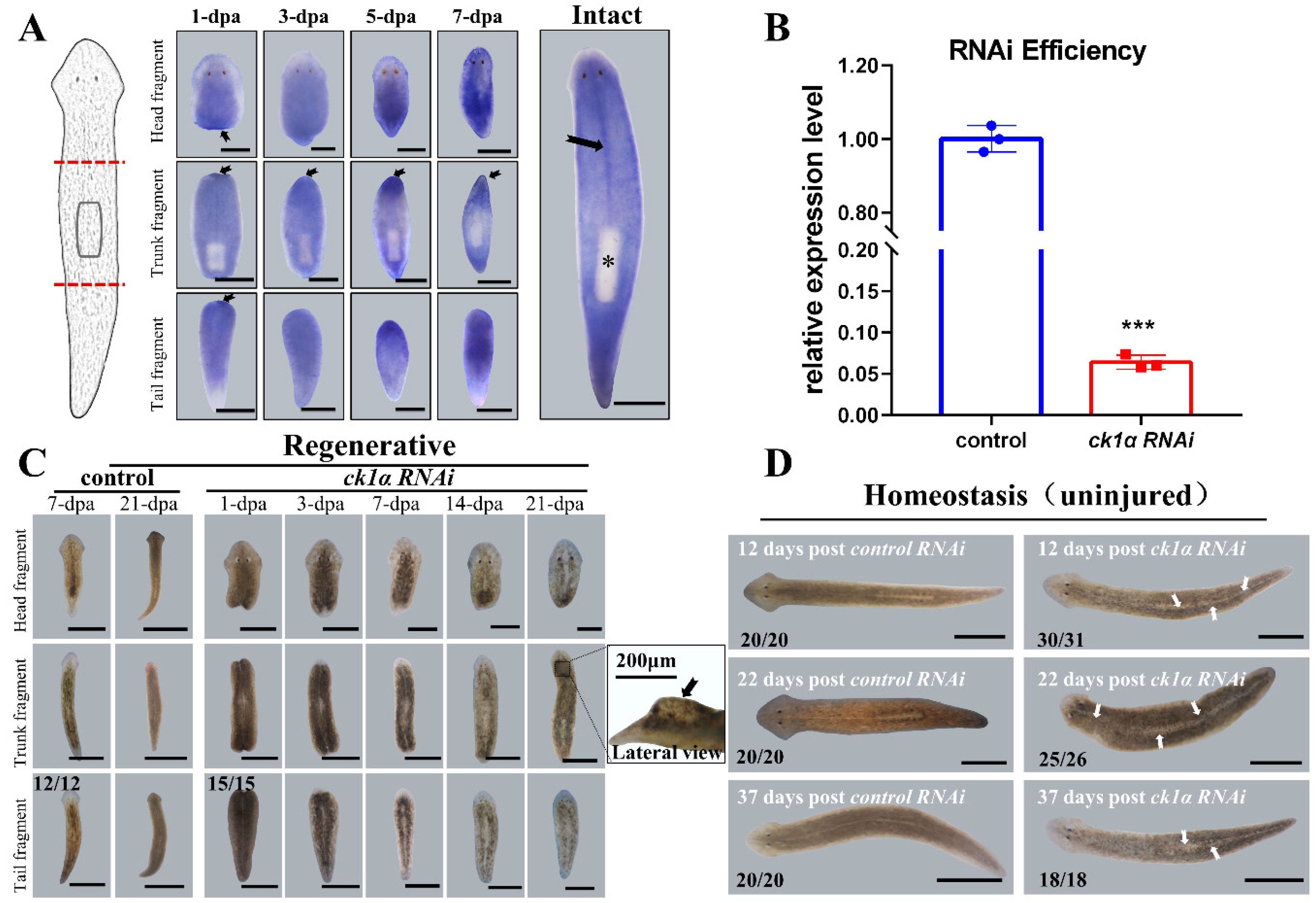
3.1. Djck1α Expresses in the Middle and Is Required for Normal Tissue Regeneration and Maintenance
3.2. Djck1α Inhibition Causes Abnormalities in Nervous, Intestine, and Epidermis Systems during Regeneration
3.3. Djck1α RNAi Results in Hyper-Proliferation
3.4. Inhibition of Djck1α Affects Tissue Turnover and Generates Medial Tissue Dilation in Intact Planarians
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lemeer, S.; Heck, A.J. The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 2009, 13, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Sikes, R.A. Chemistry and pharmacology of anticancer drugs. Br. J. Cancer 2007, 97, 1713. [Google Scholar] [CrossRef]
- Cheong, J.K.; Virshup, D.M. Casein kinase 1: Complexity in the family. Int. J. Biochem. Cell. Biol. 2011, 43, 465–469. [Google Scholar] [CrossRef]
- Gross, S.D.; Anderson, R.A. Casein kinase I: Spatial organization and positioning of a multifunctional protein kinase family. Cell Signal 1998, 10, 699–711. [Google Scholar] [CrossRef] [PubMed]
- DeMaggio, A.J.; Lindberg, R.A.; Hunter, T.; Hoekstra, M.F. The budding yeast HRR25 gene product is a casein kinase I isoform. Proc. Natl. Acad. Sci. USA 1992, 89, 7008–7012. [Google Scholar] [CrossRef]
- Dhillon, N.; Hoekstra, M.F. Characterization of two protein kinases from Schizosaccharomyces pombe involved in the regulation of DNA repair. EMBO J. 1994, 13, 2777–2788. [Google Scholar] [CrossRef]
- Robinson, L.C.; Hubbard, E.J.; Graves, P.R.; DePaoli-Roach, A.A.; Roach, P.J.; Kung, C.; Haas, D.W.; Hagedorn, C.H.; Goebl, M.; Culbertson, M.R.; et al. Yeast casein kinase I homologues: An essential gene pair. Proc. Natl. Acad. Sci. USA 1992, 89, 28–32. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013, 23, 186–200. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Knippschild, U.; Gocht, A.; Wolff, S.; Huber, N.; Lohler, J.; Stoter, M. The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell Signal 2005, 17, 675–689. [Google Scholar] [CrossRef]
- Walczak, C.E.; Anderson, R.A.; Nelson, D.L. Identification of a family of casein kinases in Paramecium: Biochemical characterization and cellular localization. Biochem. J. 1993, 296 Pt 3, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Kearney, P.H.; Ebert, M.; Kuret, J. Molecular cloning and sequence analysis of two novel fission yeast casein kinase-1 isoforms. Biochem. Biophys. Res. Commun. 1994, 203, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.C.; Vancura, A.; Mitcheson, T.G.; Kuret, J. Two genes in Saccharomyces cerevisiae encode a membrane-bound form of casein kinase-1. Mol. Biol. Cell 1992, 3, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Plowman, G.D.; Sudarsanam, S.; Bingham, J.; Whyte, D.; Hunter, T. The protein kinases of Caenorhabditis elegans: A model for signal transduction in multicellular organisms. Proc. Natl. Acad. Sci. USA 1999, 96, 13603–13610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jia, J.; Wang, B.; Amanai, K.; Wharton, K.A., Jr.; Jiang, J. Regulation of wingless signaling by the CKI family in Drosophila limb development. Dev. Biol. 2006, 299, 221–237. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.K.; Murakami, M.S.; Cleghon, V. Protein kinases and phosphatases in the Drosophila genome. J. Cell Biol. 2000, 150, F57–F62. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef] [PubMed]
