3D Spheroid Configurations Are Possible Indictors for Evaluating the Pathophysiology of Melanoma Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. 2D Planar and 3D Spheroid Cultures of MM Cell Lines
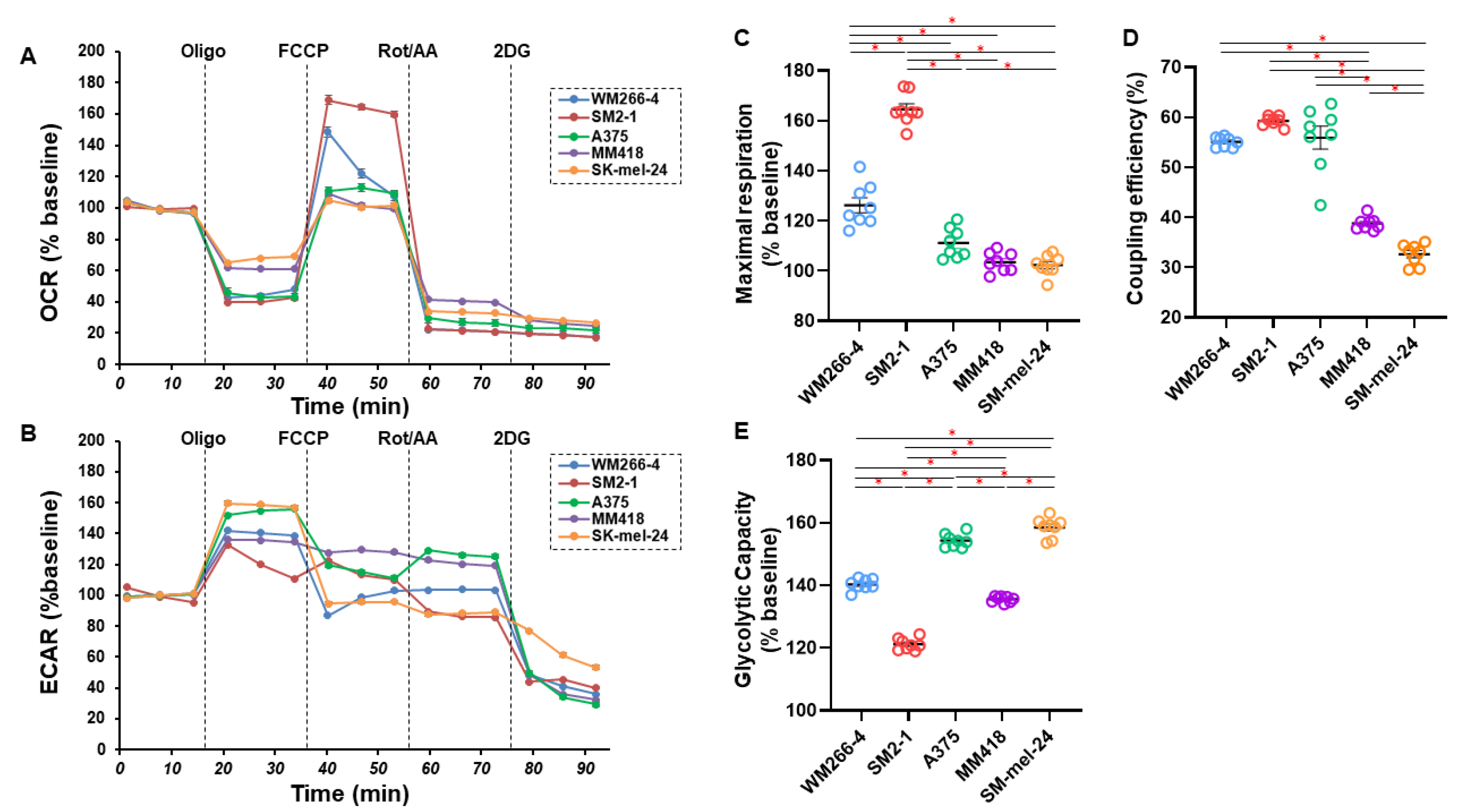
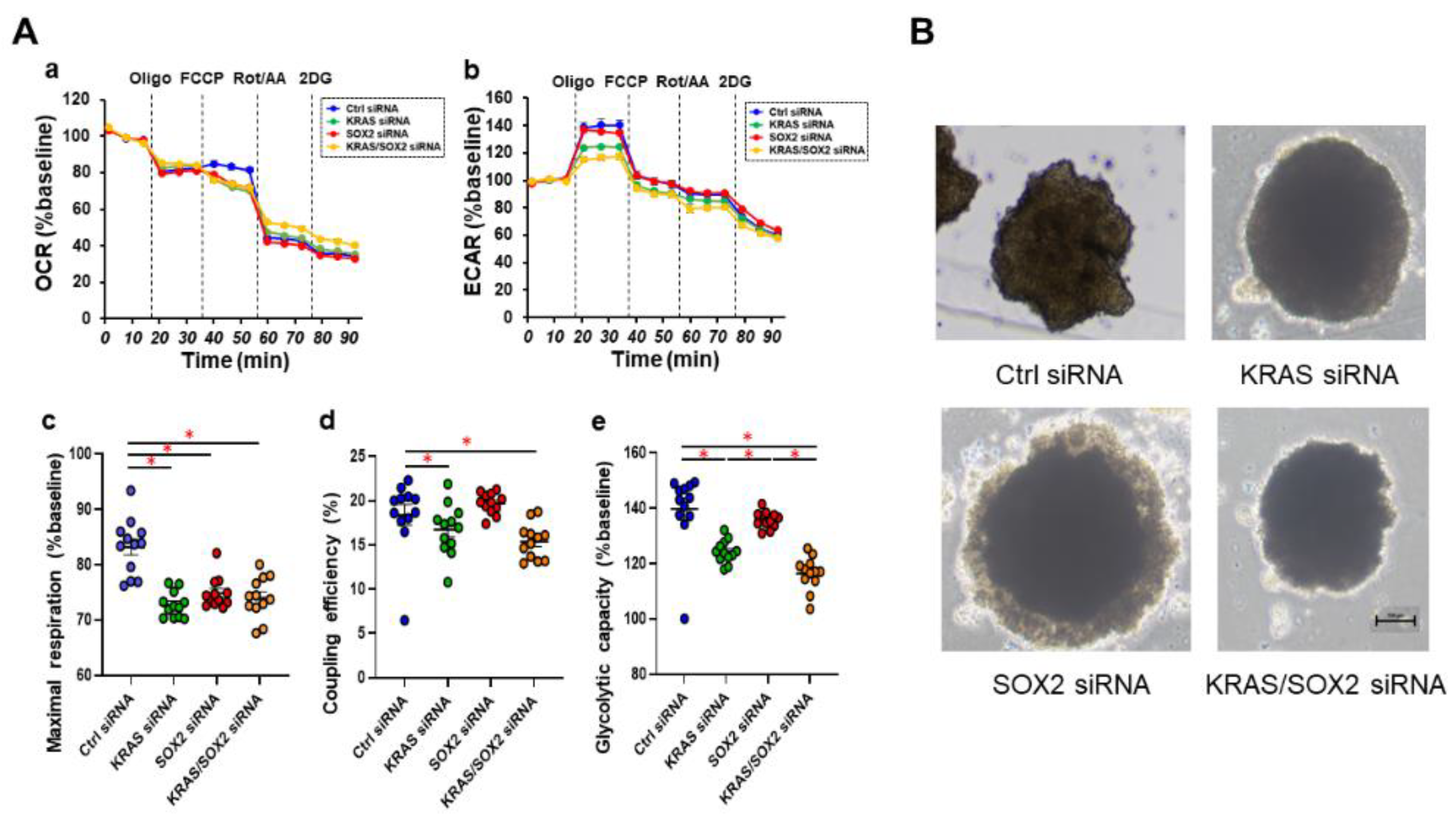
2.2. Measurement of Mitochondrial and Glycolytic Functions of Various MM Cell Lines
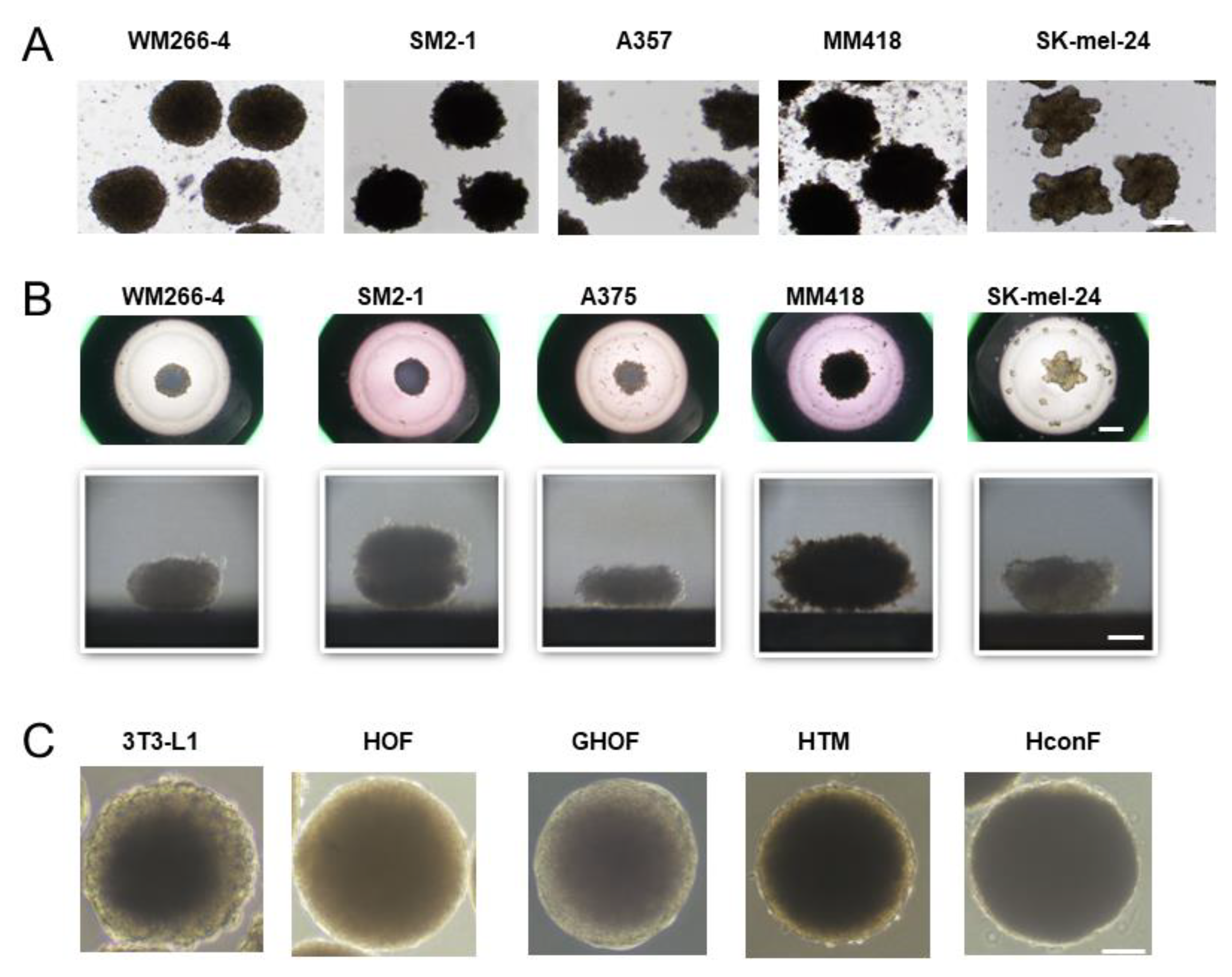
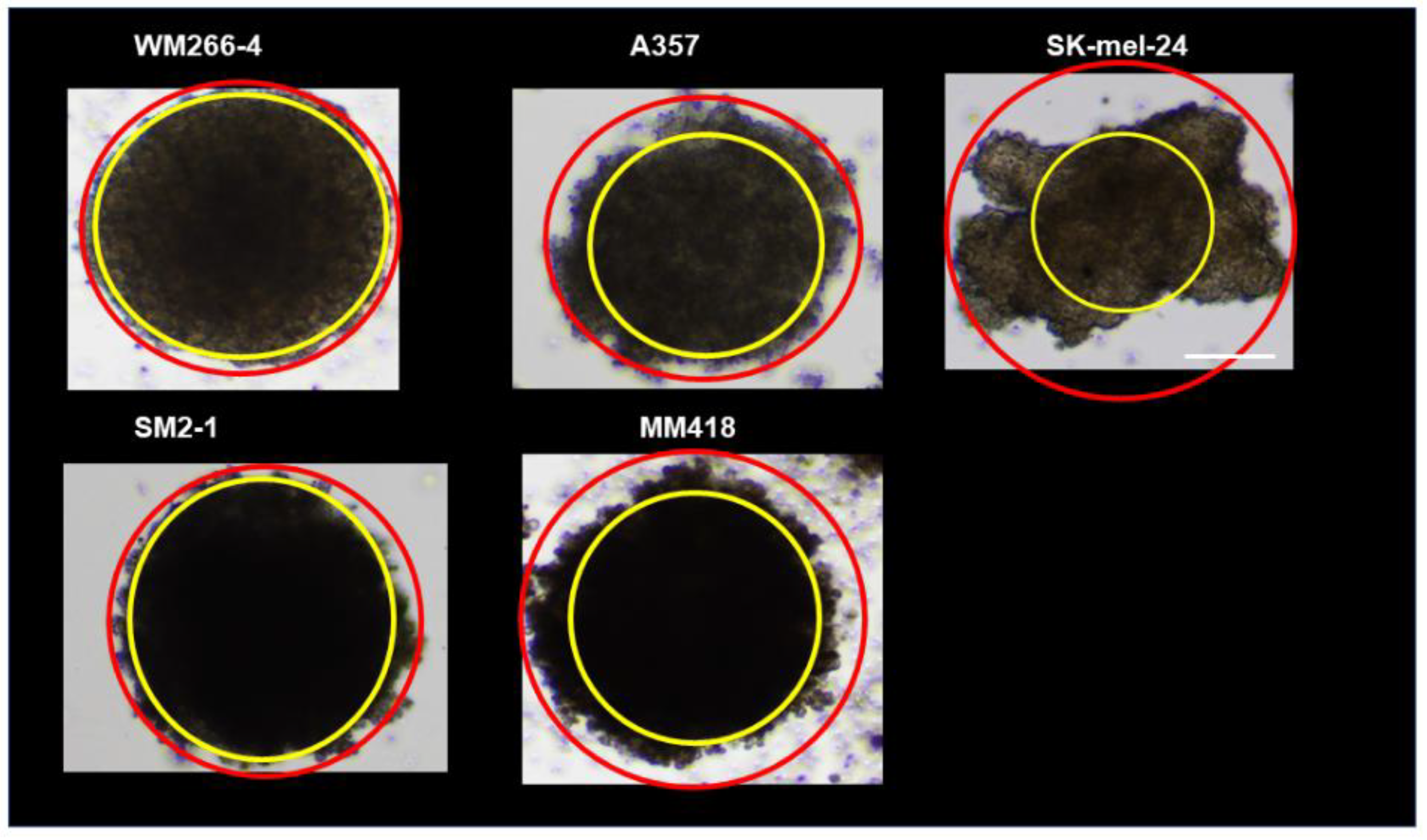
2.3. Phase Contrast Microscopy of the 3D Spheroids Derived from Various Human MM Cell Lines
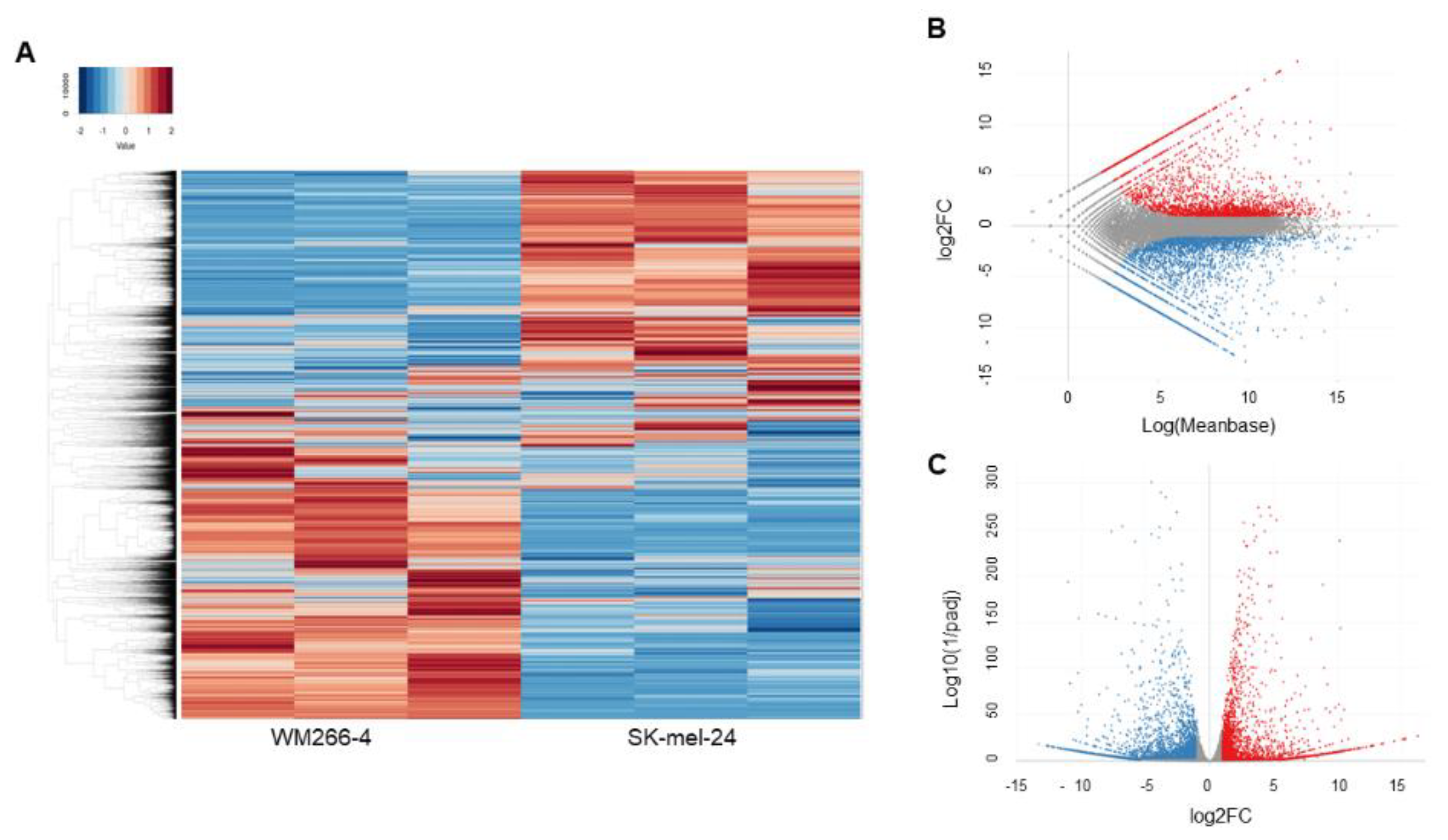
2.4. Analyses of RNA Sequence Gene Function and Metabolic Pathways
2.5. Transfection of siRNA
2.6. Other Analytical Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lo, J.A.; Fisher, D.E. The melanoma revolution: From UV carcinogenesis to a new era in therapeutics. Science 2014, 346, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Karimkhani, C.; Green, A.C.; Nijsten, T.; Weinstock, M.A.; Dellavalle, R.P.; Naghavi, M.; Fitzmaurice, C. The global burden of melanoma: Results from the Global Burden of Disease Study 2015. Br. J. Dermatol. 2017, 177, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef]
- Akbani, R.; Akdemir, K.C.; Aksoy, B.A.; Albert, M.; Ally, A.; Amin, S.B.; Arachchi, H.; Arora, A.; Auman, J.T.; Ayala BBaboud, J. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Carvajal, R.D.; Antonescu, C.R.; Wolchok, J.D.; Chapman, P.B.; Roman, R.A.; Teitcher, J.; Panageas, K.S.; Busam, K.J.; Chmielowski, B.; Lutzky, J.; et al. KIT as a therapeutic target in metastatic melanoma. JAMA 2011, 305, 2327–2334. [Google Scholar] [CrossRef]
- Postow, M.A.; Chesney, J.; Pavlick, A.C.; Robert, C.; Grossmann, K.; McDermott, D.; Linette, G.P.; Meyer, N.; Giguere, J.K.; Agarwala, S.S.; et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med. 2015, 372, 2006–2017. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef]
- Klemen, N.D.; Wang, M.; Rubinstein, J.C.; Olino, K.; Clune, J.; Ariyan, S.; Cha, C.; Weiss, S.A.; Kluger, H.M.; Sznol, M. Survival after checkpoint inhibitors for metastatic acral, mucosal and uveal melanoma. J. Immunother. Cancer 2020, 8, e000341. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef] [PubMed]
