Depletion of Paraoxonase 1 (Pon1) Dysregulates mTOR, Autophagy, and Accelerates Amyloid Beta Accumulation in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Brain Protein Extraction
2.3. Aβ Quantification
2.4. Cell Culture and Treatments
2.5. Western Blots
2.6. RNA Isolation, cDNA Synthesis, RT-qPCR Analysis
2.7. Chromatin Immunoprecipitation Assay
2.8. Confocal Microscopy, Aβ Quantification in N2a-APPswe Cells
2.9. Behavioral Testing
2.9.1. Hindlimb Test
2.9.2. Ledge Test
2.9.3. Cylinder Test
2.10. Statistical Analysis
3. Results
3.1. Pon1 Depletion Downregulates the Expression of Histone Demethylase Phf8 and Increases the H4K20me1 Epigenetic Mark in Mouse Brain
3.2. Pon1 Depletion Upregulates mTOR and Inhibits Autophagy in Mouse Brain
3.3. Pon1 Depletion Upregulates APP Protein Expression in Mouse Brain
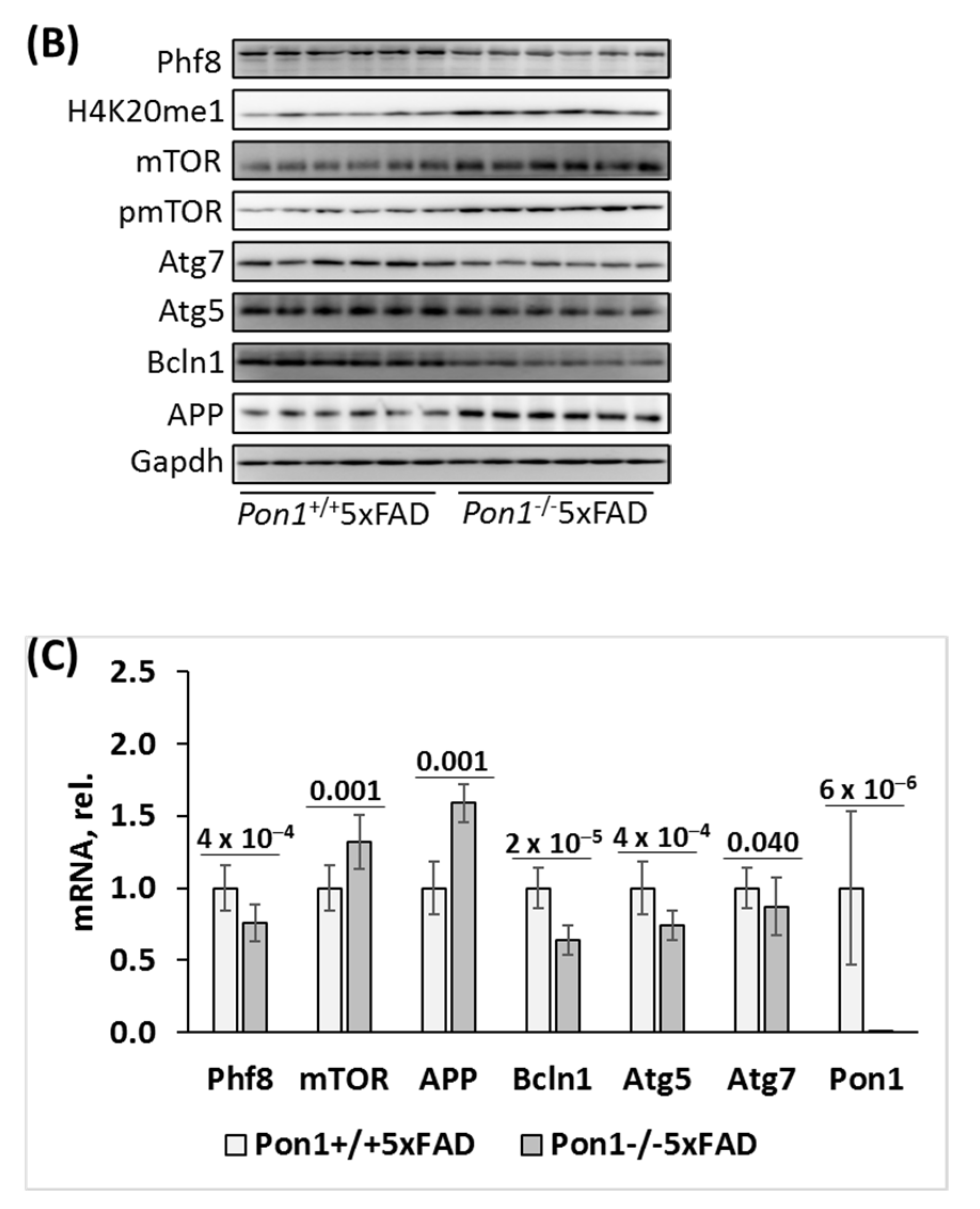
3.4. Pon1 Gene Exerts Transcriptional Control on the Expression of Phf8, mTOR, Autophagy-Related Proteins, and APP in Pon1─/─5xFAD Mice
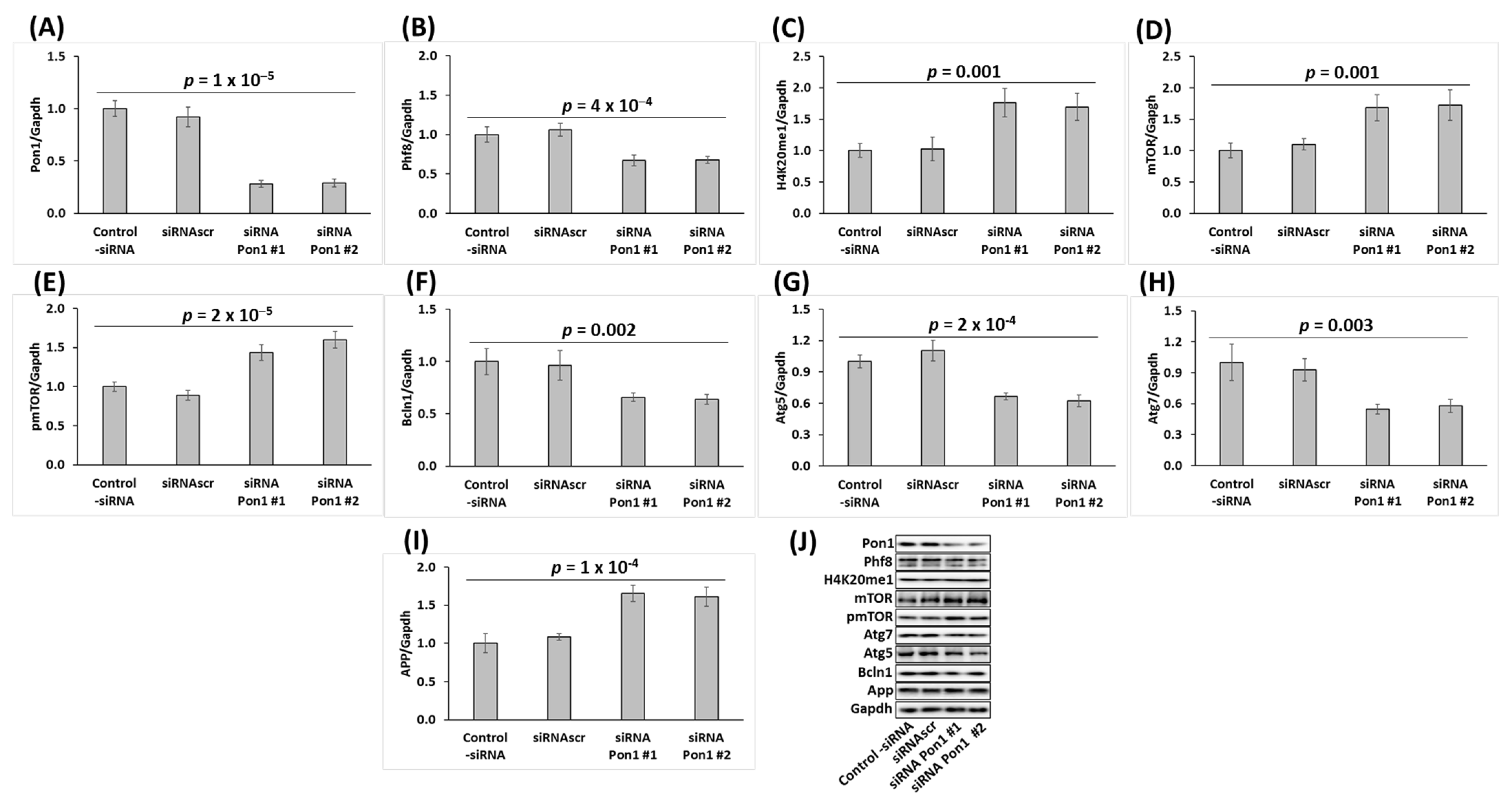
3.5. Pon1 Gene Silencing Downregulates the Histone Demethylase Phf8, Upregulates H4K20me1 Epigenetic Mark, mTOR and pmTOR, APP, and Inhibits Autophagy in Mouse Neuroblastoma N2a-APPswe Cells
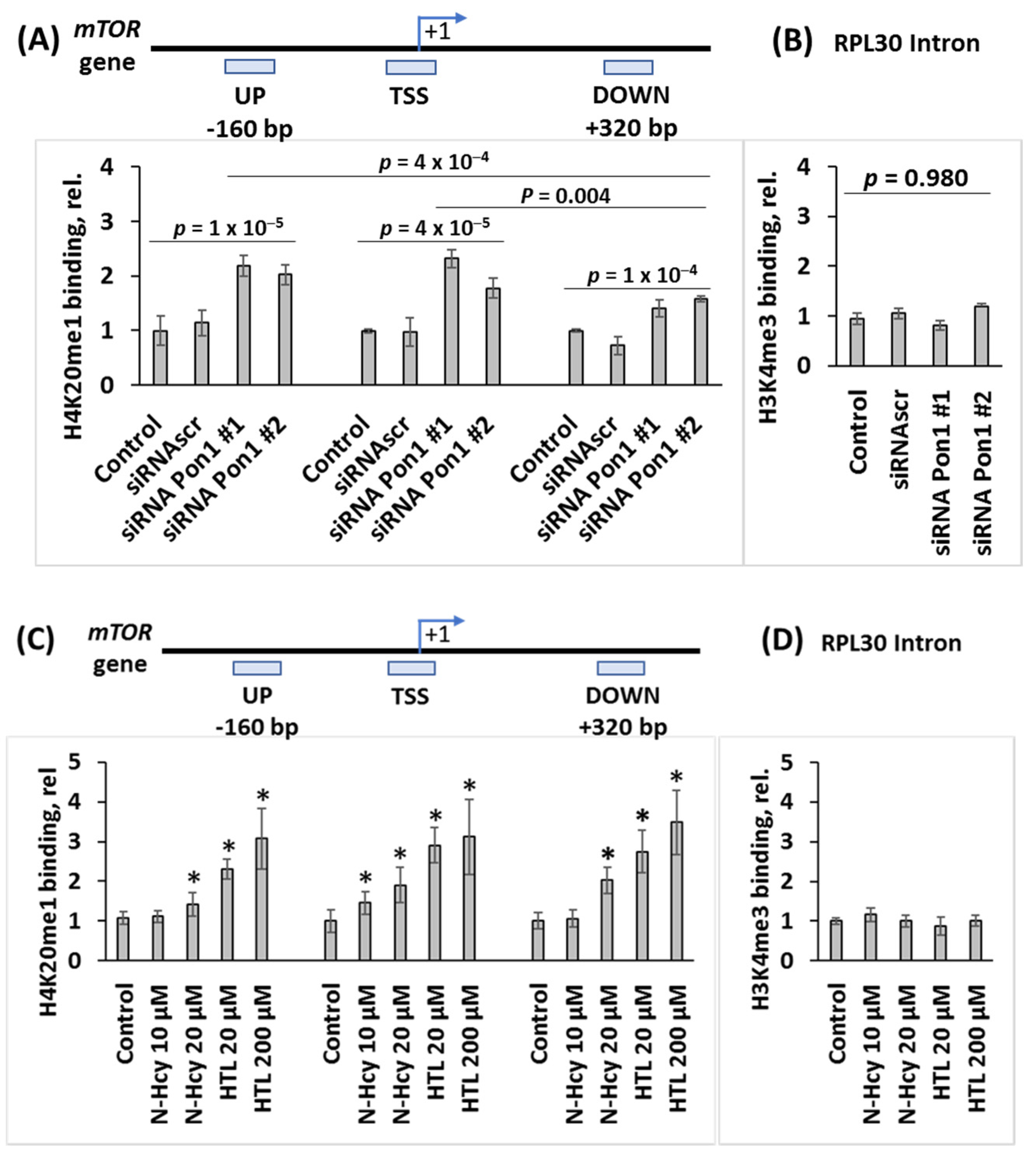
3.6. Pon1 Gene Silencing Increases H4K20me1 Biding to mTOR Promoter in N2a-APPswe Cells
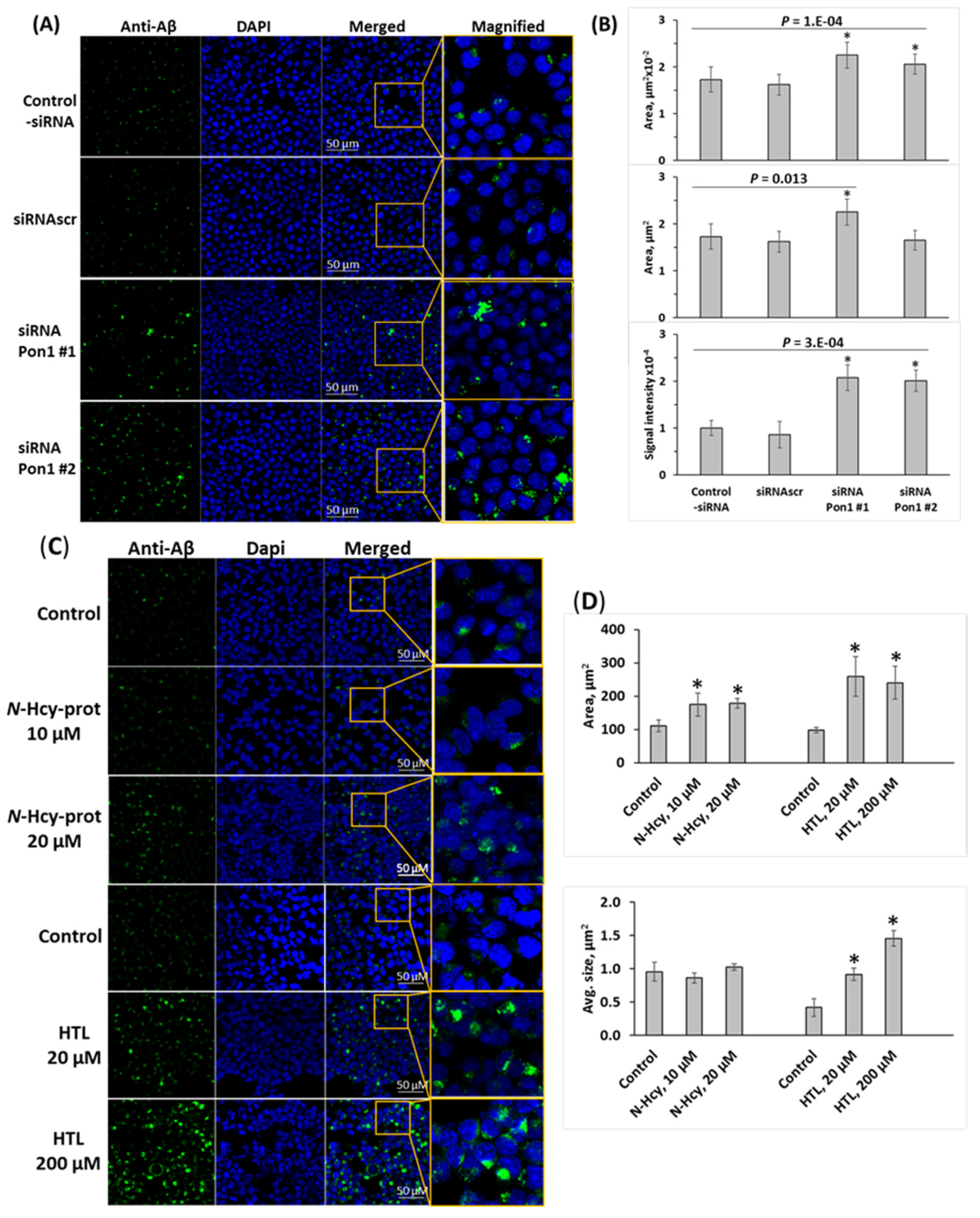
3.7. Pon1 Depletion Increases Aβ Accumulation in N2a-APPswe Cells
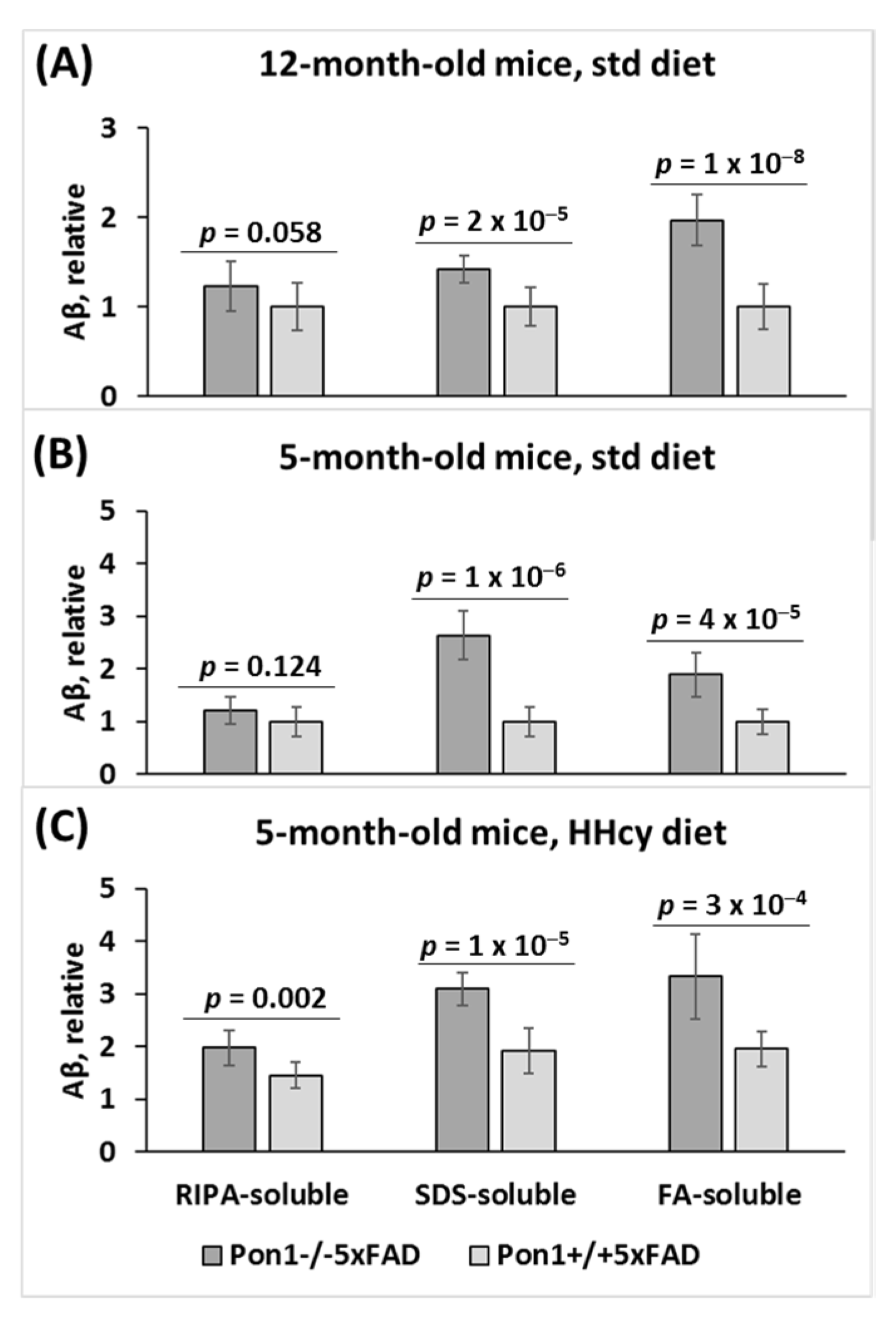
3.8. Pon1 Depletion Increases Aβ Accumulation in Pon1−/−5xFAD Mice
3.9. Pon1 Depletion Does Not Induce Sensorimotor Deficits