- Graves, P.R.; Haas, D.W.; Hagedorn, C.H.; DePaoli-Roach, A.A.; Roach, P.J. Molecular cloning, expression, and characterization of a 49-kilodalton casein kinase I isoform from rat testis. J. Biol. Chem. 1993, 268, 6394–6401. [Google Scholar] [CrossRef] [PubMed]
- Lubben, T.H.; Traugh, J.A. Cyclic nucleotide-independent protein kinases from rabbit reticulocytes. Purification and characterization of protease-activated kinase II. J. Biol. Chem. 1983, 258, 13992–13997. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; McKay, R.M.; McKay, J.P.; Graff, J.M. Casein kinase I transduces Wnt signals. Nature 1999, 401, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.; Chen, X.; Lin, S.Y.; Slack, F.J. kin-19/casein kinase Ialpha has dual functions in regulating asymmetric division and terminal differentiation in C. elegans epidermal stem cells. Cell Cycle 2010, 9, 4748–4765. [Google Scholar] [CrossRef] [PubMed]
- Bozatzi, P.; Dingwell, K.S.; Wu, K.Z.; Cooper, F.; Cummins, T.D.; Hutchinson, L.D.; Vogt, J.; Wood, N.T.; Macartney, T.J.; Varghese, J.; et al. PAWS1 controls Wnt signalling through association with casein kinase 1alpha. EMBO Rep. 2018, 19, e44807. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.E.; Wang, I.E.; Reddien, P.W. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 2011, 332, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.; Li, H.; Guo, L.; Gao, X.; McKinney, S.; Wang, Y.; Yu, Z.; Park, J.; Semerad, C.; Ross, E.; et al. Prospectively Isolated Tetraspanin(+) Neoblasts Are Adult Pluripotent Stem Cells Underlying Planaria Regeneration. Cell 2018, 173, 1593–1608 e1520. [Google Scholar] [CrossRef] [PubMed]
- Reddien, P.W.; Sanchez Alvarado, A. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 2004, 20, 725–757. [Google Scholar] [CrossRef] [PubMed]
- Salo, E. The power of regeneration and the stem-cell kingdom: Freshwater planarians (Platyhelminthes). Bioessays 2006, 28, 546–559. [Google Scholar] [CrossRef]
- Hyman, L.H. The Invertebrates: Platyhelminthes and Rhynchocoela:The Acoelomate Bilateria. Q. Rev. Biol. 1951, 2, 550. [Google Scholar]
- Reddien, P.W. The Cellular and Molecular Basis for Planarian Regeneration. Cell 2018, 175, 327–345. [Google Scholar] [CrossRef]
- Newmark, P.A.; Sanchez Alvarado, A. Not your father’s planarian: A classic model enters the era of functional genomics. Nat. Rev. Genet. 2002, 3, 210–219. [Google Scholar] [CrossRef]
- Agata, K. Regeneration and gene regulation in planarians. Curr. Opin. Genet. Dev. 2003, 13, 492–496. [Google Scholar] [CrossRef]