- van Elsas, A.; Hurwitz, A.A.; Allison, J.P. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J. Exp. Med. 1999, 190, 355–366. [Google Scholar]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Boucherit, N.; Gorvel, L.; Olive, D. 3D Tumor Models and Their Use for the Testing of Immunotherapies. Front. Immunol. 2020, 11, 603640. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Ono, K.; Hata, S.; Suzuki, T.; Kumamoto, H.; Sasano, H. The advantages of co-culture over mono cell culture in simulating in vivo environment. J. Steroid Biochem. Mol. Biol. 2012, 131, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Soon, R.H.; Lim, C.T.; Khoo, B.L.; Han, J. Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab A Chip 2019, 19, 369–386. [Google Scholar] [CrossRef]
- Yamada, K.M.; Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef]
- Zanoni, M.; Pignatta, S.; Arienti, C.; Bonafè, M.; Tesei, A. Anticancer drug discovery using multicellular tumor spheroid models. Expert Opin. Drug Discov. 2019, 14, 289–301. [Google Scholar] [CrossRef]
- Santini, M.T.; Rainaldi, G. Three-dimensional spheroid model in tumor biology. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 1999, 67, 148–157. [Google Scholar] [CrossRef]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Weaver, V.M.; Petersen, O.W.; Larabell, C.A.; Dedhar, S.; Briand, P.; Lupu, R.; Bissell, M.J. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: A different perspective in epithelial biology. Proc. Natl. Acad. Sci. USA 1998, 95, 14821–14826. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, K.A.; Mohana-Kumaran, N.; Haass, N.K. Modeling Melanoma In Vitro and In Vivo. Healthcare 2013, 2, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Haass, N.K. Microenvironment-Driven Dynamic Heterogeneity and Phenotypic Plasticity as a Mechanism of Melanoma Therapy Resistance. Front. Oncol. 2018, 8, 173. [Google Scholar] [CrossRef]
- Hikage, F.; Atkins, S.; Kahana, A.; Smith, T.J.; Chun, T.H. HIF2A-LOX Pathway Promotes Fibrotic Tissue Remodeling in Thyroid-Associated Orbitopathy. Endocrinology 2019, 160, 20–35. [Google Scholar] [CrossRef]