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perla-Kajan, J.; Jakubowski, H. Paraoxonase 1 and homocysteine metabolism. Amino Acids 2012, 43, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Marsillach, J.; Mackness, B.; Mackness, M.; Riu, F.; Beltran, R.; Joven, J.; Camps, J. Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free Radic. Biol. Med. 2008, 45, 146–157. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Cole, T.B.; Marsillach, J.; Furlong, C.E. Paraoxonase 1 (PON1) as a genetic determinant of susceptibility to organophosphate toxicity. Toxicology 2013, 307, 115–122. [Google Scholar] [CrossRef]
- Shih, D.M.; Gu, L.; Xia, Y.R.; Navab, M.; Li, W.F.; Hama, S.; Castellani, L.W.; Furlong, C.E.; Costa, L.G.; Fogelman, A.M.; et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 1998, 394, 284–287. [Google Scholar] [CrossRef]
- Shih, D.M.; Xia, Y.R.; Wang, X.P.; Miller, E.; Castellani, L.W.; Subbanagounder, G.; Cheroutre, H.; Faull, K.F.; Berliner, J.A.; Witztum, J.L.; et al. Combined serum paraoxonase knockout/apolipoprotein E knockout mice exhibit increased lipoprotein oxidation and atherosclerosis. J. Biol. Chem. 2000, 275, 17527–17535. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, T.; Nicholls, S.J.; Topol, E.J.; Zhang, R.L.; Yang, X.; Schmitt, D.; Fu, X.M.; Shao, M.Y.; Brennan, D.M.; Ellis, S.G.; et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA-J. Am. Med. Assoc. 2008, 299, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Hartiala, J.; Fan, Y.; Wu, Y.; Stewart, A.F.; Erdmann, J.; Kathiresan, S.; Consortium, C.A.; Roberts, R.; McPherson, R.; et al. Clinical and genetic association of serum paraoxonase and arylesterase activities with cardiovascular risk. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2803–2812. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.J.; Tang, W.H.; Fan, Y.; Wu, Y.; Mann, S.; Pepoy, M.; Hazen, S.L. Diminished antioxidant activity of high-density lipoprotein-associated proteins in chronic kidney disease. J. Am. Heart Assoc. 2013, 2, e000104. [Google Scholar] [CrossRef]
- Hassan, A.; Dohi, T.; Miyauchi, K.; Ogita, M.; Kurano, M.; Ohkawa, R.; Nakamura, K.; Tamura, H.; Isoda, K.; Okazaki, S.; et al. Prognostic impact of homocysteine levels and homocysteine thiolactonase activity on long-term clinical outcomes in patients undergoing percutaneous coronary intervention. J. Cardiol. 2017, 69, 830–835. [Google Scholar] [CrossRef]
- Kunutsor, S.K.; Bakker, S.J.; James, R.W.; Dullaart, R.P. Serum paraoxonase-1 activity and risk of incident cardiovascular disease: The PREVEND study and meta-analysis of prospective population studies. Atherosclerosis 2016, 245, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Tward, A.; Xia, Y.R.; Wang, X.P.; Shi, Y.S.; Park, C.; Castellani, L.W.; Lusis, A.J.; Shih, D.M. Decreased atherosclerotic lesion formation in human serum paraoxonase transgenic mice. Circulation 2002, 106, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Borowczyk, K.; Shih, D.M.; Jakubowski, H. Metabolism and neurotoxicity of homocysteine thiolactone in mice: Evidence for a protective role of paraoxonase 1. J. Alzheimer’s Dis. JAD 2012, 30, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H. Calcium-dependent human serum homocysteine thiolactone hydrolase. A protective mechanism against protein N-homocysteinylation. J. Biol. Chem. 2000, 275, 3957–3962. [Google Scholar] [CrossRef]