- Sánchez Alvarado, A. Planarian Regeneration: Its End Is Its Beginning. Cell 2006, 124, 241–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddien, P.W.; Oviedo, N.J.; Jennings, J.R.; Jenkin, J.C.; Sánchez Alvarado, A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 2005, 310, 1327–1330. [Google Scholar] [CrossRef] [PubMed]
- Lapan, S.W.; Reddien, P.W. dlx and sp6-9 Control optic cup regeneration in a prototypic eye. PLoS Genet. 2011, 7, e1002226. [Google Scholar] [CrossRef] [PubMed]
- Witchley, J.N.; Mayer, M.; Wagner, D.E.; Owen, J.H.; Reddien, P.W. Muscle cells provide instructions for planarian regeneration. Cell Rep. 2013, 4, 633–641. [Google Scholar] [CrossRef]
- Scimone, M.L.; Cote, L.E.; Rogers, T.; Reddien, P.W. Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife 2016, 5, e12845. [Google Scholar] [CrossRef]
- Fincher, C.T.; Wurtzel, O.; de Hoog, T.; Kravarik, K.M.; Reddien, P.W. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 2018, 360, eaaq1736. [Google Scholar] [CrossRef]
- Reddien, P.W. Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 2011, 27, 277–285. [Google Scholar] [CrossRef]
- Umesono, Y.; Tasaki, J.; Nishimura, Y.; Hrouda, M.; Kawaguchi, E.; Yazawa, S.; Nishimura, O.; Hosoda, K.; Inoue, T.; Agata, K. The molecular logic for planarian regeneration along the anterior-posterior axis. Nature 2013, 500, 73–76. [Google Scholar] [CrossRef]
- Yazawa, S.; Umesono, Y.; Hayashi, T.; Tarui, H.; Agata, K. Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 22329–22334. [Google Scholar] [CrossRef]
- Gurley, K.A.; Rink, J.C.; Sanchez Alvarado, A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 2008, 319, 323–327. [Google Scholar] [CrossRef]
- Petersen, C.P.; Reddien, P.W. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 2008, 319, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, C.; Saito, Y.; Ogawa, K.; Agata, K. Wnt signaling is required for antero-posterior patterning of the planarian brain. Dev. Biol. 2007, 306, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Adell, T.; Salo, E.; Boutros, M.; Bartscherer, K. Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development 2009, 136, 905–910. [Google Scholar] [CrossRef]
- Gurley, K.A.; Elliott, S.A.; Simakov, O.; Schmidt, H.A.; Holstein, T.W.; Sanchez Alvarado, A. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 2010, 347, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.D.; Salo, E.; Cebria, F. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev. Biol. 2007, 311, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Gavino, M.A.; Reddien, P.W. A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr. Biol. 2011, 21, 294–299. [Google Scholar] [CrossRef]
- Cebria, F.; Guo, T.; Jopek, J.; Newmark, P.A. Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev. Biol. 2007, 307, 394–406. [Google Scholar] [CrossRef]