- Watanabe, M.; Ida, Y.; Furuhashi, M.; Tsugeno, Y.; Ohguro, H.; Hikage, F. Screening of the Drug-Induced Effects of Prostaglandin EP2 and FP Agonists on 3D Cultures of Dexamethasone-Treated Human Trabecular Meshwork Cells. Biomedicines 2021, 9, 930. [Google Scholar] [CrossRef]
- Oouchi, Y.; Watanabe, M.; Ida, Y.; Ohguro, H.; Hikage, F. Rosiglitasone and ROCK Inhibitors Modulate Fibrogenetic Changes in TGF-β2 Treated Human Conjunctival Fibroblasts (HconF) in Different Manners. Int. J. Mol. Sci. 2021, 22, 7335. [Google Scholar] [CrossRef]
- Ida, Y.; Hikage, F.; Itoh, K.; Ida, H.; Ohguro, H. Prostaglandin F2α agonist-induced suppression of 3T3-L1 cell adipogenesis affects spatial formation of extra-cellular matrix. Sci. Rep. 2020, 10, 7958. [Google Scholar] [CrossRef]
- Marconi, A.; Quadri, M.; Saltari, A.; Pincelli, C. Progress in melanoma modelling in vitro. Exp. Dermatol. 2018, 27, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Furney, S.J.; Turajlic, S.; Fenwick, K.; Lambros, M.B.; MacKay, A.; Ricken, G.; Mitsopoulos, C.; Kozarewa, I.; Hakas, J.; Zvelebil, M.; et al. Genomic characterisation of acral melanoma cell lines. Pigment. Cell Melanoma Res. 2012, 25, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Ohguro, H.; Ida, Y.; Hikage, F.; Umetsu, A.; Ichioka, H.; Watanabe, M.; Furuhashi, M. STAT3 Is the Master Regulator for the Forming of 3D Spheroids of 3T3-L1 Preadipocytes. Cells 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Akama, T.; Leung, B.M.; Labuz, J.; Takayama, S.; Chun, T.H. Designing 3-D Adipospheres for Quantitative Metabolic Study. Methods Mol. Biol. 2017, 1566, 177–183. [Google Scholar] [PubMed]
- Wang, L.; Leite de Oliveira, R.; Huijberts, S.; Bosdriesz, E.; Pencheva, N.; Brunen, D.; Bosma, A.; Song, J.Y.; Zevenhoven, J.; Los-de Vries, G.T.; et al. An Acquired Vulnerability of Drug-Resistant Melanoma with Therapeutic Potential. Cell 2018, 173, 1413–1425.e14. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhou, Y.; Jiang, H.; Chen, Z.; Lu, B. Analysis of core genes for colorectal cancer prognosis based on immune and stromal scores. PeerJ 2021, 9, e12452. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Vander Heiden, M.G.; McCormick, F. The Metabolic Landscape of RAS-Driven Cancers from biology to therapy. Nat. Cancer 2021, 2, 271–283. [Google Scholar] [CrossRef]