- Perla-Kajan, J.; Jakubowski, H. Paraoxonase 1 protects against protein N-homocysteinylation in humans. FASEB J. 2010, 24, 931–936. [Google Scholar] [CrossRef]
- Perla-Kajan, J.; Borowczyk, K.; Glowacki, R.; Nygard, O.; Jakubowski, H. Paraoxonase 1 Q192R genotype and activity affect homocysteine thiolactone levels in humans. FASEB J. 2018, 32, 6019–6024. [Google Scholar] [CrossRef] [PubMed]
- Menini, T.; Gugliucci, A. Paraoxonase 1 in neurological disorders. Redox. Rep. 2014, 19, 49–58. [Google Scholar] [CrossRef]
- Marsillach, J.; Adorni, M.P.; Zimetti, F.; Papotti, B.; Zuliani, G.; Cervellati, C. HDL Proteome and Alzheimer’s Disease: Evidence of a Link. Antioxidants 2020, 9, 1224. [Google Scholar] [CrossRef]
- de la Torre, J.C. Alzheimer disease as a vascular disorder: Nosological evidence. Stroke 2002, 33, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Erlich, P.M.; Lunetta, K.L.; Cupples, L.A.; Abraham, C.R.; Green, R.C.; Baldwin, C.T.; Farrer, L.A. Serum paraoxonase activity is associated with variants in the PON gene cluster and risk of Alzheimer disease. Neurobiol. Aging 2012, 33, 1015.e7–1015.e23. [Google Scholar] [CrossRef] [PubMed]
- Bednarska-Makaruk, M.E.; Krzywkowski, T.; Graban, A.; Lipczynska-Lojkowska, W.; Bochynska, A.; Rodo, M.; Wehr, H.; Ryglewicz, D.K. Paraoxonase 1 (PON1) gene-108C>T and p.Q192R polymorphisms and arylesterase activity of the enzyme in patients with dementia. Folia Neuropathol. 2013, 51, 111–119. [Google Scholar] [CrossRef]