- Tian, Q.N.; Bao, Z.X.; Lu, P.; Qin, Y.F.; Chen, S.J.; Liang, F.; Mai, J.; Zhao, J.M.; Zhu, Z.Y.; Zhang, Y.Z.; et al. Differential expression of microRNA patterns in planarian normal and regenerative tissues. Mol. Biol. Rep. 2012, 39, 2653–2658. [Google Scholar] [CrossRef]
- Tian, Q.; Sun, Y.; Gao, T.; Li, J.; Fang, H.; Zhang, S. Djnedd4L Is Required for Head Regeneration by Regulating Stem Cell Maintenance in Planarians. Int. J. Mol. Sci. 2021, 22, 11707. [Google Scholar] [CrossRef]
- Tian, Q.; Guo, Q.; Guo, Y.; Luo, L.; Kristiansen, K.; Han, Z.; Fang, H.; Zhang, S. Whole-genome sequence of the planarian Dugesia japonica combining Illumina and PacBio data. Genomics 2022, 114, 110293. [Google Scholar] [CrossRef]
- Rouhana, L.; Weiss, J.A.; Forsthoefel, D.J.; Lee, H.; King, R.S.; Inoue, T.; Shibata, N.; Agata, K.; Newmark, P.A. RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: Methodology and dynamics. Dev. Dyn. 2013, 242, 718–730. [Google Scholar] [CrossRef] [Green Version]
- Cebria, F.; Newmark, P.A. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 2005, 132, 3691–3703. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhao, G.; Ni, J.; Guo, Y.; Zhang, Y.; Tian, Q.; Zhang, S. Down-regulate of Djrfc2 causes tissues hypertrophy during planarian regeneration. Biochem. Biophys. Res. Commun. 2017, 493, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Zhang, Y.; Song, G.; Wu, M.; Chen, G. Identification of Autophagy-Related Gene 7 and Autophagic Cell Death in the Planarian Dugesia japonica. Front. Physiol. 2018, 9, 1223. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego Calif.) 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.J., 3rd; Hou, X.; Romanova, E.V.; Lambrus, B.G.; Miller, C.M.; Saberi, A.; Sweedler, J.V.; Newmark, P.A. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010, 8, e1000509. [Google Scholar] [CrossRef] [PubMed]
- Cebria, F.; Kobayashi, C.; Umesono, Y.; Nakazawa, M.; Mineta, K.; Ikeo, K.; Gojobori, T.; Itoh, M.; Taira, M.; Sanchez Alvarado, A.; et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 2002, 419, 620–624. [Google Scholar] [CrossRef]
- Cebria, F.; Newmark, P.A. Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development 2007, 134, 833–837. [Google Scholar] [CrossRef]
- Pearson, B.J.; Sánchez Alvarado, A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 2010, 137, 213–221. [Google Scholar] [CrossRef]
- Tazaki, A.; Kato, K.; Orii, H.; Agata, K.; Watanabe, K. The body margin of the planarian Dugesia japonica: Characterization by the expression of an intermediate filament gene. Dev. Genes Evol. 2002, 212, 365–373. [Google Scholar] [CrossRef]
- Tu, K.C.; Cheng, L.C.; TK Vu, H.; Lange, J.J.; McKinney, S.A.; Seidel, C.W.; Sanchez Alvarado, A. Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. Elife 2015, 4, e10501. [Google Scholar] [CrossRef]
- Tian, Q.; Zhao, G.; Sun, Y.; Yuan, D.; Guo, Q.; Zhang, Y.; Liu, J.; Zhang, S. Exportin-1 is required for the maintenance of the planarian epidermal lineage. Int. J. Biol. Macromol. 2019, 126, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Oderberg, I.M.; Reddien, P.W. Planarian Epidermal Stem Cells Respond to Positional Cues to Promote Cell-Type Diversity. Dev. Cell 2017, 40, 491–504.e495. [Google Scholar] [CrossRef]