- Zhang, S.; Xiong, X.; Sun, Y. Functional characterization of SOX2 as an anticancer target. Signal Transduct. Target. Ther. 2020, 5, 135. [Google Scholar] [CrossRef]
- Kolch, W. Meaningful relationships: The regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000, 351 Pt 2, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Hauschild, A.; Grob, J.J.; Demidov, L.V.; Jouary, T.; Gutzmer, R.; Millward, M.; Rutkowski, P.; Blank, C.U.; Miller, W.H., Jr.; Kaempgen, E.; et al. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet 2012, 380, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.B.; Kefford, R.; Pavlick, A.C.; Infante, J.R.; Ribas, A.; Sosman, J.A.; Fecher, L.A.; Millward, M.; McArthur, G.A.; Hwu, P.; et al. Phase II study of the MEK1/MEK2 inhibitor Trametinib in patients with metastatic BRAF-mutant cutaneous melanoma previously treated with or without a BRAF inhibitor. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Chen, X.; Zheng, M.; Liu, X.; Wang, H.; Lou, L. Pharmacologic characterization of SHR8443, a novel dual inhibitor of phosphatidylinositol 3-kinase and mammalian target of rapamycin. Oncotarget 2017, 8, 107977–107990. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.Z.; Wang, H.B.; Chen, X.L.; Cao, W.T.; Fu, L.; Li, Y.; Quan, H.T.; Xie, C.Y.; Lou, L.G. Aberrant modulation of ribosomal protein S6 phosphorylation confers acquired resistance to MAPK pathway inhibitors in BRAF-mutant melanoma. Acta Pharmacol. Sin. 2019, 40, 268–278. [Google Scholar] [CrossRef]
- Patton, E.E.; Mueller, K.L.; Adams, D.J.; Anandasabapathy, N.; Aplin, A.E.; Bertolotto, C.; Bosenberg, M.; Ceol, C.J.; Burd, C.E.; Chi, P.; et al. Melanoma models for the next generation of therapies. Cancer Cell 2021, 39, 610–631. [Google Scholar] [CrossRef]
- Rebecca, V.W.; Somasundaram, R.; Herlyn, M. Pre-clinical modeling of cutaneous melanoma. Nat. Commun. 2020, 11, 2858. [Google Scholar] [CrossRef]
- Mery, B.; Vallard, A.; Rowinski, E.; Magne, N. High-throughput sequencing in clinical oncology: From past to present. Swiss Med. Wkly. 2019, 149, w20057. [Google Scholar] [CrossRef]