- Dantoine, T.F.; Debord, J.; Merle, L.; Lacroix-Ramiandrisoa, H.; Bourzeix, L.; Charmes, J.P. Paraoxonase 1 activity: A new vascular marker of dementia? Ann. N. Y. Acad. Sci. 2002, 977, 96–101. [Google Scholar] [CrossRef]
- Paragh, G.; Balla, P.; Katona, E.; Seres, I.; Egerhazi, A.; Degrell, I. Serum paraoxonase activity changes in patients with Alzheimer’s disease and vascular dementia. Eur. Arch. Psychiatry Clin. Neurosci. 2002, 252, 63–67. [Google Scholar] [CrossRef]
- Bednarz-Misa, I.; Berdowska, I.; Zboch, M.; Misiak, B.; Zielinski, B.; Placzkowska, S.; Fleszar, M.; Wisniewski, J.; Gamian, A.; Krzystek-Korpacka, M. Paraoxonase 1 decline and lipid peroxidation rise reflect a degree of brain atrophy and vascular impairment in dementia. Adv. Clin. Exp. Med. 2020, 29, 71–78. [Google Scholar] [CrossRef]
- Perla-Kajan, J.; Wloczkowska, O.; Ziola-Frankowska, A.; Frankowski, M.; Smith, A.D.; de Jager, C.A.; Refsum, H.; Jakubowski, H. Paraoxonase 1, B Vitamins Supplementation, and Mild Cognitive Impairment. J. Alzheimers Dis. 2021, 81, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Aluganti Narasimhulu, C.; Mitra, C.; Bhardwaj, D.; Burge, K.Y.; Parthasarathy, S. Alzheimer’s Disease Markers in Aged ApoE-PON1 Deficient Mice. J. Alzheimers Dis. 2019, 67, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.G.; Marsillach, J.; Reverte, I.; Mackness, B.; Mackness, M.; Joven, J.; Camps, J.; Colomina, M.T. Paraoxonase-1 and -3 Protein Expression in the Brain of the Tg2576 Mouse Model of Alzheimer’s Disease. Antioxidants 2021, 10(3), 339. [Google Scholar] [CrossRef]