- Sakai, T.; Kato, K.; Watanabe, K.; Orii, H. Planarian pharynx regeneration revealed by the expression of myosin heavy chain-A. Int. J. Dev. Biol. 2002, 46, 329–332. [Google Scholar] [PubMed]
- Fuchs, E.; Chen, T. A matter of life and death: Self-renewal in stem cells. EMBO Rep. 2013, 14, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.Y.T.; Pearson, B.J. Yorkie is required to restrict the injury responses in planarians. PLoS Genet. 2017, 13, e1006874. [Google Scholar] [CrossRef]
- Rossi, L.; Salvetti, A.; Lena, A.; Batistoni, R.; Deri, P.; Pugliesi, C.; Loreti, E.; Gremigni, V. DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Dev. Genes Evol. 2006, 216, 335–346. [Google Scholar] [CrossRef]
- Jiang, J. CK1 in Developmental Signaling: Hedgehog and Wnt. Curr. Top. Dev. Biol. 2017, 123, 303–329. [Google Scholar] [CrossRef]
- Janovska, P.; Normant, E.; Miskin, H.; Bryja, V. Targeting Casein Kinase 1 (CK1) in Hematological Cancers. Int. J. Mol. Sci. 2020, 21, 9026. [Google Scholar] [CrossRef]
- Albornoz, A.; Yáñez, J.M.; Foerster, C.; Aguirre, C.; Pereiro, L.; Burzio, V.; Moraga, M.; Reyes, A.E.; Antonelli, M. The CK1 gene family: Expression patterning in zebrafish development. Biol. Res. 2007, 40, 251–266. [Google Scholar] [CrossRef]
- Li, D.J.; McMann, C.L.; Reddien, P.W. Nuclear receptor NR4A is required for patterning at the ends of the planarian anterior-posterior axis. Elife 2019, 8, e42015. [Google Scholar] [CrossRef] [PubMed]
- Bonar, N.A.; Gittin, D.I.; Petersen, C.P. Src acts with WNT/FGFRL signaling to pattern the planarian anteroposterior axis. Development 2022, 149, dev200125. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.D.; Neto, A.; Maeso, I.; Gomez-Skarmeta, J.L.; Salo, E.; Cebria, F. Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr. Biol. 2011, 21, 300–305. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Sun, Y.; Guo, Y.; Ma, M.; Zhang, S.; Tian, Q. Djck1α Is Required for Proper Regeneration and Maintenance of the Medial Tissues in Planarians. Cells 2023, 12, 473. https://doi.org/10.3390/cells12030473
Huang Y, Sun Y, Guo Y, Ma M, Zhang S, Tian Q. Djck1α Is Required for Proper Regeneration and Maintenance of the Medial Tissues in Planarians. Cells. 2023; 12(3):473. https://doi.org/10.3390/cells12030473
Chicago/Turabian StyleHuang, Yongding, Yujia Sun, Yajun Guo, Mengwen Ma, Shoutao Zhang, and Qingnan Tian. 2023. "Djck1α Is Required for Proper Regeneration and Maintenance of the Medial Tissues in Planarians" Cells 12, no. 3: 473. https://doi.org/10.3390/cells12030473
APA StyleHuang, Y., Sun, Y., Guo, Y., Ma, M., Zhang, S., & Tian, Q. (2023). Djck1α Is Required for Proper Regeneration and Maintenance of the Medial Tissues in Planarians. Cells, 12(3), 473. https://doi.org/10.3390/cells12030473