- Groner, B.; von Manstein, V. Jak Stat signaling and cancer: Opportunities, benefits and side effects of targeted inhibition. Mol. Cell Endocrinol. 2017, 451, 1–14. [Google Scholar] [CrossRef]
- Valle-Mendiola, A.; Soto-Cruz, I. Energy Metabolism in Cancer: The Roles of STAT3 and STAT5 in the Regulation of Metabolism-Related Genes. Cancers 2020, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Poli, V.; Camporeale, A. STAT3-Mediated Metabolic Reprograming in Cellular Transformation and Implications for Drug Resistance. Front. Oncol. 2015, 5, 121. [Google Scholar] [CrossRef] [PubMed]
- Albi, E.; Curcio, F.; Lazzarini, A.; Floridi, A.; Cataldi, S.; Lazzarini, R.; Loreti, E.; Ferri, I.; Ambesi-Impiombato, F.S. A firmer understanding of the effect of hypergravity on thyroid tissue: Cholesterol and thyrotropin receptor. PLoS ONE 2014, 9, e98250. [Google Scholar] [CrossRef] [PubMed]
- Cavey, T.; Pierre, N.; Nay, K.; Allain, C.; Ropert, M.; Loréal, O.; Derbré, F. Simulated microgravity decreases circulating iron in rats: Role of inflammation-induced hepcidin upregulation. Exp. Physiol. 2017, 102, 291–298. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Shi, J.; Martin, B.L.; Crawford, H.C.; Petrenko, O.; Reich, N.C. STAT3 is a master regulator of epithelial identity and KRAS-driven tumorigenesis. Genes Dev. 2018, 32, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.W.; Yang, S.T.; Chien, M.H.; Hua, K.T.; Wu, C.J.; Hsiao, S.M.; Lin, H.; Hsiao, M.; Su, J.L.; Wei, L.H. The STAT3-miRNA-92-Wnt Signaling Pathway Regulates Spheroid Formation and Malignant Progression in Ovarian Cancer. Cancer Res. 2017, 77, 1955–1967. [Google Scholar] [CrossRef]
- Gkountela, S.; Castro-Giner, F.; Szczerba, B.M.; Vetter, M.; Landin, J.; Scherrer, R.; Krol, I.; Scheidmann, M.C.; Beisel, C.; Stirnimann, C.U.; et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019, 176, 98–112.e14. [Google Scholar] [CrossRef] [PubMed]
- Hüser, L.; Sachindra, S.; Granados, K.; Federico, A.; Larribère, L.; Novak, D.; Umansky, V.; Altevogt, P.; Utikal, J. SOX2-mediated upregulation of CD24 promotes adaptive resistance toward targeted therapy in melanoma. Int. J. Cancer 2018, 143, 3131–3142. [Google Scholar] [CrossRef]