- Suszynska-Zajczyk, J.; Luczak, M.; Marczak, L.; Jakubowski, H. Inactivation of the paraoxonase 1 gene affects the expression of mouse brain proteins involved in neurodegeneration. J. Alzheimers Dis. 2014, 42, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Tanzi, R.E. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006296. [Google Scholar] [CrossRef]
- Blatter, M.C.; James, R.W.; Messmer, S.; Barja, F.; Pometta, D. Identification of a distinct human high-density lipoprotein subspecies defined by a lipoprotein-associated protein, K-45. Identity of K-45 with paraoxonase. Eur. J. Biochem. 1993, 211, 871–879. [Google Scholar] [CrossRef]
- Sikora, M.; Bretes, E.; Perla-Kajan, J.; Lewandowska, I.; Marczak, L.; Jakubowski, H. Genetic Attenuation of Paraoxonase 1 Activity Induces Proatherogenic Changes in Plasma Proteomes of Mice and Humans. Antioxidants 2020, 9, 1198. [Google Scholar] [CrossRef]
- Khayati, K.; Antikainen, H.; Bonder, E.M.; Weber, G.F.; Kruger, W.D.; Jakubowski, H.; Dobrowolski, R. The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J. 2017, 31, 598–609. [Google Scholar] [CrossRef]
- Yates, S.C.; Zafar, A.; Hubbard, P.; Nagy, S.; Durant, S.; Bicknell, R.; Wilcock, G.; Christie, S.; Esiri, M.M.; Smith, A.D.; et al. Dysfunction of the mTOR pathway is a risk factor for Alzheimer’s disease. Acta Neuropathol. Commun. 2013, 1, 3. [Google Scholar] [CrossRef]
- Chen, X.; Wang, S.; Zhou, Y.; Han, Y.; Li, S.; Xu, Q.; Xu, L.; Zhu, Z.; Deng, Y.; Yu, L.; et al. Phf8 histone demethylase deficiency causes cognitive impairments through the mTOR pathway. Nat. Commun. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L.; et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 2006, 26, 10129–10140. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; Beckett, T.L.; Ding, Q.; Patel, E.; Markesbery, W.R.; St Clair, D.K.; LeVine, H., 3rd; Keller, J.N. Abeta solubility and deposition during AD progression and in APPxPS-1 knock-in mice. Neurobiol. Dis. 2007, 27, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Sadleir, K.R.; Eimer, W.A.; Cole, S.L.; Vassar, R. Abeta reduction in BACE1 heterozygous null 5XFAD mice is associated with transgenic APP level. Mol. Neurodegener. 2015, 10, 1–16. [Google Scholar] [CrossRef]
- Thinakaran, G.; Teplow, D.B.; Siman, R.; Greenberg, B.; Sisodia, S.S. Metabolism of the “Swedish” amyloid precursor protein variant in neuro2a (N2a) cells. Evidence that cleavage at the “beta-secretase” site occurs in the golgi apparatus. J. Biol. Chem. 1996, 271, 9390–9397. [Google Scholar] [CrossRef]
- Gurda, D.; Handschuh, L.; Kotkowiak, W.; Jakubowski, H. Homocysteine thiolactone and N-homocysteinylated protein induce pro-atherogenic changes in gene expression in human vascular endothelial cells. Amino. Acids 2015, 47, 1319–1339. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lieu, C.A.; Chinta, S.J.; Rane, A.; Andersen, J.K. Age-related behavioral phenotype of an astrocytic monoamine oxidase-B transgenic mouse model of Parkinson’s disease. PLoS ONE 2013, 8, e54200. [Google Scholar] [CrossRef]
- Guyenet, S.J.; Furrer, S.A.; Damian, V.M.; Baughan, T.D.; La Spada, A.R.; Garden, G.A. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J. Vis. Exp. 2010, 39, e1787. [Google Scholar]
- Witucki, Ł.; Jakubowski, H. Deletion of bleomycin hydrolase gene in mice induces biochemical and behavioral hallmarks of Alzheimer’s disease. J. Alzheimer’s Dis. 2023. submitted. [Google Scholar]