- Hüser, L.; Altevogt, P.; Utikal, J. Role of STAT3 dependent SOX2 and CD24 expression in melanoma cell adaptive resistance towards targeted therapies. Oncotarget 2019, 10, 1662–1663. [Google Scholar] [CrossRef]
- Hodgkinson, C.A.; Moore, K.J.; Nakayama, A.; Steingrímsson, E.; Copeland, N.G.; Jenkins, N.A.; Arnheiter, H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 1993, 74, 395–404. [Google Scholar] [CrossRef]
- Cimadamore, F.; Shah, M.; Amador-Arjona, A.; Navarro-Peran, E.; Chen, C.; Huang, C.T.; Terskikh, A.V. SOX2 modulates levels of MITF in normal human melanocytes, and melanoma lines in vitro. Pigment. Cell Melanoma Res. 2012, 25, 533–536. [Google Scholar] [CrossRef]
- Bulle, A.; Lim, K.H. Beyond just a tight fortress: Contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Signal Transduct. Target. Ther. 2020, 5, 249. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.B.; Kim, E.M.; Byun, H.; Chang, H.K.; Jeong, K.; Aman, Z.M.; Choi, Y.S.; Park, J.; Shin, H. Engineering spheroids potentiating cell-cell and cell-ECM interactions by self-assembly of stem cell microlayer. Biomaterials 2018, 165, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, I.; Ambati, R.; Gundamaraju, R. Exploring the Crosstalk between Inflammation and Epithelial-Mesenchymal Transition in Cancer. Mediat. Inflamm. 2021, 2021, 9918379. [Google Scholar] [CrossRef] [PubMed]
- Dilshat, R.; Fock, V.; Kenny, C.; Gerritsen, I.; Lasseur, R.M.J.; Travnickova, J.; Eichhoff, O.M.; Cerny, P.; Möller, K.; Sigurbjörnsdóttir, S.; et al. MITF reprograms the extracellular matrix and focal adhesion in melanoma. eLife 2021, 10, e63093. [Google Scholar] [CrossRef]

| Categories | Diseases or Functions Annotation | p-Value | Activations z-Score |
|---|---|---|---|
| Cancer, Organismal Injury and Abnormalities | Melanoma | 6.25 × 10−118 | 2.669 |
| Cancer, Organismal Injury and Abnormalities | Tumorigenesis of reproductive tract | 2.00 × 10−89 | 2.574 |
| Cancer, Organismal Injury and Abnormalities | Female genital neoplasm | 1.05 × 10−88 | 2.574 |
| Cancer, Gastrointestinal Disease | Upper gastrointestinal tract cancer | 1.05 × 10−63 | 2.425 |
| Cancer, Endocrine System Disorders | Gonadal tumor | 2.76 × 10−38 | 2.733 |
| Cancer, Endocrine System Disorders | Ovarian tumor | 4.69 × 10−34 | 2.703 |
| Cancer, Organismal Injury and Abnormalities | Non-hematological solid tumor | 4.67 × 10−288 | 0.135 |
| Cancer, Organismal Injury and Abnormalities | Non-hematologic malignant neoplasm | 1.31 × 10−286 | −0.425 |
| Cancer, Organismal Injury and Abnormalities | Non-melanoma solid tumor | 1.12 × 10−283 | −0.257 |
| Cancer, Organismal Injury and Abnormalities | Tumorigenesis of tissue | 9.79 × 10−279 | −1.779 |
| Cancer, Organismal Injury and Abnormalities | Carcinoma | 1.06 × 10−278 | −1.242 |
| Cancer, Organismal Injury and Abnormalities | Epithelial neoplasm | 6.80 × 10−278 | −1.482 |
| Cancer, Organismal Injury and Abnormalities | Cancer | 6.13 × 10−277 | 1.348 |
| Cancer, Organismal Injury and Abnormalities | Malignant solid tumor | 1.06 × 10−276 | 0.222 |
| Cancer, Organismal Injury and Abnormalities | Solid tumor | 2.01 × 10−276 | 1.229 |
| Up-Stream Regulator | Expr Log Ratio | Molecule Type | Activation z-Score |
|---|---|---|---|
| IL1B | ↑ 8668 | cytokine | 6.412 |
| KRAS | ↑ 1.404 | enzyme | 1.277 |
| FGF2 | ↑ 1.127 | growth factor | 1.731 |
| JUN | ↑ 2.294 | transcription regulator | 1.925 |
| EGFR | ↑ 5.897 | kinase | 0.324 |
| SOX2 | ↑ 1.519 | transcription regulator | −0.283 |
| AGT | ↓ −4.28 | growth factor | 2.246 |
| ESR2 | ↓ −7.881 | ligand-dependent nuclear receptor | 1.231 |
| EGF | ↓ −1.523 | growth factor | 2.162 |
| Master Regulator | Expr Log Ratio | Molecule Type | Activations z-Score |
|---|---|---|---|
| KLP9 | 1.444 | transcription regulator | 1.477 |
| KRAS | 1.404 | enzyme | 1.27 |
| SOX2 | 1.519 | transcription regulator | 2.01 |
| TP63 | 8.374 | transcription regulator | 0.371 |
| SMYD3 | −1.272 | enzyme | −0.614 |
| WM266-4 | SM2-1 | A375 | MM418 | SK-mel-24 | ||
|---|---|---|---|---|---|---|
| KRAS | 2D | cont | (↑) | → | (↑) | (↑) |
| 3D | cont | ↑↑ | ↑ | ↑↑ | ↑↑ | |
| SOX2 | 2D | cont | ↓↓ | ↓↓ | ↓↓ | ↓↓ |
| 3D | cont | ↓↓ | → | ↓↓ | ↑↑ | |
| STAT3 | 2D | cont | (↓) | (↓) | → | ↑ |
| 3D | cont | ↓↓ | → | ↑↑ | ↑↑ | |
| BRAF | 2D | cont | → | → | (↑) | (↑) |
| 3D | cont | (↓) | → | ↑ | ↑ | |
| FOS | 2D | cont | → | → | ↑↑ | → |
| 3D | cont | → | ↓↓ | → | ↓↓ | |
| MITF | 2D | cont | → | ↓↓ | ↓↓ | ↓↓ |
| 3D | cont | → | ↓↓ | → | ↓↓ | |