- Jakubowski, H.; Perla-Kajan, J.; Finnell, R.H.; Cabrera, R.M.; Wang, H.; Gupta, S.; Kruger, W.D.; Kraus, J.P.; Shih, D.M. Genetic or nutritional disorders in homocysteine or folate metabolism increase protein N-homocysteinylation in mice. Faseb. J. 2009, 23, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, T.P.; Robertson, A.; Chipman, P.H.; Rafuse, V.F.; Brown, R.E. Motor function deficits in the 12 month-old female 5xFAD mouse model of Alzheimer’s disease. Behav. Brain Res. 2018, 337, 256–263. [Google Scholar] [CrossRef]
- O’Leary, T.P.; Mantolino, H.M.; Stover, K.R.; Brown, R.E. Age-related deterioration of motor function in male and female 5xFAD mice from 3 to 16 months of age. Genes Brain Behav. 2020, 19, e12538. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Witucki, Ł.; Jakubowski, H. Depletion of Paraoxonase 1 (Pon1) Dysregulates mTOR, Autophagy, and Accelerates Amyloid Beta Accumulation in Mice. Cells 2023, 12, 746. https://doi.org/10.3390/cells12050746
Witucki Ł, Jakubowski H. Depletion of Paraoxonase 1 (Pon1) Dysregulates mTOR, Autophagy, and Accelerates Amyloid Beta Accumulation in Mice. Cells. 2023; 12(5):746. https://doi.org/10.3390/cells12050746
Chicago/Turabian StyleWitucki, Łukasz, and Hieronim Jakubowski. 2023. "Depletion of Paraoxonase 1 (Pon1) Dysregulates mTOR, Autophagy, and Accelerates Amyloid Beta Accumulation in Mice" Cells 12, no. 5: 746. https://doi.org/10.3390/cells12050746
APA StyleWitucki, Ł., & Jakubowski, H. (2023). Depletion of Paraoxonase 1 (Pon1) Dysregulates mTOR, Autophagy, and Accelerates Amyloid Beta Accumulation in Mice. Cells, 12(5), 746. https://doi.org/10.3390/cells12050746