| PCG1a | 2D | cont | ↓↓ | ↓↓ | ↓↓ | ↓↓ |
| 3D | cont | ↓↓ | ↓↓ | ↓↓ | ↓↓ | |
| COL4 | 2D | cont | ↓↓ | ↓↓ | ↓↓ | ↑↑ |
| 3D | cont | ↓↓ | ↓↓ | ↓↓ | ↓↓ | |
| COL6 | 2D | cont | ↓↓ | ↓↓ | ↓↓ | ↓↓ |
| 3D | cont | ↓↓ | → | ↓↓ | ↓↓ | |
| FN | 2D | cont | ↓↓ | ↓ | ↑ | ↓↓ |
| 3D | cont | → | ↓↓ | ↓↓ | ↓↓ | |
| aSMA | 2D | cont | ↓↓ | ↓↓ | ↓↓ | ↓ |
| 3D | cont | ↓↓ | ↓↓ | → | ↓ | |
| ZO1 | 2D | cont | ↓↓ | ↓↓ | ↓↓ | ↓↓ |
| 3D | cont | ↓↓ | → | ↓↓ | ↓ |
| A375 | A375DT | ||
|---|---|---|---|
| KRAS | 2D | cont | ↑↑ |
| 3D | cont | ↑↑ | |
| SOX2 | 2D | cont | ↑↑ |
| 3D | cont | ↑↑ | |
| STAT3 | 2D | cont | ↑↑ |
| 3D | cont | ↑↑ | |
| BRAF | 2D | cont | (↑) |
| 3D | cont | ↑ | |
| FOS | 2D | cont | ↑↑ |
| 3D | cont | ↑↑ | |
| MITF | 2D | cont | ↑↑ |
| 3D | cont | ↑↑ | |
| PCG1a | 2D | cont | ↑↑ |
| 3D | cont | ↓↓ | |
| COL4 | 2D | cont | (↑) |
| 3D | cont | ↓↓ | |
| COL6 | 2D | cont | ↑↑ |
| 3D | cont | ↑↑ | |
| FN | 2D | cont | ↓↓ |
| 3D | cont | ↑↑ | |
| αSMA | 2D | cont | ↓↓ |
| 3D | cont | ↓↓ | |
| ZO1 | 2D | cont | ↑↑ |
| 3D | cont | ↑↑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohguro, H.; Watanabe, M.; Sato, T.; Hikage, F.; Furuhashi, M.; Okura, M.; Hida, T.; Uhara, H. 3D Spheroid Configurations Are Possible Indictors for Evaluating the Pathophysiology of Melanoma Cell Lines. Cells 2023, 12, 759. https://doi.org/10.3390/cells12050759
Ohguro H, Watanabe M, Sato T, Hikage F, Furuhashi M, Okura M, Hida T, Uhara H. 3D Spheroid Configurations Are Possible Indictors for Evaluating the Pathophysiology of Melanoma Cell Lines. Cells. 2023; 12(5):759. https://doi.org/10.3390/cells12050759
Chicago/Turabian StyleOhguro, Hiroshi, Megumi Watanabe, Tatsuya Sato, Fumihito Hikage, Masato Furuhashi, Masae Okura, Tokimasa Hida, and Hisashi Uhara. 2023. "3D Spheroid Configurations Are Possible Indictors for Evaluating the Pathophysiology of Melanoma Cell Lines" Cells 12, no. 5: 759. https://doi.org/10.3390/cells12050759
APA StyleOhguro, H., Watanabe, M., Sato, T., Hikage, F., Furuhashi, M., Okura, M., Hida, T., & Uhara, H. (2023). 3D Spheroid Configurations Are Possible Indictors for Evaluating the Pathophysiology of Melanoma Cell Lines. Cells, 12(5), 759. https://doi.org/10.3390/cells12050759









